

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.2 - phase: 0
(68 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
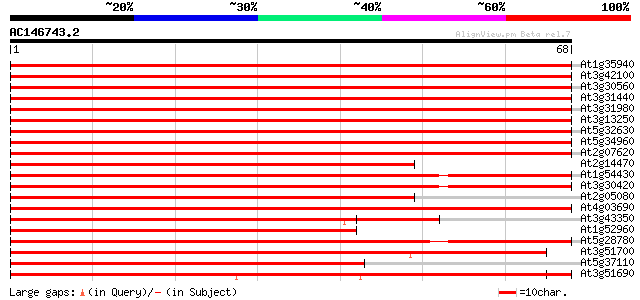
Score E
Sequences producing significant alignments: (bits) Value
At1g35940 hypothetical protein 99 5e-22
At3g42100 putative protein 97 1e-21
At3g30560 hypothetical protein 97 1e-21
At3g31440 hypothetical protein 97 2e-21
At3g31980 hypothetical protein 94 1e-20
At3g13250 hypothetical protein 94 1e-20
At5g32630 putative protein 93 3e-20
At5g34960 putative protein 92 5e-20
At2g07620 putative helicase 87 2e-18
At2g14470 pseudogene 85 6e-18
At1g54430 hypothetical protein 79 3e-16
At3g30420 hypothetical protein 79 4e-16
At2g05080 putative helicase 77 1e-15
At4g03690 hypothetical protein 77 2e-15
At3g43350 putative protein 72 4e-14
At1g52960 hypothetical protein 72 4e-14
At5g28780 putative protein 71 9e-14
At3g51700 unknown protein 69 4e-13
At5g37110 putative helicase 67 2e-12
At3g51690 putative protein 63 2e-11
>At1g35940 hypothetical protein
Length = 1678
Score = 98.6 bits (244), Expect = 5e-22
Identities = 47/68 (69%), Positives = 57/68 (83%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQSLEH+G+YLP P+FSHGQLYVA+SRVTS+ GLKILI D D +NVV
Sbjct: 1610 MTINKSQGQSLEHIGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGKLQKQTTNVV 1669
Query: 61 YREVFRNL 68
++EVF+N+
Sbjct: 1670 FKEVFQNI 1677
>At3g42100 putative protein
Length = 1752
Score = 97.1 bits (240), Expect = 1e-21
Identities = 47/68 (69%), Positives = 57/68 (83%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQSLE VG+YLP P+FSHGQLYVA+SRVTS+ GLKILI D D + +NVV
Sbjct: 1684 MTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGNMQKQTTNVV 1743
Query: 61 YREVFRNL 68
++EVF+N+
Sbjct: 1744 FKEVFQNI 1751
>At3g30560 hypothetical protein
Length = 1473
Score = 97.1 bits (240), Expect = 1e-21
Identities = 45/68 (66%), Positives = 56/68 (82%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQSL +VG+YLP P+FSHGQLYVA+SRV S+GGLK+LI D + +NVV
Sbjct: 1406 MTINKSQGQSLGNVGIYLPKPVFSHGQLYVAMSRVKSKGGLKVLITDSKGKQKNETTNVV 1465
Query: 61 YREVFRNL 68
++E+FRNL
Sbjct: 1466 FKEIFRNL 1473
>At3g31440 hypothetical protein
Length = 536
Score = 96.7 bits (239), Expect = 2e-21
Identities = 46/68 (67%), Positives = 57/68 (83%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQSLEHVG+YLP P+FSHGQLYVA+SRVTS+ GLKILI D + +N+V
Sbjct: 468 MTINKSQGQSLEHVGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKNGKLQKQTTNIV 527
Query: 61 YREVFRNL 68
++EVF+N+
Sbjct: 528 FKEVFQNI 535
>At3g31980 hypothetical protein
Length = 1099
Score = 94.4 bits (233), Expect = 1e-20
Identities = 43/68 (63%), Positives = 56/68 (82%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQSL+ VG+YLP P+FSHGQLYVA+SRVTS+ GLK+LI D + + NVV
Sbjct: 1030 MTINKSQGQSLKEVGIYLPKPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQTMNVV 1089
Query: 61 YREVFRNL 68
++E+F+N+
Sbjct: 1090 FKEIFQNI 1097
>At3g13250 hypothetical protein
Length = 1419
Score = 94.4 bits (233), Expect = 1e-20
Identities = 46/68 (67%), Positives = 56/68 (81%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQSLE VG+YLP P+FSHGQLYVA+SRVTS+ GLKILI D + +NVV
Sbjct: 1352 MTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKTGLKILILDKEGKIQKQTTNVV 1411
Query: 61 YREVFRNL 68
++EVF+N+
Sbjct: 1412 FKEVFQNI 1419
>At5g32630 putative protein
