

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.5 + phase: 0
(377 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
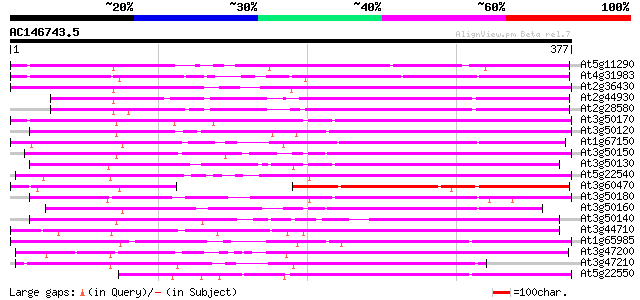
Score E
Sequences producing significant alignments: (bits) Value
At5g11290 putative protein 189 2e-48
At4g31983 putative protein 185 4e-47
At2g36430 unknown protein (At2g36430) 178 4e-45
At2g44930 unknown protein 165 4e-41
At2g28580 hypothetical protein 157 1e-38
At3g50170 putative protein 154 8e-38
At3g50120 putative protein 154 8e-38
At1g67150 hypothetical protein 154 8e-38
At3g50150 putative protein 151 6e-37
At3g50130 putative protein 144 1e-34
At5g22540 unknown protein 136 2e-32
At3g60470 putative protein 136 2e-32
At3g50180 putative protein 135 4e-32
At3g50160 putative protein 135 4e-32
At3g50140 putative protein 133 1e-31
At3g44710 unknown protein 125 5e-29
At1g65985 putative protein 125 5e-29
At3g47200 unknown protein 123 1e-28
At3g47210 unknown protein 113 1e-25
At5g22550 unknown protein 99 4e-21
>At5g11290 putative protein
Length = 422
Score = 189 bits (480), Expect = 2e-48
Identities = 128/387 (33%), Positives = 196/387 (50%), Gaps = 45/387 (11%)
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACF 60
MED KLRYL+SF + RT L D + + EE R CY+E + +SD++VK+++ DA F
Sbjct: 70 MEDHKLRYLQSF-IPRTALSLEDLVRVARTWEERARFCYTEDVRLSSDEYVKMLIVDASF 128
Query: 61 IIEYFLRS---LEWPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDG 117
++E LRS + D + K + DV D++LLENQLP+FV+E +F L
Sbjct: 129 LVELLLRSQFDVYRGMLDRIYGKQKMIVDVNHDVMLLENQLPYFVVEGMFGLLH------ 182
Query: 118 EVSSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHN---- 173
V +H LP T H H++++ M S + +
Sbjct: 183 -------VDYH--------RELPPLTR-------IIHNHFKKFWMSIPSFSRSISDSKIC 220
Query: 174 -LTDLLRVFYLPPDM-LPKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGAL 231
DLLR +LP + P +++ + SA ++ AGVKL + L++ FA G L
Sbjct: 221 HFVDLLRSIHLPLVLSFPGGSMRMMDSVLSAKEIQNAGVKLQPADNNTCALDISFANGVL 280
Query: 232 TIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMIN 291
TIP+ ++ TE+L RN++ FEQCH Y Y L I + D + ++ GI++N
Sbjct: 281 TIPKIKINDITESLYRNIILFEQCH-RLDAYFIHYMRFLSCFIRSPMDAELFIDHGIIVN 339
Query: 292 TLGDSNAVAKMINNLCLNVVQENINGG--YISLCRKLNCFYEDPSHKYKAIFIHDYFSTP 349
G++ V+++ N ++++E G Y ++ L P +K+KA DYF P
Sbjct: 340 RFGNAEDVSRLFN----SILKETSYSGFYYKTVYGNLQAHCNAPWNKWKATLRRDYFHNP 395
Query: 350 WKITSFAAAIVLLLLTLIQATCSIISL 376
W S AA VLLLLT +QA CSI++L
Sbjct: 396 WSAASVVAACVLLLLTFVQAICSILAL 422
>At4g31983 putative protein
Length = 414
Score = 185 bits (469), Expect = 4e-47
Identities = 128/382 (33%), Positives = 202/382 (52%), Gaps = 37/382 (9%)
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACF 60
MED K RYL SF + RT+ L D + + E+ RSCY+E ++ SD+FV++++ D F
Sbjct: 64 MEDQKYRYLLSF-IPRTNSSLEDLVRLARTWEQNARSCYAEDVKLHSDEFVEMLVVDGSF 122
Query: 61 IIEYFLRSLEWP----QEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCID 116
++E LRS +P + D + + DV D+IL+ENQLP+FV++++F L
Sbjct: 123 LVELLLRS-HYPRLRGENDRIFGNSMMITDVCRDMILIENQLPFFVVKEIFLLLLNYYQQ 181
Query: 117 GEVSSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTD 176
G S +A +F +FL I +++I +P+ + D
Sbjct: 182 G-TPSIIQLAQRHFS-YFLSRI-----------------DDEKFITEPE-------HFVD 215
Query: 177 LLRVFYLPPDMLPKREKQIVK--HLFSASQLVEAGVKLHVGQDYQSVLELKFAKGALTIP 234
LLR YLP + K E VK + A++L AGV+ + +L++ FA G L IP
Sbjct: 216 LLRSCYLPQFPI-KLEYTTVKVDNAPEATELHTAGVRFKPAETSSCLLDISFADGVLKIP 274
Query: 235 RFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLG 294
V TE+L +N++ FEQC + ++ DY +LL I + D D L+ GI++N LG
Sbjct: 275 TIVVDDLTESLYKNIIGFEQCRCSNKNFL-DYIMLLGCFIKSPTDADLLIHSGIIVNYLG 333
Query: 295 DSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITS 354
+S V+ + N++ V+ + + L L + P +++KAI DYF PW + S
Sbjct: 334 NSVDVSNLFNSISKEVIYDR-RFYFSMLSENLQAYCNTPWNRWKAILRRDYFHNPWAVAS 392
