

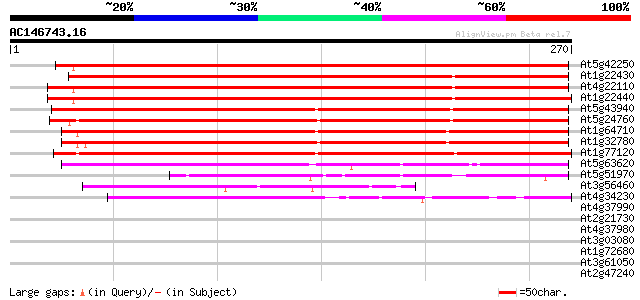
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.16 - phase: 2 /pseudo
(270 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g42250 alcohol dehydrogenase 362 e-101
At1g22430 putative alcohol dehydrogenase 310 5e-85
At4g22110 alcohol dehydrogenase like protein 307 3e-84
At1g22440 Very similar to alcohol dehydrogenase 295 1e-80
At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III (pir||S71... 258 3e-69
At5g24760 alcohol dehydrogenase - like protein 248 3e-66
At1g64710 alcohol dehydrogenase (EC 1.1.1.1) like protein 234 5e-62
At1g32780 alcohol dehydrogenase like protein 232 1e-61
At1g77120 alcohol dehydrogenase 222 2e-58
At5g63620 alcohol dehydrogenase-like protein 97 1e-20
At5g51970 sorbitol dehydrogenase-like protein 58 5e-09
At3g56460 quinone reductase-like protein 43 2e-04
At4g34230 cinnamyl alcohol dehydrogenase like protein 42 3e-04
At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2 36 0.025
At2g21730 putative cinnamyl-alcohol dehydrogenase 35 0.032
At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1 32 0.27
At3g03080 putative NADP-dependent oxidoreductase 32 0.27
At1g72680 putative cinnamyl-alcohol dehydrogenase 32 0.27
At3g61050 CaLB protein 31 0.61
At2g47240 putative acyl-CoA synthetase 31 0.61
>At5g42250 alcohol dehydrogenase
Length = 390
Score = 362 bits (930), Expect = e-101
Identities = 171/248 (68%), Positives = 210/248 (83%), Gaps = 1/248 (0%)
Query: 23 LPRNGNT-RFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTG 81
+PR N+ RFTDLNGE ++HF+ +SSFSEYTV+D+ANV+KID IPP RACLL CGVSTG
Sbjct: 133 MPRYDNSSRFTDLNGETLFHFLNVSSFSEYTVLDVANVVKIDSSIPPSRACLLSCGVSTG 192
Query: 82 VGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTD 141
VGAAW TA VE GS+V IFGLGS+GLAVAEGARLCGA+RIIGVD+N KF++G+KFG+T+
Sbjct: 193 VGAAWETAKVEKGSTVVIFGLGSIGLAVAEGARLCGASRIIGVDINPTKFQVGQKFGVTE 252
Query: 142 FVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPG 201
FV+ C VS++I EMTDGGADYCFECVG +SLV EAYA CR+GWGKTI LGVDKPG
Sbjct: 253 FVNSMTCEKNRVSEVINEMTDGGADYCFECVGSSSLVQEAYACCRQGWGKTITLGVDKPG 312
Query: 202 ARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINK 261
+++ L S +VLH GK LMGS++GGLK K+H+PILLKRY+ EL+LD+FVTHE++F++IN
Sbjct: 313 SQICLDSFDVLHHGKILMGSLFGGLKAKTHIPILLKRYLSNELELDKFVTHEMKFEEIND 372
Query: 262 AFDLLSKG 269
AF LL +G
Sbjct: 373 AFQLLLEG 380
>At1g22430 putative alcohol dehydrogenase
Length = 388
Score = 310 bits (794), Expect = 5e-85
Identities = 147/241 (60%), Positives = 194/241 (79%), Gaps = 1/241 (0%)
Query: 29 TRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRT 88
+RF D +GE+I+HF+F+SSFSEYTVVDIA+++KI P+IP +A LL CGVSTG+GAAW+
Sbjct: 138 SRFKDSSGEVIHHFLFVSSFSEYTVVDIAHLVKISPEIPVDKAALLSCGVSTGIGAAWKV 197
Query: 89 AGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEEC 148
A VE GS++AIFGLG+VGLAVAEGARL GA +IIG+D N +KFE+GKKFG TDF++ C
