

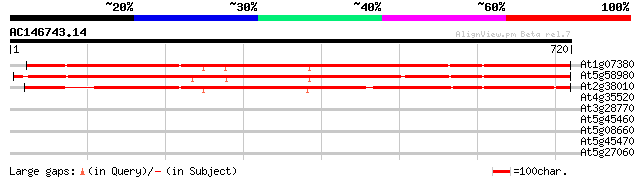
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.14 + phase: 0
(720 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g07380 unknown protein 813 0.0
At5g58980 random slug protein - like 783 0.0
At2g38010 hypothetical protein 756 0.0
At4g35520 putative protein 33 0.43
At3g28770 hypothetical protein 30 3.6
At5g45460 unknown protein 30 6.2
At5g08660 putative protein 30 6.2
At5g45470 putative protein 29 8.1
At5g27060 resistance protein - like 29 8.1
>At1g07380 unknown protein
Length = 779
Score = 813 bits (2101), Expect = 0.0
Identities = 429/752 (57%), Positives = 527/752 (70%), Gaps = 60/752 (7%)
Query: 22 TYGEYLIGVGSYDMTGPAADVNMMGYANIEQNTAGIHFRLRARTFIVAENLQGPRFVFVN 81
++ EYLIG+GSYD+TGPAADVNMMGYAN+EQ +GIHFRLRARTFIV+E QG R VFVN
Sbjct: 33 SHSEYLIGLGSYDITGPAADVNMMGYANMEQVASGIHFRLRARTFIVSEP-QGKRVVFVN 91
Query: 82 LDAGMASQLLTIKLLERLKSRFGNLYTEENVAISGIHTHAGPGGYLQYVVYSVTSLGFVT 141
LDA MASQ++ +K++ERLK+R+G+LYTE+NV ISGIHTHAGPGGYLQYVVY VTSLGFV
Sbjct: 92 LDACMASQIVKLKVIERLKARYGDLYTEQNVGISGIHTHAGPGGYLQYVVYIVTSLGFVR 151
Query: 142 QSFDAIANAVEQSIIQAHNNLKPGSIFINTGDVKEASINRSPSAYLLNPAEERSRYPSNV 201
QSFDA+ + +E SIIQAH NL+PGSIF+N G++ +A +NRSPSAYL NP++ERS++ NV
Sbjct: 152 QSFDALVDGIENSIIQAHENLRPGSIFLNNGELLDAGVNRSPSAYLNNPSKERSKHKYNV 211
Query: 202 DTQMTLLKFVDSASGKSKGSFSWFATHGTSMSNNNKLISGDNKDIG-------------- 247
D +MTLLKFVD G GSF+WFATHGTSMS N LISGDNK
Sbjct: 212 DKEMTLLKFVDDQWGPV-GSFNWFATHGTSMSRTNSLISGDNKGAASRFMEDWYEQNTAE 270
Query: 248 ----------------------------ELVQIAQLIKATGGKDCNEKSSQASKVR---- 275
EL+++A ++ GK SS A +VR
Sbjct: 271 RSYSEEFISDEIPRRVSSLIENHQDSHHELLELASYFESQPGKPVTRISSSARRVRSALR 330
Query: 276 KNDGSLFVGAFCQSNVGDVSPNVLGAFCIDSGKPCDFNHSSCNGNDLLCVGRGPGYPNEI 335
K D FV AFCQ+N GDVSPNVLGAFC+D+G PCDFNHS+C G + +C GRGPGYP+E
Sbjct: 331 KADKPGFVSAFCQTNCGDVSPNVLGAFCLDTGLPCDFNHSTCGGKNEMCYGRGPGYPDEF 390
Query: 336 LSTKIIGERQFRSAVELFGSASEELTGKIDYRHVYLNFTNIEVELDNK----KVVKTCPA 391
ST+IIGERQF+ A+ELF ASE+L GK+DYRHVY++F+ + V L K +VVKTCPA
Sbjct: 391 ESTRIIGERQFKMALELFNKASEQLQGKVDYRHVYVDFSQLNVTLPKKDGKSEVVKTCPA 450
Query: 392 ALGPGFAAGTTDGPGVFGFQQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEM 451
A+G FAAGTTDGPG F F QGD + +PFW+ VR+ LK P + Q+DC PKP+LL +GEM
Sbjct: 451 AMGFAFAAGTTDGPGAFDFTQGDDKGNPFWRLVRNVLKTPDKKQIDCHYPKPILLDTGEM 510