Query: 355 FAAAIVLLLLTLIQATCSIISL 376
AA++LLLLT IQ+ CSI++L
Sbjct: 393 VFAALLLLLLTFIQSVCSILAL 414
>At2g36430 unknown protein (At2g36430)
Length = 448
Score = 178 bits (452), Expect = 4e-45
Identities = 118/387 (30%), Positives = 190/387 (48%), Gaps = 32/387 (8%)
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACF 60
+E+ K RYL + L D + +K EE+ R CYSE I S++F ++++ D CF
Sbjct: 82 IEEHKWRYLNVLLTRTQNLTLEDYMKSVKNVEEVARECYSETIHMDSEEFNEMMVLDGCF 141
Query: 61 IIEYFLRS---LEWPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDG 117
++E F + + + DPL++ W+ D + LENQ+P+FVLE LFNLT +
Sbjct: 142 LLELFRKVNNLVPFEPNDPLVAMAWVLPFFYRDFLCLENQIPFFVLETLFNLTRGDNENE 201
Query: 118 EVSSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDL 177
+S +AF +F N + + F E ++ +L DL
Sbjct: 202 TNASLQSLAFAFFN---------NMMHRTEEDLARFKE-------------LRAKHLLDL 239
Query: 178 LRVFYLPPDML-------PKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGA 230
LR ++P L P +EK + S S+L AG+KL +D +S L ++F G
Sbjct: 240 LRSSFIPESELHTPPATNPGKEKMPSHIIHSISKLRRAGIKLRELKDAESFLVVRFRHGT 299
Query: 231 LTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMI 290
+ +P V + + L N VA+EQCH + T Y LLD L +T +DV+ L ++ I+
Sbjct: 300 IEMPAITVDDFMSSFLENCVAYEQCHVACSMHFTTYATLLDCLTNTYKDVEYLCDQNIIE 359
Query: 291 NTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPW 350
N G +AK +N+L +V + L ++N +Y+ H A F YF++PW
Sbjct: 360 NYFGTDTELAKFVNSLGRDVAFDITQCYLKDLFEEVNEYYKSSWHVEWATFKFTYFNSPW 419
Query: 351 KITSFAAAIVLLLLTLIQATCSIISLF 377
S AA+VLL+L++IQ ++ +
Sbjct: 420 SFVSALAALVLLVLSVIQTIYTVFQAY 446
>At2g44930 unknown protein
Length = 515
Score = 165 bits (417), Expect = 4e-41
Identities = 117/363 (32%), Positives = 185/363 (50%), Gaps = 40/363 (11%)
Query: 28 IKKSEEIIRSCYSEAIEQTS-DDFVKIILTDACFIIEYFLRS-----------LEWPQED 75
I+ SE+ +R Y+E+ + DFV +IL D+ FI+ F+++ + + +ED
Sbjct: 162 IETSEKFVRESYAESTDWIKPQDFVDMILLDSVFILGVFIQAETTQNIKKKEEILFHEED 221
Query: 76 PLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFF-DVAFHYFKVHF 134
+ +P L + DLILLENQLP+ +LE L++P + + F D+ F+
Sbjct: 222 IIFQEPCLITTILEDLILLENQLPYVLLE---KLSKPFFANLKTKETFRDIILRAFR--- 275
Query: 135 LQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLLRVFYLPPDMLPKREKQ 194
L+ + TN ++FT + + + +Q+ PP+
Sbjct: 276 LKGEIKEGTNFRHFTDLFRRVRVETLCLTKEQIKSAKDK----------PPE-------- 317
Query: 195 IVKHLFSASQLVEAGVK-LHVGQDYQSVLELKFAKGALTIPRFEVCHWTETLLRNVVAFE 253
I+K L +A +L AGV + + + L + F +G L IP F TE ++RN++A E
Sbjct: 318 IIKSLHNADKLDSAGVDFVRLERKNDLSLVITFERGILEIPCFLADDNTERIMRNLMALE 377
Query: 254 QCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQE 313
QCHYP Y+ +Y LDFLIDT QDVD LV+KG++ N LG +VA+M+N LCL +V
Sbjct: 378 QCHYPLTAYVCNYIAFLDFLIDTDQDVDLLVKKGVIKNWLGHQASVAEMVNKLCLGLV-- 435
Query: 314 NINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQATCSI 373
+ Y + +LN YE ++ A YF W T+ AA V+L+LTLI S+
Sbjct: 436 DFGSHYYGIADRLNKHYESRRNRSIATLRRVYFKDLWTGTATIAAAVILVLTLIGTVASV 495
Query: 374 ISL 376
+ +
Sbjct: 496 LQV 498
>At2g28580 hypothetical protein
Length = 468
Score = 157 bits (396), Expect = 1e-38
Identities = 114/357 (31%), Positives = 189/357 (52%), Gaps = 33/357 (9%)
Query: 28 IKKSEEIIRSCYSEA-IEQTSDDFVKIILTDACFIIEYFLRS---LEWPQEDPLL---SK 80
I+ +E+ IR Y+E+ I + DFV++IL D+ FI+ +F+++ L + +++ +L S+
Sbjct: 119 IEINEKFIRDSYAESTIWINTKDFVEMILHDSVFILLFFIQTGSTLNFNKKEDILFNQSR 178
Query: 81 PWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFFDVAFHYFKVHFLQSILP 140
+ DLILLENQLP+ +LE LF + E +F D+ F+ + +
Sbjct: 179 LINATAILEDLILLENQLPYALLEKLFEPFSSNVNTKE--TFRDITLRAFR---FEGKIK 233
Query: 141 NETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLLRVFYLPPDMLPKREKQIVKHLF 200
E ++FT + + +Q+++ + P + ++I ++
Sbjct: 234 EEVRFQHFTDLFRCVRVSTLSLTEEQINIAKNE---------------PPKSRKI---MY 275
Query: 201 SASQLVEAGVK-LHVGQDYQSVLELKFAKGALTIPRFEVCHWTETLLRNVVAFEQCHYPF 259