Sbjct: 198 ANVEEGSTIAIFGLGAVGLAVAEGARLRGAAKIIGIDTNSDKFELGKKFGFTDFINPTLC 257
Query: 149 GNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSS 208
G K +S++I EMT+GG DY FECVG+ASL++EA+ S R G GKT++LG++K A +SL S
Sbjct: 258 GEKKISEVIKEMTEGGVDYSFECVGLASLLNEAFISTRTGTGKTVMLGMEKHAAPISLGS 317
Query: 209 SEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSK 268
++L G+ + GS++GGLK K +PIL+ Y+ KEL LD F+THEL FK+INKAF LL +
Sbjct: 318 FDLLR-GRVICGSLFGGLKSKLDIPILVDHYLKKELNLDSFITHELNFKEINKAFALLEE 376
Query: 269 G 269
G
Sbjct: 377 G 377
>At4g22110 alcohol dehydrogenase like protein
Length = 389
Score = 307 bits (787), Expect = 3e-84
Identities = 154/252 (61%), Positives = 194/252 (76%), Gaps = 2/252 (0%)
Query: 19 FLLGLPRNGNT-RFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCG 77
FL R G T RF D GE IYHF+F+SSFSEYTVVDIA+++KI P IP +A LL CG
Sbjct: 128 FLSNTRRYGMTSRFKDSFGEDIYHFLFVSSFSEYTVVDIAHLVKISPDIPVDKAALLSCG 187
Query: 78 VSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKF 137
VSTG+GAAW+ A VE GS+VA+FGLG+VGLAV EGARL GA +IIGVD+N EKFE+GKKF
Sbjct: 188 VSTGIGAAWKVANVEKGSTVAVFGLGAVGLAVGEGARLRGAGKIIGVDLNPEKFELGKKF 247
Query: 138 GLTDFVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGV 197
G TDF++ CG +S++I EMT GG DY FECVG+ SL+ EA++S R G GKT+VLG+
Sbjct: 248 GFTDFINSTLCGENKISEVIKEMTGGGVDYSFECVGLPSLLTEAFSSTRTGSGKTVVLGI 307
Query: 198 DKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFK 257
DK +SL S ++L G+ + GS++GGLKPK +PIL+ Y+ KEL LD F+THEL+F+
Sbjct: 308 DKHLTPVSLGSFDLLR-GRHVCGSLFGGLKPKLDIPILVDHYLKKELNLDSFITHELKFE 366
Query: 258 DINKAFDLLSKG 269
+INKAFDLL +G
Sbjct: 367 EINKAFDLLVQG 378
>At1g22440 Very similar to alcohol dehydrogenase
Length = 386
Score = 295 bits (756), Expect = 1e-80
Identities = 147/253 (58%), Positives = 197/253 (77%), Gaps = 2/253 (0%)
Query: 19 FLLGLPRNGNT-RFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCG 77
+L R G T RF D GE I+HF+F+SSF+EYTVVDIA+++KI P+IP A LL C
Sbjct: 125 YLSNTRRYGMTSRFKDSRGEDIHHFIFVSSFTEYTVVDIAHLVKISPEIPVDIAALLSCS 184
Query: 78 VSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKF 137
V+TG+GAAW+ A VE GS+V IFGLG+VGLAVAEG RL GA +IIGVD+N KFEIGK+F
Sbjct: 185 VATGLGAAWKVADVEEGSTVVIFGLGAVGLAVAEGVRLRGAAKIIGVDLNPAKFEIGKRF 244
Query: 138 GLTDFVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGV 197
G+TDFV+ CG K++S++I EMTD GADY FEC+G+ASL+ EA+ S R G GKTIVLG+
Sbjct: 245 GITDFVNPALCGEKTISEVIREMTDVGADYSFECIGLASLMEEAFKSTRPGSGKTIVLGM 304
Query: 198 DKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFK 257
++ +SL S ++L G+++ G+++GGLKPK +PIL+ RY+ KEL L++ +THEL F+
Sbjct: 305 EQKALPISLGSYDLLR-GRTVCGTLFGGLKPKLDIPILVDRYLKKELNLEDLITHELSFE 363
Query: 258 DINKAFDLLSKGD 270
+INKAF LL++G+
Sbjct: 364 EINKAFHLLAEGN 376
>At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III
(pir||S71244)
Length = 379
Score = 258 bits (658), Expect = 3e-69
Identities = 121/249 (48%), Positives = 178/249 (70%), Gaps = 2/249 (0%)