Query: 452 FDPYPWAPAILPIQILRLGKLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHV 511
PY WAP+IL +Q+LR+G+L ILSVPGEFTTMAGRRLR AVK L ++ N + + E HV
Sbjct: 511 TKPYDWAPSILSLQVLRIGQLFILSVPGEFTTMAGRRLRYAVKTQLKNSGNKDLSGEIHV 570
Query: 512 VIAGLTNTYSQYIATFEEYHQQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYGNG 571
VIAGL N YSQY+ TFEEY QRYE ASTLYGPHTLS YIQEF KL++++ D G
Sbjct: 571 VIAGLANGYSQYVTTFEEYQVQRYEGASTLYGPHTLSGYIQEFKKLSKSLVL-DMPVQPG 629
Query: 572 TSPPDLLSVQKSFLLDPFGDTTPDGIKLGDIKEDIAFPGSGYFTKGD-KPSATFWSANPR 630
PPDLL Q SFL DTTP G GD+ D+ P + +G+ + + F SA PR
Sbjct: 630 PQPPDLLDKQLSFLTPVMMDTTPSGDSFGDVISDV--PKNLSLKRGNGQVTVVFRSACPR 687
Query: 631 YDLLTEGTYAVVERLQ--GERWISVQDDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAI 687
DLLTEGT+ +VERL+ + W V DDDDL L F+W + S A +EW IP A
Sbjct: 688 NDLLTEGTFTLVERLEQKDKTWTPVYDDDDLCLRFKWSRHKKLSSRSQATVEWRIPESAS 747
Query: 688 SGVYRLKHFGASKKTIVSPINYFTGASSAFAV 719
GVYR+ HFGA+KK + +++FTG+SSAF V
Sbjct: 748 PGVYRITHFGAAKK-LFGSVHHFTGSSSAFVV 778
>At5g58980 random slug protein - like
Length = 733
Score = 783 bits (2023), Expect = 0.0
Identities = 422/732 (57%), Positives = 518/732 (70%), Gaps = 31/732 (4%)
Query: 6 IITVLGLVCFWSWMQSTYGEYLIGVGSYDMTGPAADVNMMGYANIEQNTAGIHFRLRART 65
++ +L L C +S +YL+G+GSYD+TGPAADVNMMGYAN+EQ +G+HFRLRAR
Sbjct: 14 LLFLLRLTCIFS-----DSDYLMGLGSYDITGPAADVNMMGYANMEQVASGVHFRLRARA 68
Query: 66 FIVAENLQGPRFVFVNLDAGMASQLLTIKLLERLKSRFGNLYTEENVAISGIHTHAGPGG 125
FIVAE + R FVNLDAGMASQL+TIK++ERLK R+G LYTEENVAISG HTHAGPGG
Sbjct: 69 FIVAEPYK-KRIAFVNLDAGMASQLVTIKVIERLKQRYGELYTEENVAISGTHTHAGPGG 127
Query: 126 YLQYVVYSVTSLGFVTQSFDAIANAVEQSIIQAHNNLKPGSIFINTGDVKEASINRSPSA 185
YLQY++Y VTSLGFV QSF+A+ + +EQSIIQAH NL+PGSI IN G++ +A +NRSPSA
Sbjct: 128 YLQYILYLVTSLGFVHQSFNALVDGIEQSIIQAHENLRPGSILINKGELLDAGVNRSPSA 187
Query: 186 YLLNPAEERSRYPSNVDTQMTLLKFVDSASGK-SKGSFSWFATHGTSMS---NNNKLISG 241
YL NPA ERS+Y +VD +MTL+KFVD G ++ WF S + + +S
Sbjct: 188 YLNNPAHERSKYEYDVDKEMTLVKFVDDQWGPVARIMEDWFERENGCRSVDVESPRRVSS 247
Query: 242 DNKD--IGELVQIAQLIKATGGKDCNEKSSQASKVRKN----DGSLFVGAFCQSNVGDVS 295
D +L+++A + +TGGK SS A +VR D FV AFCQ+N GDVS
Sbjct: 248 IISDPYDQDLMEMASSLLSTGGKTVTRMSSVARRVRSRFRHADKPRFVSAFCQTNCGDVS 307
Query: 296 PNVLGAFCIDSGKPCDFNHSSCNGNDLLCVGRGPGYPNEILSTKIIGERQFRSAVELFGS 355
PNVLGAFCID+G PC+FN S+C G + C GRGPGYP+E ST+IIGERQF+ A +LF
Sbjct: 308 PNVLGAFCIDTGLPCEFNQSTCGGKNEQCYGRGPGYPDEFESTRIIGERQFKKAADLFTK 367