+A +L AGV ++V ++ L + F G L +P F V TE ++RN++A EQCHYP
Sbjct: 276 NADKLDSAGVNFVNVDEENDLSLVITFKDGILKMPCFTVEDNTERVVRNLMALEQCHYPR 335
Query: 260 QTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQENINGGY 319
T++ DY LDFLI+T QDVD L +KGI+ N LG +V +M+N LCL +V + Y
Sbjct: 336 TTFVCDYISFLDFLINTDQDVDLLAKKGIVKNWLGHQGSVTEMVNKLCLGLV--DFGSHY 393
Query: 320 ISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQATCSIISL 376
+ LN Y++ ++ YF W T+ AA+VLL+LTLIQ SI+ +
Sbjct: 394 SDIVENLNKHYDNRLNRSVGTLRRVYFKDLWTGTATIAAVVLLVLTLIQTVASILQV 450
>At3g50170 putative protein
Length = 541
Score = 154 bits (389), Expect = 8e-38
Identities = 117/391 (29%), Positives = 190/391 (47%), Gaps = 19/391 (4%)
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACF 60
ME K R L L+R + + +++ EE R+CY I + ++F ++++ D CF
Sbjct: 150 MERHKWRALNKV-LKRLKQRIEMYTNAMRELEEKARACYEGPISLSRNEFTEMLVLDGCF 208
Query: 61 IIEYFLRSLE------WPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNL----- 109
++E F ++E + + DP+ + L ++ D+I+LENQLP FVL+ L L
Sbjct: 209 VLELFRGTVEGFTEIGYARNDPVFAMRGLMHSIQRDMIMLENQLPLFVLDRLLELQLGTQ 268
Query: 110 TEPSCIDGEVSSFFDVAFHYFKVHFL--QSILPNETNNKNFTINYFHE-HYQQYIMKPDQ 166
+ + FFD + QS L N T+ E H +
Sbjct: 269 NQTGIVAHVAVKFFDPLMPTGEALTKPDQSKLMNWLEKSLDTLGDKGELHCLDVFRRSLL 328
Query: 167 VSMQLHNLTDLLRVFYLPPDMLPKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKF 226
S N LL+ ++ KR++Q+V ++L EAGVK + +++F
Sbjct: 329 QSSPTPNTRSLLKRLTRNTRVVDKRQQQLVH---CVTELREAGVKFRKRKT-DRFWDIEF 384
Query: 227 AKGALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEK 286
G L IP+ + T++L N++AFEQCH +IT Y I +D LI++S+DV L
Sbjct: 385 KNGYLEIPKLLIHDGTKSLFSNLIAFEQCHIESSNHITSYIIFMDNLINSSEDVSYLHYC 444
Query: 287 GIMINTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYF 346
GI+ + LG + VA + N LC VV + + L +N +Y + KA H YF
Sbjct: 445 GIIEHWLGSDSEVADLFNRLCQEVVFDPKDSHLSRLSGDVNRYYNRKWNVLKATLTHKYF 504
Query: 347 STPWKITSFAAAIVLLLLTLIQATCSIISLF 377
+ PW SF+AA++LLLLTL Q+ ++ + +
Sbjct: 505 NNPWAYFSFSAAVILLLLTLCQSFYAVYAYY 535
>At3g50120 putative protein
Length = 531
Score = 154 bits (389), Expect = 8e-38
Identities = 110/384 (28%), Positives = 192/384 (49%), Gaps = 28/384 (7%)
Query: 14 LERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFIIEYFLRSLE--- 70
L+RT++G+ I +++ EE R+CY + +S++F+++++ D CF++E F ++E
Sbjct: 152 LKRTNQGIKMYIDAMRELEEKARACYEGPLSLSSNEFIEMLVLDGCFVLELFRGAVEGFT 211
Query: 71 ---WPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFFDVAF 127
+ + DP+ + ++ D+++LENQLP FVL L L G + VA
Sbjct: 212 ELGYARNDPVFAMRGSMHSIQRDMVMLENQLPLFVLNRLLELQL-----GTRNQTGLVA- 265
Query: 128 HYFKVHFLQSILP-NETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLT----DLLRVFY 182
+ F ++P +E K+ + + P +LH L LLR
Sbjct: 266 -QLAIRFFDPLMPTDEPLTKSGQSKLENSLARDKSFDPFADMGELHCLDVFRRSLLRSSP 324
Query: 183 LPPDMLPKR---------EKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGALTI 233
P L ++ +K+ + + ++L EAG+K + +++F G L I
Sbjct: 325 KPEPRLTRKRWSRNTRVADKRRQQLIHCVTELKEAGIKFR-RRKTDRFWDMQFKNGYLEI 383
Query: 234 PRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTL 293
PR + T++L N++AFEQCH IT Y I +D LID+ +DV L GI+ + L
Sbjct: 384 PRLLIHDGTKSLFLNLIAFEQCHIDSSNDITSYIIFMDNLIDSHEDVSYLHYCGIIEHWL 443
Query: 294 GDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKIT 353
G + VA + N LC VV + + L ++N +Y+ + ++A H YF+ PW I
Sbjct: 444 GSDSEVADLFNRLCQEVVFDTEDSYLSRLSIEVNRYYDHKWNAWRATLKHKYFNNPWAIV 503
Query: 354 SFAAAIVLLLLTLIQATCSIISLF 377
SF AA++LL+LT Q+ ++ + +
Sbjct: 504 SFCAAVILLVLTFSQSFYAVYAYY 527
>At1g67150 hypothetical protein
Length = 408
Score = 154 bits (389), Expect = 8e-38
Identities = 128/382 (33%), Positives = 180/382 (46%), Gaps = 41/382 (10%)
Query: 1 MEDLKLRYLKSFF--LERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQT-SDDFVKIILTD 57
MED K+ Y +F L+ L D IK E IR+ Y+ + SD FV++IL D