Query: 21 LGLPRNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVST 80
+G+ N +NG+ IYHFM S+FS+YTVV +V KIDP P + CLLGCGV T
Sbjct: 122 VGIMMNDRKSRFSVNGKPIYHFMGTSTFSQYTVVHDVSVAKIDPTAPLDKVCLLGCGVPT 181
Query: 81 GVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLT 140
G+GA W TA VEPGS+VAIFGLG+VGLAVAEGA+ GA+RIIG+D++ +K+E KKFG+
Sbjct: 182 GLGAVWNTAKVEPGSNVAIFGLGTVGLAVAEGAKTAGASRIIGIDIDSKKYETAKKFGVN 241
Query: 141 DFVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKP 200
+FV+ ++ +K + ++I+++TDGG DY FEC+G S++ A C KGWG ++++GV
Sbjct: 242 EFVNPKD-HDKPIQEVIVDLTDGGVDYSFECIGNVSVMRAALECCHKGWGTSVIVGVAAS 300
Query: 201 GARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDIN 260
G +S +++ G+ G+ +GG K ++ VP L+++YM+KE+++DE++TH L +IN
Sbjct: 301 GQEISTRPFQLV-TGRVWKGTAFGGFKSRTQVPWLVEKYMNKEIKVDEYITHNLTLGEIN 359
Query: 261 KAFDLLSKG 269
KAFDLL +G
Sbjct: 360 KAFDLLHEG 368
>At5g24760 alcohol dehydrogenase - like protein
Length = 381
Score = 248 bits (632), Expect = 3e-66
Identities = 126/256 (49%), Positives = 172/256 (66%), Gaps = 9/256 (3%)
Query: 20 LLGLPRNG------NTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACL 73
+LG+ R G TRF+ + G+ +YH+ +SSFSEYTVV +K+DP P H+ CL
Sbjct: 118 VLGMERKGLMHSDQKTRFS-IKGKPVYHYCAVSSFSEYTVVHSGCAVKVDPLAPLHKICL 176
Query: 74 LGCGVSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEI 133
L CGV+ G+GAAW A V+ GSSV IFGLG+VGL+VA+GA+L GA +I+GVD+N K E
Sbjct: 177 LSCGVAAGLGAAWNVADVQKGSSVVIFGLGTVGLSVAQGAKLRGAAQILGVDINPAKAEQ 236
Query: 134 GKKFGLTDFVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTI 193
K FG+TDF++ + ++ + Q+I MT GGAD+ FECVG + A SC GWG T+
Sbjct: 237 AKTFGVTDFINSNDL-SEPIPQVIKRMTGGGADFSFECVGDTGIATTALQSCSDGWGMTV 295
Query: 194 VLGVDKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHE 253
LGV K +S L GKSL G+++GG KPKS +P L+ +YM+KE+ +DEF+TH
Sbjct: 296 TLGVPKAKPEVSAHYGLFL-SGKSLKGTLFGGWKPKSDLPSLIDKYMNKEIMIDEFITHN 354
Query: 254 LEFKDINKAFDLLSKG 269
L F +INKAF L+ +G
Sbjct: 355 LSFDEINKAFVLMREG 370
>At1g64710 alcohol dehydrogenase (EC 1.1.1.1) like protein
Length = 380
Score = 234 bits (596), Expect = 5e-62
Identities = 117/245 (47%), Positives = 170/245 (68%), Gaps = 3/245 (1%)
Query: 26 NGNTRF-TDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGA 84
+G TRF T + + IYHF+ S+FSEYTV+D A VLK+DP P + LL CGVSTGVGA
Sbjct: 129 DGKTRFFTSKDNKPIYHFLNTSTFSEYTVIDSACVLKVDPLFPLEKISLLSCGVSTGVGA 188
Query: 85 AWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVH 144
AW A ++P S+VAIFGLG+VGLAVAEGAR GA++IIG+D+N +KF++G++ G+++F++
Sbjct: 189 AWNVADIQPASTVAIFGLGAVGLAVAEGARARGASKIIGIDINPDKFQLGREAGISEFIN 248
Query: 145 GEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARL 204
+E +K+V + ++E+T+GG +Y FEC G + EA+ S G G T++LGV L
Sbjct: 249 PKE-SDKAVHERVMEITEGGVEYSFECAGSIEALREAFLSTNSGVGVTVMLGVHASPQLL 307
Query: 205 SLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFD 264
+ E L G+S+ SV+GG KPK+ +P + + + L LD F++H+L F DIN+A