Query: 356 ASEELTGKIDYRHVYLNFTNIEVELDNK----KVVKTCPAALGPGFAAGTTDGPGVFGFQ 411
ASEE+ GK+DYRH Y++F+ +EV ++ + +VVKTCPAA+G GFAAGTTDGPG F F+
Sbjct: 368 ASEEIQGKVDYRHAYVDFSQLEVTINGQNGGSEVVKTCPAAMGFGFAAGTTDGPGAFDFK 427
Query: 412 QGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDPYPWAPAILPIQILRLGK 471
QGD + +PFW+ VR+ LK P++ QV CQ PKP+LL +GEM PY WAP+ILP+QILR+G+
Sbjct: 428 QGDDQGNPFWRLVRNLLKNPTEEQVRCQRPKPILLDTGEMKQPYDWAPSILPVQILRIGQ 487
Query: 472 LIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLTNTYSQYIATFEEYH 531
L+IL VPGEFTTMAGRRLR+AVK L SNG E VVIAGLTN+YSQYIATFEEY
Sbjct: 488 LVILCVPGEFTTMAGRRLRDAVKTVLKEGSNG---REFSVVIAGLTNSYSQYIATFEEYQ 544
Query: 532 QQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYGNGTSPPDLLSVQKSFLLDPFGD 591
QRYE ASTLYGPHTLS YIQEF KLA + + G PPDLL Q S L D
Sbjct: 545 VQRYEGASTLYGPHTLSGYIQEFKKLANDLLSA-QTTDPGPQPPDLLHKQISLLTPVVAD 603
Query: 592 TTPDGIKLGDIKEDIAFPGSGYFTKG-DKPSATFWSANPRYDLLTEGTYAVVER-LQG-E 648
TP G GD+ D+ P F KG D F SANPR DL+TEGT+A+VER L+G E
Sbjct: 604 MTPIGTAFGDVTSDV--PRLSKFRKGADIVRVQFRSANPRNDLMTEGTFALVERWLEGRE 661
Query: 649 RWISVQDDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAISGVYRLKHFGASKKTIVSPI 707
W+ V DDDD L F+W + S A IEW IP A GVYR+ HFG S KT +S I
Sbjct: 662 TWVPVYDDDDFCLRFKWSRPFKLSTQSTATIEWRIPETASPGVYRITHFG-SAKTPISSI 720
Query: 708 NYFTGASSAFAV 719
++F+G+SSAF V
Sbjct: 721 HHFSGSSSAFVV 732
>At2g38010 hypothetical protein
Length = 715
Score = 756 bits (1952), Expect = 0.0
Identities = 403/744 (54%), Positives = 492/744 (65%), Gaps = 91/744 (12%)
Query: 19 MQSTYGEYLIGVGSYDMTGPAADVNMMGYANIEQNTAGIHFRLRARTFIVAENLQGPRFV 78
+ T YLIGVGSYD+TGPAADVNMMGYAN +Q +GIHFRLRAR FIVAE
Sbjct: 19 LSRTVYAYLIGVGSYDITGPAADVNMMGYANSDQIASGIHFRLRARAFIVAEP------- 71
Query: 79 FVNLDAGMASQLLTIKLLERLKSRFGNLYTEENVAISGIHTHAGPGGYLQYVVYSVTSLG 138
E+NVAISGIHTHAGPGGYLQYV Y VTSLG
Sbjct: 72 -----------------------------QEKNVAISGIHTHAGPGGYLQYVTYIVTSLG 102
Query: 139 FVTQSFDAIANAVEQSIIQAHNNLKPGSIFINTGDVKEASINRSPSAYLLNPAEERSRYP 198
FV QSFD + N +EQSI+QAH +L+PGS F+N GD+ +A +NRSPS+YL NPA ERS+Y
Sbjct: 103 FVRQSFDVVVNGIEQSIVQAHESLRPGSAFVNKGDLLDAGVNRSPSSYLNNPAAERSKYK 162
Query: 199 SNVDTQMTLLKFVDSASGKSKGSFSWFATHGTSMSNNNKLISGDNKDIG----------- 247
+VD +MTL+KFVDS G + GSF+WFATHGTSMS N LISGDNK
Sbjct: 163 YDVDKEMTLVKFVDSQLGPT-GSFNWFATHGTSMSRTNSLISGDNKGAAARFMEDWFENG 221