Length = 856
Score = 92.8 bits (229), Expect = 3e-20
Identities = 45/68 (66%), Positives = 55/68 (80%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQSLE G+YLP +FSHGQLYVA+SRVTS+ GLKILI D D D +NVV
Sbjct: 788 MTINKSQGQSLEQAGLYLPKLVFSHGQLYVALSRVTSKSGLKILILDKDGDIQKQTTNVV 847
Query: 61 YREVFRNL 68
++E+F+N+
Sbjct: 848 FKELFQNI 855
>At5g34960 putative protein
Length = 1033
Score = 92.0 bits (227), Expect = 5e-20
Identities = 45/68 (66%), Positives = 54/68 (79%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTIN SQGQSLEHVG+YLP +FSHGQLYVA+SRVTS+ GLK LI D D +NVV
Sbjct: 965 MTINTSQGQSLEHVGLYLPKAVFSHGQLYVALSRVTSKKGLKFLILDKDGKLQKQTTNVV 1024
Query: 61 YREVFRNL 68
++EVF+N+
Sbjct: 1025 FKEVFQNI 1032
>At2g07620 putative helicase
Length = 1241
Score = 87.0 bits (214), Expect = 2e-18
Identities = 41/68 (60%), Positives = 53/68 (77%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
M I KSQGQSL+ V +YLP P+FSHGQLYVA+SRVTS+ GLK+LI D + + NVV
Sbjct: 1172 MRIKKSQGQSLKEVEIYLPRPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQTMNVV 1231
Query: 61 YREVFRNL 68
++E+F+NL
Sbjct: 1232 FKEIFQNL 1239
>At2g14470 pseudogene
Length = 1265
Score = 85.1 bits (209), Expect = 6e-18
Identities = 40/49 (81%), Positives = 45/49 (91%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDD 49
MTINKS+GQSLEHVG+YLP P+FSHGQLYVA+SRVTS+ GLKILI D D
Sbjct: 1214 MTINKSEGQSLEHVGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKD 1262
>At1g54430 hypothetical protein
Length = 1639
Score = 79.3 bits (194), Expect = 3e-16
Identities = 40/68 (58%), Positives = 48/68 (69%), Gaps = 1/68 (1%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTINKSQGQ+L V +YLP P+FSHGQLYVA+SRVTS GL +L + +N+V
Sbjct: 1565 MTINKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLDTSKKKEG-KYVTNIV 1623
Query: 61 YREVFRNL 68
YREVF L
Sbjct: 1624 YREVFNGL 1631
>At3g30420 hypothetical protein
Length = 837
Score = 79.0 bits (193), Expect = 4e-16
Identities = 39/68 (57%), Positives = 48/68 (70%), Gaps = 1/68 (1%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MT+NKSQGQ+L V +YLP P+FSHGQLYVA+SRVTS GL +L + +N+V
Sbjct: 763 MTVNKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLDTSKKKEG-KYVTNIV 821
Query: 61 YREVFRNL 68
YREVF L
Sbjct: 822 YREVFNGL 829
>At2g05080 putative helicase
Length = 1219
Score = 77.4 bits (189), Expect = 1e-15
Identities = 37/49 (75%), Positives = 43/49 (87%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDD 49
MTINKSQGQSL+ VG++LP P FSH QLYVAISRVTS+ GLKILI +D+
Sbjct: 1131 MTINKSQGQSLKRVGIFLPRPCFSHSQLYVAISRVTSKSGLKILIVNDE 1179
>At4g03690 hypothetical protein
Length = 570
Score = 76.6 bits (187), Expect = 2e-15
Identities = 37/68 (54%), Positives = 49/68 (71%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
M INKSQ QSL +VG+ L P+FSHGQLYVA+SRV S+ LK+LI D +NV+
Sbjct: 502 MMINKSQRQSLANVGINLLKPVFSHGQLYVAMSRVKSKARLKVLITDSKGKQKKETTNVI 561
Query: 61 YREVFRNL 68
++E+F+NL
Sbjct: 562 FKEIFQNL 569
>At3g43350 putative protein
Length = 830
Score = 72.4 bits (176), Expect = 4e-14
Identities = 34/42 (80%), Positives = 38/42 (89%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLK 42
MTINKSQGQ+LE VG+YLP P+FSHGQLYVAISRVTS+ G K
Sbjct: 787 MTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTGTK 828
Score = 69.7 bits (169), Expect = 3e-13