Sbjct: 53 MEDQKVLYKDAFTKRLDTDIAILEDMKTTIKDEEVNIRASYAVSTAWILSDIFVELILND 112
Query: 58 ACFIIEYFLRSLEWPQE--DPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFN-LTEPSC 114
+ FI+E+ LR E ++ D ++ KP+ V DL LLENQLP+F L L N +T+ C
Sbjct: 113 SIFIVEFILRMYESHEKIGDMIVDKPFYTSTVLDDLTLLENQLPYFCLSKLLNPITKRFC 172
Query: 115 IDGEVSSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNL 174
D S V F V+ I N+N N+F
Sbjct: 173 GD---QSLDQVILQLFSVNDSGMI------NENTKFNHF--------------------- 202
Query: 175 TDLLRVFY---LPPDMLPKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGAL 231
TDL R Y L ++ K + + SA+ L GV V + S L + F +G L
Sbjct: 203 TDLFRCVYKESLGQNLELNNSKSQIVDMKSANMLYSVGVDFKVVKREYS-LNVSFEEGCL 261
Query: 232 TIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMIN 291
+P F + +LRNV+A+EQCH P + T+Y ++FLI + +DV L G++ N
Sbjct: 262 ILPSFPADESSNIILRNVIAYEQCHDPENAFTTNYINFMNFLITSDEDVAILTSAGVLTN 321
Query: 292 TLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWK 351
+G S+ V KM+NNL + V+ N Y + KLN + + A YFS W
Sbjct: 322 GVGRSSMVLKMVNNLAIGVLLSN-QSQYHDIVEKLNIHHSSRRKRIWAKLRKVYFSDLWT 380
Query: 352 ITSFAAAIVLLLLTLIQATCSI 373
T+ AAI LLLLTL SI
Sbjct: 381 TTATLAAISLLLLTLAGTVASI 402
>At3g50150 putative protein
Length = 509
Score = 151 bits (381), Expect = 6e-37
Identities = 106/389 (27%), Positives = 193/389 (49%), Gaps = 41/389 (10%)
Query: 11 SFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIE-QTSDDFVKIILTDACFIIEYFLRSL 69
+ + RT + I +K+ EE R+CY I+ + S++F ++++ D CF++E F ++
Sbjct: 136 NMIMARTKHNIEMYIDAMKELEEEARACYQGPIDMKNSNEFTEMLVLDGCFVLELFKGTI 195
Query: 70 E------WPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFF 123
+ + + DP+ +K L ++ D+I+LENQLP FVL+ L L + +
Sbjct: 196 QGFQKIGYARNDPVFAKRGLMHSIQRDMIMLENQLPLFVLDRLLGLQTGT--PNQTGIVA 253
Query: 124 DVAFHYFKVHFLQSILPNET---------------NNKNFTINYFHEHYQQYIMKPDQVS 168
+VA +FK S + ++ N ++ FH +++ + +
Sbjct: 254 EVAVRFFKTLMPTSEVLTKSERSLDSQEKSDELGDNGGLHCLDVFHRS----LIQSSETT 309
Query: 169 MQLHNLTDLLRVFYLPPDMLPKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAK 228
Q D+ V ++++Q++ ++L AGV + ++ + +++F
Sbjct: 310 NQGTPYEDMSMV---------EKQQQLIH---CVTELRGAGVNF-MRKETGQLWDIEFKN 356
Query: 229 GALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGI 288
G L IP+ + T++L N++AFEQCH IT Y I +D LI++SQDV L GI
Sbjct: 357 GYLKIPKLLIHDGTKSLFSNLIAFEQCHTQSSNNITSYIIFMDNLINSSQDVSYLHHDGI 416
Query: 289 MINTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFST 348
+ + LG + VA + N LC V+ + +G L R++N +Y + KA YF+
Sbjct: 417 IEHWLGSDSEVADLFNRLCKEVIFDPKDGYLSQLSREVNRYYSRKWNSLKATLRQKYFNN 476
Query: 349 PWKITSFAAAIVLLLLTLIQATCSIISLF 377
PW SF+AA++LL LT Q+ ++ + +
Sbjct: 477 PWAYFSFSAAVILLFLTFFQSFFAVYAYY 505
>At3g50130 putative protein
Length = 564
Score = 144 bits (362), Expect = 1e-34
Identities = 111/379 (29%), Positives = 178/379 (46%), Gaps = 37/379 (9%)
Query: 14 LERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFIIEYF------LR 67
+ RT + I +K+ E+ R+CY I+ +S+ F ++++ D CF++E F
Sbjct: 188 MARTKHDIEMYIDAMKELEDRARACYEGPIDLSSNKFSEMLVLDGCFVLELFRGADEGFS 247
Query: 68 SLEWPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFFDVAF 127
L + + DP+ + ++ D+++LENQLP FVL L + + S
Sbjct: 248 ELGYDRNDPVFAMRGSMHSIQRDMVMLENQLPLFVLNRLLEIQLGKRHQTGLVS------ 301
Query: 128 HYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLLRVFYLPPDM 187
V F ++P + + + + I D+ +LH L D+ R L P
Sbjct: 302 -RLAVRFFDPLMPTDEPLTKTDDSLEQDKFFNPIADKDKG--ELHCL-DVFRRNLLRPCS 357
Query: 188 LP-----------------KREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGA 230
P KR++Q++ ++L EAG+K + +++F G
Sbjct: 358 NPEPRLSRMRWSWRTRVADKRQQQLIH---CVTELREAGIKFRTRKT-DRFWDIRFKNGY 413
Query: 231 LTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMI 290
L IP+ + T++L N++AFEQCH IT Y I +D LID+S+DV L GI+