Sbjct: 308 PIHPME-LFQGRSITASVFGGFKPKTQLPFFITQCLQGLLNLDLFISHQLPFHDINEAMQ 366
Query: 265 LLSKG 269
LL +G
Sbjct: 367 LLHQG 371
>At1g32780 alcohol dehydrogenase like protein
Length = 394
Score = 232 bits (592), Expect = 1e-61
Identities = 127/252 (50%), Positives = 165/252 (65%), Gaps = 10/252 (3%)
Query: 26 NGNTRF-TDLN-------GEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCG 77
+G TRF T +N + IYHF+ S+F+EYTV+D A V+KIDP P + LL CG
Sbjct: 130 DGGTRFSTTINKDGGSSQSQPIYHFLNTSTFTEYTVLDSACVVKIDPNSPLKQMSLLSCG 189
Query: 78 VSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKF 137
VSTGVGAAW A V+ G S A+FGLGSVGLAVAEGAR GA+RIIGVD N KFE GK
Sbjct: 190 VSTGVGAAWNIANVKEGKSTAVFGLGSVGLAVAEGARARGASRIIGVDANASKFEKGKLM 249
Query: 138 GLTDFVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGV 197
G+TDF++ ++ K V Q+I E+T GG DY FEC G ++ EA+ S GWG T+++G+
Sbjct: 250 GVTDFINPKDL-TKPVHQMIREITGGGVDYSFECTGNVDVLREAFLSTHVGWGSTVLVGI 308
Query: 198 DKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFK 257
L L E L DG+ + GSV+GG KPKS +P ++ M ++L+ F+T+EL F+
Sbjct: 309 YPTPRTLPLHPME-LFDGRRITGSVFGGFKPKSQLPNFAQQCMKGVVKLEPFITNELPFE 367
Query: 258 DINKAFDLLSKG 269
IN AF LL G
Sbjct: 368 KINDAFQLLRDG 379
>At1g77120 alcohol dehydrogenase
Length = 379
Score = 222 bits (565), Expect = 2e-58
Identities = 109/249 (43%), Positives = 166/249 (65%), Gaps = 3/249 (1%)
Query: 22 GLPRNGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTG 81
G+ +G +RF+ +NG+ IYHF+ S+FSEYTVV V KI+P P + C++ CG+STG
Sbjct: 124 GMIHDGESRFS-INGKPIYHFLGTSTFSEYTVVHSGQVAKINPDAPLDKVCIVSCGLSTG 182
Query: 82 VGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTD 141
+GA A + G SVAIFGLG+VGL AEGAR+ GA+RIIGVD N ++F+ K+FG+T+
Sbjct: 183 LGATLNVAKPKKGQSVAIFGLGAVGLGAAEGARIAGASRIIGVDFNSKRFDQAKEFGVTE 242
Query: 142 FVHGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPG 201
V+ ++ +K + Q+I EMTDGG D EC G + +A+ GWG +++GV
Sbjct: 243 CVNPKD-HDKPIQQVIAEMTDGGVDRSVECTGSVQAMIQAFECVHDGWGVAVLVGVPSKD 301
Query: 202 ARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINK 261
L++ ++L G+ +G KPK+ +P ++++YM+KEL+L++F+TH + F +INK
Sbjct: 302 DAFKTHPMNFLNE-RTLKGTFFGNYKPKTDIPGVVEKYMNKELELEKFITHTVPFSEINK 360
Query: 262 AFDLLSKGD 270
AFD + KG+
Sbjct: 361 AFDYMLKGE 369
>At5g63620 alcohol dehydrogenase-like protein
Length = 427
Score = 96.7 bits (239), Expect = 1e-20
Identities = 67/245 (27%), Positives = 117/245 (47%), Gaps = 7/245 (2%)
Query: 26 NGNTRFTDLNGEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAA 85
+G TR + + + + +EY V + + +P + +LGC V T GA
Sbjct: 178 DGETRLFLRHDDSPVYMYSMGGMAEYCVTPAHGLAPLPESLPYSESAILGCAVFTAYGAM 237
Query: 86 WRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHG 145
A + PG S+A+ G+G VG + + AR GA+ II VDV +K + K G T V+
Sbjct: 238 AHAAEIRPGDSIAVIGIGGVGSSCLQIARAFGASDIIAVDVQDDKLQKAKTLGATHIVN- 296
Query: 146 EECGNKSVSQIIIEMTDG-GADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARL 204