Query: 248 --------------------------ELVQIAQLIKATGGKDCNEKSSQASKVRKNDGSL 281
L+ IA K++ G ++ ++VR
Sbjct: 222 QKNSVSSRNIPRRVSTIVSDFSRNQSRLLDIAATYKSSRGHSVDKSLDVKTRVRNGSKRK 281
Query: 282 FVGAFCQSNVGDVSPNVLGAFCIDSGKPCDFNHSSCNGNDLLCVGRGPGYPNEILSTKII 341
FV AFCQSN GDVSPN LG FCID+G PCDFNHS+CNG + LC GRGPGYP+E ST+II
Sbjct: 282 FVSAFCQSNCGDVSPNTLGTFCIDTGLPCDFNHSTCNGQNELCYGRGPGYPDEFESTRII 341
Query: 342 GERQFRSAVELFGSASEELTGKIDYRHVYLNFTNIEVEL----DNKKVVKTCPAALGPGF 397
GE+QF+ AVELF A+E+L GKI Y+H YL+F+N++V + + VKTCPAA+G GF
Sbjct: 342 GEKQFKMAVELFNKATEKLQGKIGYQHAYLDFSNLDVTVPKAGGGSETVKTCPAAMGFGF 401
Query: 398 AAGTTDGPGVFGFQQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDPYPW 457
AAGTTDGPG F F+QGD + + FW+ VR+ L+ P QV CQ PKP+LL +GEM +PY W
Sbjct: 402 AAGTTDGPGAFDFKQGDDQGNVFWRLVRNVLRTPGPEQVQCQKPKPILLDTGEMKEPYDW 461
Query: 458 APAILPIQILRLGKLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLT 517
A ILR+G+L+ILSVPGEFTTMAGRRLR+A+K LIS+ EF+N HVVIAGLT
Sbjct: 462 A-------ILRIGQLVILSVPGEFTTMAGRRLRDAIKSFLISSDPKEFSNNMHVVIAGLT 514
Query: 518 NTYSQYIATFEEYHQQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYGNGTSPPDL 577
NTYSQYIATFEEY QRYE ASTLYG HTL+AYIQEF KLA A+ G + G PPDL
Sbjct: 515 NTYSQYIATFEEYEVQRYEGASTLYGRHTLTAYIQEFKKLATALVNGLTL-PRGPQPPDL 573
Query: 578 LSVQKSFLLDPFGDTTPDGIKLGDIKEDIAFPGSGYFTKGDKPSATFWSANPRYDLLTEG 637
L Q S L D+TP G+K GD+K D+ P F +G + +ATFWS PR DL+TEG
Sbjct: 574 LDKQISLLSPVVVDSTPLGVKFGDVKADV--PPKSTFRRGQQVNATFWSGCPRNDLMTEG 631
Query: 638 TYAVVERL-QGERWISVQDDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAISGVYRLKH 695
++AVVE L +G +W V DDDD SL F+W + S A IEW +P A++GVYR++H
Sbjct: 632 SFAVVETLREGGKWAPVYDDDDFSLKFKWSRPAKLSSESQATIEWRVPESAVAGVYRIRH 691
Query: 696 FGASKKTIVSPINYFTGASSAFAV 719
+GAS K++ I+ F+G+SSAF V
Sbjct: 692 YGAS-KSLFGSISSFSGSSSAFVV 714
>At4g35520 putative protein
Length = 1151
Score = 33.5 bits (75), Expect = 0.43
Identities = 22/67 (32%), Positives = 30/67 (43%), Gaps = 6/67 (8%)
Query: 265 NEKSSQASKVRKNDGSLFVGAFCQSNVGDVSPNVLGAFCIDSGKPCD------FNHSSCN 318
N SS S + ++ G+ V + C+ NV D NV G C D KP D N N
Sbjct: 205 NPSSSAFSLLMRDAGTEAVNSLCKVNVTDGMLNVSGFECADDWKPTDGQQTGRRNRLQSN 264
Query: 319 GNDLLCV 325
+LC+
Sbjct: 265 PGYILCI 271
>At3g28770 hypothetical protein
Length = 2081
Score = 30.4 bits (67), Expect = 3.6
Identities = 32/135 (23%), Positives = 52/135 (37%), Gaps = 31/135 (22%)