Sbjct: 414 LEIPKLLIHDGTKSLFSNLIAFEQCHIDSSNDITSYIIFMDNLIDSSEDVRYLHYCGIIE 473
Query: 291 NTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPW 350
+ LG+ VA + N LC V + N L K++ Y + KAI H YF+ PW
Sbjct: 474 HWLGNDYEVADLFNRLCQEVAFDPQNSYLSQLSNKVDRNYSRKWNVLKAILKHKYFNNPW 533
Query: 351 KITSFAAAIVLLLLTLIQA 369
SF AA+VLL+LTL Q+
Sbjct: 534 AYFSFFAALVLLVLTLFQS 552
>At5g22540 unknown protein
Length = 440
Score = 136 bits (343), Expect = 2e-32
Identities = 100/389 (25%), Positives = 179/389 (45%), Gaps = 42/389 (10%)
Query: 5 KLRYLKSFFLERTHKGL--GDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFII 62
K R+LK F + KG + + + E +IR YSE + S++ V++++ D CFI+
Sbjct: 72 KRRFLKFFVAKMEEKGFVPQELVKAVSSLEGVIRGSYSEDLGLDSENLVQMMVLDGCFIL 131
Query: 63 EYFL---RSLEWPQ-EDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGE 118
F +E+ +DP+ PW+ ++ DL+LLENQ+P+ +L+ LF ++ G
Sbjct: 132 TLFFVVSGKVEYTNLDDPIFRMPWILPSIRADLLLLENQVPYVLLQTLFETSKLVTCSG- 190
Query: 119 VSSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLL 178
++AF +F S+ ET ++ +HY ++ +L DL+
Sbjct: 191 ---LNEIAFEFFNY----SLQKPET--------FWEKHY----------GLEAKHLLDLI 225
Query: 179 RVFYLPPDMLPKREKQIVKHLF----------SASQLVEAGVKLHVGQDYQSVLELKFAK 228
R ++P + + K F SA +L G+K ++ S+L++ ++
Sbjct: 226 RKTFVPVPSQRRIKDHSSKSSFNDHEYLGFVLSAKKLHLRGIKFKPRKNTDSILDISYSN 285
Query: 229 GALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGI 288
G L IP + +T ++ N VAFEQ + +IT Y + LI+ D L E+ I
Sbjct: 286 GVLHIPPVVMDDFTASIFLNCVAFEQLYADSSNHITSYVAFMACLINEESDASFLSERRI 345
Query: 289 MINTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFST 348
+ N G + V++ + ++ + + +N + H + A FIH +F +
Sbjct: 346 LENYFGTEDEVSRFYKRIGKDIALDLEKSYLAKVFEGVNEYTSQGFHVHCAEFIHTHFDS 405
Query: 349 PWKITSFAAAIVLLLLTLIQATCSIISLF 377
PW S AA++LLL +Q + S F
Sbjct: 406 PWTFASSFAALLLLLFAALQVFFAAYSYF 434
>At3g60470 putative protein
Length = 540
Score = 136 bits (342), Expect = 2e-32
Identities = 76/188 (40%), Positives = 114/188 (60%), Gaps = 6/188 (3%)
Query: 191 REKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGALTIPRFEVCHWTETLLRNVV 250
+E I+KH+++A++L AGVK D S+ ++ F G L IP V E LRN++
Sbjct: 354 KELPIIKHMYNAAKLQSAGVKFKAVTDEFSI-DVTFENGCLKIPCLWVPDDAEITLRNIM 412
Query: 251 AFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGD--SNAVAKMINNLCL 308
A EQCHYPF+ Y+ ++ LDFLIDT +DVD L+E GI +N G+ S AVA+M+N L
Sbjct: 413 ALEQCHYPFKAYVCNFVSFLDFLIDTDKDVDLLMENGI-LNNWGEKSSVAVAEMVNTLFS 471
Query: 309 NVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQ 368
VV+ Y + ++N +YE+P ++ + I YF W+ T+ AA +LL++TLIQ
Sbjct: 472 GVVES--RSYYAGIASRVNAYYENPVNRSRTILGRQYFGNLWRGTATVAAALLLVMTLIQ 529
Query: 369 ATCSIISL 376
SI+ +
Sbjct: 530 TVASILQV 537
Score = 70.9 bits (172), Expect = 1e-12
Identities = 52/126 (41%), Positives = 70/126 (55%), Gaps = 15/126 (11%)
Query: 1 MEDLKLRYLKSFFLERT--HKGLGDCIGYIKKSEEIIRSCYSEAIEQT-SDDFVKIILTD 57
ME+ K YL F ER K + + IK+ E+IIR+ YSE+ S FV +IL D
Sbjct: 85 MEEHKKIYLARFS-ERLAGQKTIDELRRRIKEEEDIIRASYSESTAWIESPKFVDMILHD 143
Query: 58 ACFIIEYFLRSLEWP-----------QEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDL 106
+ FIIE+ LRS ++ DP+ +P L+ V DLILLENQLP+F+LE L
Sbjct: 144 SVFIIEFLLRSKDYTLLEDQGSCIKKTWDPIFEQPCLQTTVDEDLILLENQLPYFILEKL 203
Query: 107 FNLTEP 112
F+ P
Sbjct: 204 FDPIVP 209
>At3g50180 putative protein
Length = 588
Score = 135 bits (340), Expect = 4e-32
Identities = 115/383 (30%), Positives = 181/383 (47%), Gaps = 44/383 (11%)
Query: 14 LERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFIIEY-------FL 66
L+RT++G+ + + + EE R+CY +I +S++F +++L D CFI+E FL
Sbjct: 227 LKRTNQGIEVFLDAMIELEEKARACYEGSIVLSSNEFTEMLLLDGCFILELLQGVNEGFL 286
Query: 67 RSLEWPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFFDVA 126
+ L + DP+ + ++ D+I+LENQLP FVL L L + +
Sbjct: 287 K-LGYDHNDPVFAVRGSMHSIQRDMIMLENQLPLFVLNRLLEL--------QPGTQNQTG 337