+ + I E+T G G D E +G + S + G GK +++G+ + G+
Sbjct: 297 --AAKEDAVERIREITGGMGVDVAVEALGKPQTFMQCTLSVKDG-GKAVMIGLSQAGSVG 353
Query: 205 SLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFD 264
+ + ++ ++GS YGG + + +P ++K L V+ + +F+D KAF
Sbjct: 354 EIDINRLVRRKIKVIGS-YGG-RARQDLPKVVKLAESGIFNLTNAVSSKYKFEDAGKAFQ 411
Query: 265 LLSKG 269
L++G
Sbjct: 412 DLNEG 416
>At5g51970 sorbitol dehydrogenase-like protein
Length = 364
Score = 58.2 bits (139), Expect = 5e-09
Identities = 53/198 (26%), Positives = 90/198 (44%), Gaps = 16/198 (8%)
Query: 78 VSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKF 137
+S GV A R A V P ++V + G G +GL AR RI+ VDV+ + + K+
Sbjct: 167 LSVGVHAC-RRAEVGPETNVLVMGAGPIGLVTMLAARAFSVPRIVIVDVDENRLAVAKQL 225
Query: 138 GLTDFV----HGEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTI 193
G + V + E+ G++ V QI M D F+C G + A A+ R G GK
Sbjct: 226 GADEIVQVTTNLEDVGSE-VEQIQKAM-GSNIDVTFDCAGFNKTMSTALAATRCG-GKVC 282
Query: 194 VLGVDKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDEFVTHE 253
++G+ + L+ + V G + K+ P+ L+ ++ + +TH
Sbjct: 283 LVGMGHGIMTVPLTPAAARE------VDVVGVFRYKNTWPLCLEFLTSGKIDVKPLITHR 336
Query: 254 LEF--KDINKAFDLLSKG 269
F K++ AF+ ++G
Sbjct: 337 FGFSQKEVEDAFETSARG 354
>At3g56460 quinone reductase-like protein
Length = 348
Score = 42.7 bits (99), Expect = 2e-04
Identities = 42/162 (25%), Positives = 69/162 (41%), Gaps = 6/162 (3%)
Query: 36 GEIIYHFMFISSFSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVEPGS 95
G+ + F + SF+++ V D + + + + A L T A A + G
Sbjct: 93 GDRVCSFADLGSFAQFIVADQSRLFLVPERCDMVAAAALPVAFGTSHVALVHRARLTSGQ 152
Query: 96 SVAIFGL-GSVGLAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVH-GEECGNKSV 153
+ + G G VGLA + ++CGA +I V EK ++ K G+ V G E SV
Sbjct: 153 VLLVLGAAGGVGLAAVQIGKVCGAI-VIAVARGTEKIQLLKSMGVDHVVDLGTENVISSV 211
Query: 154 SQIIIEMTDGGADYCFECVGMASLVHEAYASCRKGWGKTIVL 195
+ I G D ++ VG L E+ + WG I++
Sbjct: 212 KEFIKTRKLKGVDVLYDPVG-GKLTKESMKVLK--WGAQILV 250
>At4g34230 cinnamyl alcohol dehydrogenase like protein
Length = 357
Score = 42.0 bits (97), Expect = 3e-04
Identities = 58/224 (25%), Positives = 92/224 (40%), Gaps = 18/224 (8%)
Query: 48 FSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVEPGSSVAIFGLGSVGL 107
F++ TVV V+KI + +A L C T +PG I GLG VG
Sbjct: 135 FAKATVVHQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGLKQPGLRGGILGLGGVGH 194
Query: 108 AVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVSQIIIEMTDGGADY 167
+ A+ G + N ++ E + G D+V G + S E+ D DY
Sbjct: 195 MGVKIAKAMGHHVTVISSSNKKREEALQDLGADDYVIGSDQAKMS------ELAD-SLDY 247
Query: 168 CFECVGMASLVHEAYASCRKGWGKTIVLGV-DKPGARLSLSSSEVLHDGKSLMGSVYGGL 226
+ V + + E Y S K GK I++GV + P L + ++ K + GS G +
Sbjct: 248 VIDTVPVHHAL-EPYLSLLKLDGKLILMGVINNP---LQFLTPLLMLGRKVITGSFIGSM 303
Query: 227 KPKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSKGD 270
K +L+ +K L + ++ +N AF+ L K D
Sbjct: 304 KETEE---MLEFCKEKGL---SSIIEVVKMDYVNTAFERLEKND 341