Identities = 35/53 (66%), Positives = 43/53 (81%), Gaps = 1/53 (1%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRG-GLKILINDDDDDN 52
MTINKSQGQ+LE VG+YLP P+FSHGQLYVAISRVTS+ G +++ + D N
Sbjct: 671 MTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTVGCPVMLLRNMDPN 723
Score = 69.7 bits (169), Expect = 3e-13
Identities = 35/53 (66%), Positives = 43/53 (81%), Gaps = 1/53 (1%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRG-GLKILINDDDDDN 52
MTINKSQGQ+LE VG+YLP P+FSHGQLYVAISRVTS+ G +++ + D N
Sbjct: 555 MTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTIGCPVMLLRNMDPN 607
>At1g52960 hypothetical protein
Length = 924
Score = 72.4 bits (176), Expect = 4e-14
Identities = 34/42 (80%), Positives = 38/42 (89%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLK 42
MTINKSQGQ+LE VG+YLP P+FSHGQLYVAISRVTS+ G K
Sbjct: 881 MTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTGTK 922
>At5g28780 putative protein
Length = 337
Score = 71.2 bits (173), Expect = 9e-14
Identities = 38/68 (55%), Positives = 49/68 (71%), Gaps = 2/68 (2%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILINDDDDDNIDVASNVV 60
MTI K+QGQSL+ +YLP+P+FSH QLYVA+SRVTS GL IL DD + D N+V
Sbjct: 265 MTIIKNQGQSLKSDVLYLPNPVFSHVQLYVALSRVTSPIGLTILHGDDQKN--DEVKNIV 322
Query: 61 YREVFRNL 68
Y+E + +L
Sbjct: 323 YKEFYNDL 330
>At3g51700 unknown protein
Length = 344
Score = 68.9 bits (167), Expect = 4e-13
Identities = 35/66 (53%), Positives = 47/66 (71%), Gaps = 1/66 (1%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKILIND-DDDDNIDVASNV 59
MTI++SQ Q+L VG+YLP + HGQ YVAIS+V SR GLK+LI D D + + NV
Sbjct: 267 MTIDESQRQTLSKVGIYLPRQLLFHGQRYVAISKVKSRAGLKVLITDKDGKPDQEETKNV 326
Query: 60 VYREVF 65
V++E+F
Sbjct: 327 VFKELF 332
>At5g37110 putative helicase
Length = 1307
Score = 66.6 bits (161), Expect = 2e-12
Identities = 33/43 (76%), Positives = 37/43 (85%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGLKI 43
MTINKSQGQSL+ VG++L P FSHGQLYVAISRVTS+ LKI
Sbjct: 1250 MTINKSQGQSLKRVGIFLLRPCFSHGQLYVAISRVTSKTRLKI 1292
>At3g51690 putative protein
Length = 374
Score = 63.2 bits (152), Expect = 2e-11
Identities = 33/66 (50%), Positives = 45/66 (68%), Gaps = 1/66 (1%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHG-QLYVAISRVTSRGGLKILINDDDDDNIDVASNV 59
MTI++SQ +L VG+YLP +FSHG Q++VAIS+V SR GLK+LI D D + + A N
Sbjct: 239 MTIDESQVHTLSKVGLYLPRQVFSHGRQMFVAISKVKSRAGLKVLITDKDGNPQEEAKNY 298
Query: 60 VYREVF 65
+ F
Sbjct: 299 PFTLAF 304
Score = 53.9 bits (128), Expect = 1e-08
Identities = 29/69 (42%), Positives = 47/69 (68%), Gaps = 1/69 (1%)
Query: 1 MTINKSQGQSLEHVGVYLPSPIFSHGQLYVAISRVTSRGGL-KILINDDDDDNIDVASNV 59
MTI++S+GQ+ VG+YLP +F GQ Y+AIS+V + GL + LI ++ + + A NV
Sbjct: 306 MTIDQSRGQTFSKVGLYLPKQVFFPGQRYLAISKVKAGTGLTQFLIAEEAEKSQVEAENV 365
Query: 60 VYREVFRNL 68
V +++F N+
Sbjct: 366 VLKKLFWNV 374
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,484,027
Number of Sequences: 26719
Number of extensions: 50209
Number of successful extensions: 234
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 201
Number of HSP's gapped (non-prelim): 31
length of query: 68
length of database: 11,318,596
effective HSP length: 44
effective length of query: 24
effective length of database: 10,142,960
effective search space: 243431040
effective search space used: 243431040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146743.2