Query: 127 FHYFKVHFLQSILPN-ETNNKNFTINYFHEHYQQYIMKPDQVSM-QLHNLTDLLRVFYLP 184
V F ++P ET +N P VS +LH L R P
Sbjct: 338 LVELVVRFFIPLMPTAETLTEN--------------SPPRGVSNGELHCLDVFHRSLLFP 383
Query: 185 PDMLPKREKQIV-KHLF----SASQLVEAGVKLHVGQDYQSVLELKFAKGALTIPRFEVC 239
++ KHL + ++L +AG K + + ++KF+ G L IP +
Sbjct: 384 RSSGKANYSRVADKHLQRVIPTVTELRDAGFKFKLNKT-DRFWDIKFSNGYLEIPGLLIH 442
Query: 240 HWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAV 299
T++L N++AFEQCH IT Y I +D LID+ +D+ L GI+ ++LG ++ V
Sbjct: 443 DGTKSLFLNLIAFEQCHIESSNDITSYIIFMDNLIDSPEDISYLHHCGIIEHSLGSNSEV 502
Query: 300 AKMINNLCLNVVQENINGGYIS--LCRKLNCFYEDPSHK---YKAIFIHDYFSTPWKITS 354
A M N LC VV + Y+S L C+ ++ S K K I Y PW S
Sbjct: 503 ADMFNQLCQEVVFDT-KDIYLSQLLIEVHRCYKQNYSRKLNSLKTTLILKYLDNPWAYLS 561
Query: 355 FAAAIVLLLLTLIQATCSIISLF 377
F AA++LL+LT Q+ + + F
Sbjct: 562 FFAAVILLILTFSQSYFAAYAYF 584
>At3g50160 putative protein
Length = 511
Score = 135 bits (340), Expect = 4e-32
Identities = 100/341 (29%), Positives = 164/341 (47%), Gaps = 33/341 (9%)
Query: 25 IGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFIIEYFLRSLEWPQE------DPLL 78
I +K+ EE R+CY I ++F+++++ D FIIE F + E QE DP+
Sbjct: 164 IDAMKELEEKARACYQGPINMNRNEFIEMLVLDGVFIIEIFKGTSEGFQEIGYAPNDPVF 223
Query: 79 SKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFFDVAFHYFKVHFLQSI 138
L ++ D+++LENQLPW VL+ L L P +D F F Q +
Sbjct: 224 GMRGLMQSIRRDMVMLENQLPWSVLKGLLQLQRPDVLDKVNVQLFQP--------FFQPL 275
Query: 139 LPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLLRVFYLPPDMLPKREKQIVKH 198
LP + + ++ +++ S + M+ K+ +Q++
Sbjct: 276 LPTREVLTEEGGLHCLDVLRRGLLQSSGTSDE-------------DMSMVNKQPQQLIH- 321
Query: 199 LFSASQLVEAGVKLHVGQDYQSVLELKFAKGALTIPRFEVCHWTETLLRNVVAFEQCHYP 258
++L AGV+ + ++ +++F G L IP+ + T++L N++AFEQCH
Sbjct: 322 --CVTELRNAGVEF-MRKETGHFWDIEFKNGYLKIPKLLIHDGTKSLFLNLIAFEQCHIK 378
Query: 259 FQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQENINGG 318
IT Y I +D LI++S+DV L GI+ N LG + V+ + N L V+ + N G
Sbjct: 379 SSKKITSYIIFMDNLINSSEDVSYLHHYGIIENWLGSDSEVSDLFNGLGKEVIFDP-NDG 437
Query: 319 YIS-LCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAA 358
Y+S L ++N +Y + KA H YF+ PW SF AA
Sbjct: 438 YLSALTGEVNIYYRRKWNYLKATLRHKYFNNPWAYFSFIAA 478
>At3g50140 putative protein
Length = 508
Score = 133 bits (335), Expect = 1e-31
Identities = 105/364 (28%), Positives = 170/364 (45%), Gaps = 37/364 (10%)
Query: 14 LERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFIIEYFLRSLE--- 70
++RT +G+ I +K+ EE R+CY I +S+ F ++++ D CF+++ F + E
Sbjct: 162 MKRTKQGIEMYIDAMKELEERARACYEGPIGLSSNKFTQMLVLDGCFVLDLFRGAYEGFS 221
Query: 71 ---WPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFFDVAF 127
+ + DP+ + ++ D+++LENQLP FVL L L + + + V F
Sbjct: 222 KLGYDRNDPVFAMRGSMHSIRRDMLMLENQLPLFVLNRLLELQLGTQYQTGLVAQLAVRF 281
Query: 128 H--YFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLLRVFYLPP 185
+ + + N N N N D+ +LH L D+ R L P
Sbjct: 282 FNPLMPTYMSSTKIENSQENNNKFFNPI----------ADKEKEELHCL-DVFRRSLLQP 330
Query: 186 DMLPKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGALTIPRFEVCHWTETL 245
+ P + ++ + +S LV Q V EL+ A T++L
Sbjct: 331 SLKP--DPRLSRSRWSRKPLV---ADKRQQQLLHCVTELREAG-------------TKSL 372
Query: 246 LRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINN 305
N++A+EQCH IT Y I +D LID+++D+ L I+ + LG+ + VA + N
Sbjct: 373 FSNLIAYEQCHIDSTNDITSYIIFMDNLIDSAEDIRYLHYYDIIEHWLGNDSEVADVFNR 432
Query: 306 LCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLT 365
LC V + N L K++ +Y + KA H YFS PW SF AA++LLLLT
Sbjct: 433 LCQEVAFDLENTYLSELSNKVDRYYNRKWNVLKATLKHKYFSNPWAYFSFFAAVILLLLT 492
Query: 366 LIQA 369
L Q+
Sbjct: 493 LFQS 496
>At3g44710 unknown protein
Length = 504
Score = 125 bits (313), Expect = 5e-29
Identities = 108/414 (26%), Positives = 191/414 (46%), Gaps = 51/414 (12%)