>At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2
Length = 359
Score = 35.8 bits (81), Expect = 0.025
Identities = 51/226 (22%), Positives = 98/226 (42%), Gaps = 22/226 (9%)
Query: 48 FSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVE-PGSSVAIFGLGSVG 106
+S++ V + V++I +P A L C T V + + G++ PG + + GLG +G
Sbjct: 134 YSDHMVCEEGFVIRIPDNLPLDAAAPLLCAGIT-VYSPMKYHGLDKPGMHIGVVGLGGLG 192
Query: 107 LAVAEGARLCGATRIIGVDVNHEKF-EIGKKFGLTDFVHGEECGNKSVSQIIIEMTDGGA 165
+ A+ G T++ + + +K E + G F+ + K + + M DG
Sbjct: 193 HVGVKFAKAMG-TKVTVISTSEKKRDEAINRLGADAFLVSRD--PKQIKDAMGTM-DGII 248
Query: 166 DYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSVYGG 225
D + L+ K GK +++G P L L ++ + K +MGS+ GG
Sbjct: 249 DTVSATHSLLPLL-----GLLKHKGKLVMVGA--PEKPLELPVMPLIFERKMVMGSMIGG 301
Query: 226 LK-PKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSKGD 270
+K + + + K + +++L + +N A + L K D
Sbjct: 302 IKETQEMIDMAGKHNITADIEL-------ISADYVNTAMERLEKAD 340
>At2g21730 putative cinnamyl-alcohol dehydrogenase
Length = 376
Score = 35.4 bits (80), Expect = 0.032
Identities = 56/239 (23%), Positives = 90/239 (37%), Gaps = 47/239 (19%)
Query: 48 FSEYTVVDIANVLKIDPQIPPHRACLLGC-GVSTGVGAAWRTAGVEPGSSVAIFGLGSVG 106
+S+ VVD VL I +P L C G++ + E G + + GLG +G
Sbjct: 133 YSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGVNGLGGLG 192
Query: 107 LAVAEGARLCGATRIIGVDVNHEKFEIGKKFGL--TDFVHGEECGNKSVSQI----IIEM 160
H +IGK FGL T E +++ ++ +
Sbjct: 193 ---------------------HIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVT 231
Query: 161 TD--------GGADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSEVL 212
TD G D+ + V + + S K GK + LG+ P L L ++
Sbjct: 232 TDSQKMKEAVGTMDFIIDTVSAEHALLPLF-SLLKVNGKLVALGL--PEKPLDLPIFSLV 288
Query: 213 HDGKSLMGSVYGGLKPKSHV-PILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSKGD 270
K + GS GG+K + K + +++L ++ DIN A D L+K D
Sbjct: 289 LGRKMVGGSQIGGMKETQEMLEFCAKHKIVSDIEL-------IKMSDINSAMDRLAKSD 340
>At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1
Length = 357
Score = 32.3 bits (72), Expect = 0.27
Identities = 51/229 (22%), Positives = 94/229 (40%), Gaps = 28/229 (12%)
Query: 48 FSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVE-PGSSVAIFGLGSVG 106
+S++ V +++I +P A L C T V + + G++ PG + + GLG +G
Sbjct: 134 YSDHMVCAEDFIIRIPDNLPLDGAAPLLCAGVT-VYSPMKYHGLDKPGMHIGVVGLGGLG 192
Query: 107 LAVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFVHGEECGNKSVSQIIIEMTD--GG 164
+ A+ G + ++ E + G F+ VS+ +M D G
Sbjct: 193 HVAVKFAKAMGTKVTVISTSERKRDEAVTRLGADAFL---------VSRDPKQMKDAMGT 243
Query: 165 ADYCFECVGMASLVHE--AYASCRKGWGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSV 222
D + V S H K GK +++G P L L ++ K ++GS+
Sbjct: 244 MDGIIDTV---SATHPLLPLLGLLKNKGKLVMVGA--PAEPLELPVFPLIFGRKMVVGSM 298
Query: 223 YGGLK-PKSHVPILLKRYMDKELQLDEFVTHELEFKDINKAFDLLSKGD 270
GG+K + V + K + +++L + +N A + L+K D