Query: 233 SNNNKLISGDNKDIGELVQIAQLIKATGGKDCNEKSSQASKVR--KNDGSLFVGAFCQSN 290
SNN K + G + IG+ + K+ K S+V KNDGS N
Sbjct: 271 SNNEKEVEGQGESIGD---------SAIEKNLESKEDVKSEVEAAKNDGSSMT-----EN 316
Query: 291 VGDVSPNVLGAFCIDSGKPCDFNHSSCNGNDLLCVGRGPGYPNEILSTKIIGERQFRSAV 350
+G+ N G ID+ K + G+G + + + + +S V
Sbjct: 317 LGEAQGNN-GVSTIDNEKEVE--------------GQGESIEDSDIEKNLESKEDVKSEV 361
Query: 351 ELFGSASEELTGKID 365
E +A +TGK++
Sbjct: 362 EAAKNAGSSMTGKLE 376
>At5g45460 unknown protein
Length = 703
Score = 29.6 bits (65), Expect = 6.2
Identities = 14/41 (34%), Positives = 21/41 (51%)
Query: 410 FQQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGE 450
F G+P WK + + LK S+Y +N K + L+ GE
Sbjct: 570 FVHGEPMTKELWKFIFEELKNKSKYGDSPENAKRISLARGE 610
>At5g08660 putative protein
Length = 649
Score = 29.6 bits (65), Expect = 6.2
Identities = 15/44 (34%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Query: 283 VGAFCQSNVG-DVSPNVLGAFCIDSGKPCDFNHSSCNGNDLLCV 325
+G+FC ++G + G+ D G+ DF HS NG L V
Sbjct: 1 MGSFCSKSLGINFGSEYSGSSVADDGREPDFGHSQPNGQTSLIV 44
>At5g45470 putative protein
Length = 866
Score = 29.3 bits (64), Expect = 8.1
Identities = 14/41 (34%), Positives = 21/41 (51%)
Query: 410 FQQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGE 450
F G+P WK + + LK S+Y +N K + L+ GE
Sbjct: 595 FVHGEPMTRELWKFIFEELKNKSKYGDSPENAKRISLARGE 635
>At5g27060 resistance protein - like
Length = 957
Score = 29.3 bits (64), Expect = 8.1
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Query: 346 FRSAVELFGSASEELTGKIDYRHVYLNFTNIEVELDNKKVVKTCPAAL--GPGFAAGTTD 403
F + LFGS+ EE+ I + F E E +++ ++ AA+ GPG A G
Sbjct: 868 FEDNLGLFGSSLEEVCRDIHTPASHQQFETPETEEEDEDLISWIAAAIGFGPGIAFGLMF 927
Query: 404 GPGVFGFQQGDPE--ISPFWKNVR 425
G + ++ PE ++PF +N R
Sbjct: 928 GYILVSYK---PEWFMNPFDRNNR 948
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,937,489
Number of Sequences: 26719
Number of extensions: 771160
Number of successful extensions: 1626
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1590
Number of HSP's gapped (non-prelim): 11
length of query: 720
length of database: 11,318,596
effective HSP length: 106
effective length of query: 614
effective length of database: 8,486,382
effective search space: 5210638548
effective search space used: 5210638548
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146743.14