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKS---EEIIRSCYSEAIEQTSDDFVKIILTD 57
+E+ KLR+LK F E KG+ D G IK EE IR YSE++ +++++ D
Sbjct: 85 IEEHKLRFLKIFMDEAKRKGV-DTNGLIKAVSVLEEDIRDSYSESLYSDGKKLIEMMVLD 143
Query: 58 ACFIIEYFLR-----SLEWPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEP 112
CFI+ FL S + DP+ + PW+ ++ DLILLENQ+P+F+L+ +F+ ++
Sbjct: 144 GCFILMIFLVVAGVVSHSEIENDPIFAIPWILPAIRNDLILLENQVPFFLLQTIFDRSKI 203
Query: 113 SCIDGEVSSFFDVAFHYFKVHFLQSI---LPNETNNKNFTINYFHEHYQQYIMKPDQVSM 169
+ S ++ FH+F +S L ++ N ++ Y + K ++ S
Sbjct: 204 E----KSSGLNEIIFHFFNYSLQKSNTFWLKHQKVEANHLLDLIRNIYMPDVPKEEKKSN 259
Query: 170 QLHNLTDLLRVFYLPP------DMLPKREKQIV--------------------------- 196
L + ++R +P D++ K+ KQ V
Sbjct: 260 HLLDCM-MIRKICMPRESKQELDVMLKKGKQEVDISMLEEGIPKSDSLEEEDSTTGRHHL 318
Query: 197 KHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGALTIPRFEVCHWTETLLRNVVAFEQCH 256
K + SA +L G+K + +++++++ L IP + + +L N VAFEQ +
Sbjct: 319 KMVLSARKLQLKGIKFKARKKAETLMDIRHKGKLLQIPPLILDDFLIAVLLNCVAFEQYY 378
Query: 257 -YPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQENI 315
Y + ++T Y + L+ + D L E GI+ N G + V++ + +V+ +
Sbjct: 379 SYCTKQHMTSYVAFMGCLLKSEADAMFLSEVGILENYFGSGDEVSRFFKVVGKDVLFDID 438
Query: 316 NGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQA 369
+ +N + H A F H +F +PW S AA+ L++LT+IQA
Sbjct: 439 ESYLAGIFEGVNKYTSSGWHVQWAGFKHTHFDSPWTALSSCAALTLVILTIIQA 492
>At1g65985 putative protein
Length = 456
Score = 125 bits (313), Expect = 5e-29
Identities = 117/423 (27%), Positives = 177/423 (41%), Gaps = 85/423 (20%)
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQT-SDDFVKIILTDAC 59
ME K Y +SF + + I+ E+ IR+CY E+ E S+ FV +IL D+
Sbjct: 69 MEVTKKTYFESFTKRVGEDTIEEMRKTIQTEEKNIRNCYEESTEWIPSELFVDLILQDSI 128
Query: 60 FIIEYFLRSLEWPQ---EDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCID 116
FI+E +R + + +D L V DL+LLENQLP+F+L+ LF+ + +D
Sbjct: 129 FIMELIIRLSGFDRIHSDDRNLDF----AIVMNDLLLLENQLPYFILDSLFSFSMTKYLD 184
Query: 117 GEVSSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTD 176
F + K+ KNF +LTD
Sbjct: 185 SHTLDSFILRCFGLKIE----------EKKNFM-----------------------HLTD 211
Query: 177 LLRVFYLPP-DMLPKRE------KQIVKHLFSASQLVEAGVKLHVGQDYQSV-------- 221
+ R Y D +P RE K + L +A L AGV DY +
Sbjct: 212 MYRCVYKESLDYIPPREYSLNRVKWSITELRNAENLSNAGVNFK-SYDYLDLSKQDEKLD 270
Query: 222 ---------------------------LELKFAKGALTIPRFEVCHWTETLLRNVVAFEQ 254
L ++ KG L++P F T+ +LRNV+AFEQ
Sbjct: 271 IKITRDKSDSELTKLPTNKRERVEGVSLHVELKKGCLSMPSFWANDRTDMILRNVIAFEQ 330
Query: 255 CHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQEN 314
C + + + Y L+ LI + +DV+ L EKG+++N +G + V M+N L + V E
Sbjct: 331 CQHGIIPFTSHYIQFLNALIVSDRDVELLSEKGVVMNNMGRPSLVVDMVNKLKVG-VYEG 389
Query: 315 INGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQATCSII 374
Y + L Y+ + A YFS W T+ AA++LLLLTL+ S+I
Sbjct: 390 YTSQYYDIAIDLKAHYKSRRKRCWATLRKVYFSDLWTGTATLAAVLLLLLTLVGTVASVI 449
Query: 375 SLF 377
+
Sbjct: 450 QAY 452
>At3g47200 unknown protein
Length = 476
Score = 123 bits (309), Expect = 1e-28
Identities = 109/399 (27%), Positives = 190/399 (47%), Gaps = 56/399 (14%)
Query: 5 KLRYLKSFFLERTHKGLGD--CIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFII 62
K R L+ F E K + + + + E+ IR YSE ++ T D + +++ D CFI+
Sbjct: 87 KPRLLQLFLDEAKKKDVEENVLVKAVVDLEDKIRKSYSEELK-TGHDLMFMMVLDGCFIL 145
Query: 63 EYFL---RSLEWPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEV 119
FL ++E EDP+ S PWL ++ DL+LLENQ+P+FVL+ L+ ++ G
Sbjct: 146 MVFLIMSGNIEL-SEDPIFSIPWLLSSIQSDLLLLENQVPFFVLQTLYVGSK----IGVS 200
Query: 120 SSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLLR 179
S +AFH+FK N + + +Y+ +H + Y K +L DL+R
Sbjct: 201 SDLNRIAFHFFK---------NPIDKEG---SYWEKH-RNYKAK---------HLLDLIR 238
Query: 180 VFYLP---------------------PDMLPKREKQIVKHLFSASQLVEAGVKLHVGQDY 218