Sbjct: 299 VGGIKETQEMVDLAGKHNITADIEL-------ISADYVNTAMERLAKAD 340
>At3g03080 putative NADP-dependent oxidoreductase
Length = 350
Score = 32.3 bits (72), Expect = 0.27
Identities = 48/182 (26%), Positives = 80/182 (43%), Gaps = 20/182 (10%)
Query: 73 LLGCGVSTGVGAAWRTAGVEPGSSVAIFGL-GSVGLAVAEGARLCGATRIIGVDVNHEKF 131
LLG T + + G +V + G+VG V + A++ G ++G ++EK
Sbjct: 140 LLGMPGMTAYAGFYEICSPKKGETVFVSAASGAVGQLVGQFAKIMGC-YVVGSAGSNEKV 198
Query: 132 EIGK-KFGLTDFVH--GEECGNKSVSQIIIEMTDGGADYCFECVGMASLVHEAYASCRKG 188
++ K KFG D + E N ++ + E G D FE VG L +A K
Sbjct: 199 DLLKNKFGFDDAFNYKAEPDLNAALKRCFPE----GIDIYFENVGGKML--DAVLLNMKL 252
Query: 189 WGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSVYGGLKPKSHVPILLKRYMDKELQLDE 248
G+ V G+ ++ +L E +H +L +Y ++ K V + Y DK L+ +
Sbjct: 253 HGRIAVCGMI---SQYNLEDQEGVH---NLANVIYKRIRIKGFV---VSDYFDKHLKFLD 303
Query: 249 FV 250
FV
Sbjct: 304 FV 305
>At1g72680 putative cinnamyl-alcohol dehydrogenase
Length = 355
Score = 32.3 bits (72), Expect = 0.27
Identities = 41/184 (22%), Positives = 69/184 (37%), Gaps = 19/184 (10%)
Query: 48 FSEYTVVDIANVLKIDPQIPPHRACLLGCGVSTGVGAAWRTAGVEPGSSVAIFGLGSVGL 107
+S + VV KI P A L C T R +PG S+ + GLG +G
Sbjct: 136 YSSHIVVHERYCYKIPVDYPLESAAPLLCAGITVYAPMMRHNMNQPGKSLGVIGLGGLGH 195
Query: 108 AVAEGARLCGATRIIGVDVNHEKFEIGKKFGLTDFV----HGEECGNKSVSQIIIEMTDG 163
+ + G + + +K E G +FV H + + +++ G
Sbjct: 196 MAVKFGKAFGLSVTVFSTSISKKEEALNLLGAENFVISSDHDQMKALEKSLDFLVDTASG 255
Query: 164 GADYCFECVGMASLVHEAYASCRKGWGKTIVLGVDKPGARLSLSSSEVLHDGKSLMGSVY 223
D+ F + Y S K G +++G + + +S + + + L GSV
Sbjct: 256 --DHAF----------DPYMSLLKIAGTYVLVGFP---SEIKISPANLNLGMRMLAGSVT 300
Query: 224 GGLK 227
GG K
Sbjct: 301 GGTK 304
>At3g61050 CaLB protein
Length = 510
Score = 31.2 bits (69), Expect = 0.61
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query: 73 LLGCGVSTGVGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGAT 119
++G G+ TGVG +GV G + G G+VG +++ R G T
Sbjct: 439 MVGTGIGTGVGLVG--SGVSSGVGMVGSGFGAVGSGLSKAGRFMGRT 483
>At2g47240 putative acyl-CoA synthetase
Length = 660
Score = 31.2 bits (69), Expect = 0.61
Identities = 18/43 (41%), Positives = 24/43 (54%), Gaps = 2/43 (4%)
Query: 82 VGAAWRTAGVEPGSSVAIFGLGSVGLAVAEGARLCGATRIIGV 124
+G+A R AG EPGS V I+G+ +A A C A +I V
Sbjct: 89 IGSALRAAGAEPGSRVGIYGVNCPQWIIAMEA--CAAHTLICV 129
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.141 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,203,529
Number of Sequences: 26719
Number of extensions: 263324
Number of successful extensions: 694
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 653
Number of HSP's gapped (non-prelim): 39
length of query: 270
length of database: 11,318,596
effective HSP length: 98
effective length of query: 172
effective length of database: 8,700,134
effective search space: 1496423048
effective search space used: 1496423048
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146743.16