+LP +P + + V + SA +L G+K + +
Sbjct: 239 ETFLPNTSESDKASSPHVQVQLHEGKSGNVPSVDSKAVPLILSAKRLRLQGIKFRLRRSK 298
Query: 219 Q-SVLELKFAKGALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTS 277
+ S+L ++ K L IP+ + + N VAFEQ + IT Y + + L++
Sbjct: 299 EDSILNVRLKKNKLQIPQLRFDGFISSFFLNCVAFEQFYTDSSNEITTYIVFMGCLLNNE 358
Query: 278 QDVDKL-VEKGIMINTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHK 336
+DV L +K I+ N G +N V++ + +VV E ++ + +N + + +
Sbjct: 359 EDVTFLRNDKLIIENHFGSNNEVSEFFKTISKDVVFEVDTSYLNNVFKGVNEYTKKWYNG 418
Query: 337 YKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQATCSIIS 375
A F H +F +PW S A + ++LLT++Q+T +I+S
Sbjct: 419 LWAGFRHTHFESPWTFLSSCAVLFVILLTMLQSTVAILS 457
>At3g47210 unknown protein
Length = 474
Score = 113 bits (283), Expect = 1e-25
Identities = 89/333 (26%), Positives = 157/333 (46%), Gaps = 48/333 (14%)
Query: 5 KLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACFIIEY 64
KLR+L FL + E+ IR YSE +E + + V +++ D CFI+
Sbjct: 134 KLRFLH-LFLRTASVDRDVLFNAVVDWEDEIRKSYSEGLEGSPHELVYMMILDGCFILML 192
Query: 65 FL---RSLE-WPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTE---PSCIDG 117
L R +E + EDP+L+ PW+ ++ DL+LLENQ+P+FVL+ LF+ +E P ++
Sbjct: 193 LLIVSRKIELYESEDPILTIPWILPSIQSDLLLLENQVPFFVLQTLFDKSEIGVPGDLNR 252
Query: 118 EVSSFFDVAFHYFKVHFLQSILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDL 177
SFF+++ + ++++ ++NF + L DL
Sbjct: 253 MAFSFFNLSMDKPERYWVK--------HRNFNAKH---------------------LLDL 283
Query: 178 LRVFYLPPDMLP----------KREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFA 227
+R+ +LP D K+ + L SA++L G+ + S+L+++
Sbjct: 284 IRMSFLPMDGYEDFQLTKGKSRKKSSSGLTLLLSATRLSLQGIDFSLRSGADSMLDIRLK 343
Query: 228 KGALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKG 287
K L IP + + ++L N VAFEQ + +IT Y + + L++ +D L +
Sbjct: 344 KNRLQIPVLRLDGFIISILLNCVAFEQFYAKSTNHITSYVVFMGCLLNGKEDATFLSRRR 403
Query: 288 IMINTLGDSNAVAKMINNLCLNVVQENINGGYI 320
I+ N G V+K +C +VV + I+ Y+
Sbjct: 404 IIENYFGSEKEVSKFFKTICKDVVFD-IHASYL 435
>At5g22550 unknown protein
Length = 443
Score = 99.0 bits (245), Expect = 4e-21
Identities = 88/330 (26%), Positives = 149/330 (44%), Gaps = 31/330 (9%)
Query: 74 EDPLLSKPWLRCDVKLDLILLENQLPWFVLEDLFN---LTEPSCIDGEVSSFFDVAF--- 127
+DP+ W+ ++ DL+LLENQ+P F+L+ L L + ++ FFD +
Sbjct: 113 KDPIFKLRWILPTLRSDLLLLENQVPLFLLKVLLETSKLAPSTSLNMLAFKFFDYSIKKP 172
Query: 128 --------HYFKVHFLQSIL-------PNETNNKNFTINYFHEHYQQYIMKPDQVSMQLH 172
+ H L I P T + IN F+ ++Y ++ L
Sbjct: 173 EGFWEKHNNLRAKHLLDLIRKTFIPAPPPSTTPRQCCINIFNGP-REYSRTETSKNICLG 231
Query: 173 NLTDLLRVFYL----PPDMLPKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAK 228
++ + PP P + + + SA +L G+K ++ ++ L++ F
Sbjct: 232 KISCSKEITGAQTSSPP---PPPRRPFLGLIVSARKLRLRGIKFMRKENVETPLDISFKS 288
Query: 229 GALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGI 288
G + IP + LL N VAFEQ + T IT + I + LI+T D L+EKGI
Sbjct: 289 GLVEIPLLVFDDFISNLLINCVAFEQFNMSCSTEITSFVIFMGCLINTEDDATFLIEKGI 348
Query: 289 MINTLGDSNAVAKMINNLCLNVVQENINGGYIS-LCRKLNCFYEDPSHKYKAIFIHDYFS 347
+ N G V+ N+ + + +I+ ++S + +N + H + A F + +F+
Sbjct: 349 LENYFGTGEEVSLFFKNIGKD-ISFSISKSFLSNVFEGVNEYTSQGYHVHWAGFKYTHFN 407
Query: 348 TPWKITSFAAAIVLLLLTLIQATCSIISLF 377
TPW S AA+VLLLLT+ QA + + F
Sbjct: 408 TPWTFLSSCAALVLLLLTIFQAFFAAYAYF 437
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,330,084
Number of Sequences: 26719
Number of extensions: 350748
Number of successful extensions: 1123
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1021
Number of HSP's gapped (non-prelim): 52
length of query: 377
length of database: 11,318,596
effective HSP length: 101
effective length of query: 276
effective length of database: 8,619,977
effective search space: 2379113652
effective search space used: 2379113652
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146743.5


