

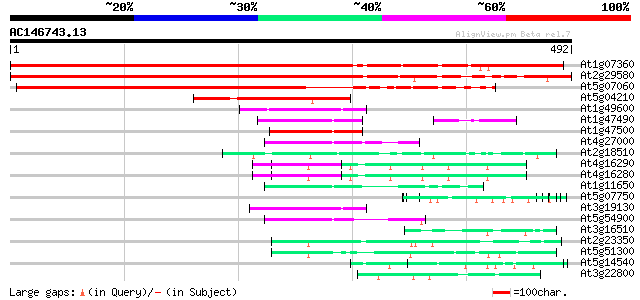
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.13 + phase: 0
(492 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g07360 unknown protein 679 0.0
At2g29580 putative RNA-binding protein 673 0.0
At5g07060 RNA-binding protein-like 463 e-130
At5g04210 RNA binding protein - like 107 2e-23
At1g49600 DNA binding protein ACBF, putative 69 8e-12
At1g47490 unknown protein 66 4e-11
At1g47500 unknown protein 66 5e-11
At4g27000 putative DNA binding protein 65 7e-11
At2g18510 putative spliceosome associated protein 63 4e-10
At4g16290 FCA delta protein 62 6e-10
At4g16280 FCA gamma protein 62 6e-10
At1g11650 putative DNA binding protein 62 6e-10
At5g07750 putative protein 62 7e-10
At3g19130 nuclear acid binding protein, putative 62 7e-10
At5g54900 unknown protein 61 2e-09
At3g16510 unknown protein 60 4e-09
At2g23350 putative poly(A) binding protein 59 8e-09
At5g51300 unknown protein 58 1e-08
At5g14540 unknown protein 57 2e-08
At3g22800 hypothetical protein 57 2e-08
>At1g07360 unknown protein
Length = 481
Score = 679 bits (1751), Expect = 0.0
Identities = 341/491 (69%), Positives = 380/491 (76%), Gaps = 17/491 (3%)
Query: 1 MAHRLLRDHEADGWERSDFPIICESCLGDSPYVRMTKAEYDKECKICTRPFTVFRWRPGR 60
MAHR+LRDHEADGWERSDFPIICESCLGD+PYVRMTKA YDKECKICTRPFTVFRWRPGR
Sbjct: 1 MAHRILRDHEADGWERSDFPIICESCLGDNPYVRMTKANYDKECKICTRPFTVFRWRPGR 60
Query: 61 DARYKKSEICQTCSKLKNVCQVCLLDLEYGLPVQVRDTALSISSNDAIPKSDVNREYFAE 120
DARYKK+EICQTC KLKNVCQVCLLDLEYGLPVQVRDTAL+IS++D+IPKSDVNREYFAE
Sbjct: 61 DARYKKTEICQTCCKLKNVCQVCLLDLEYGLPVQVRDTALNISTHDSIPKSDVNREYFAE 120
Query: 121 EHDRKARAGIDYESSFGKARPNDTILKLQRTTPYYKRNRAHICSFYTRGECTRGAECPYR 180
EHDRKARAG+DYESSFGK RPNDTILKLQRTTPYYKRNRAH+CSF+ RGECTRGAECPYR
Sbjct: 121 EHDRKARAGLDYESSFGKMRPNDTILKLQRTTPYYKRNRAHVCSFFIRGECTRGAECPYR 180
Query: 181 HEMPVTGELSQQNIKDRYYGVNDPVAMKLLGKAGEMSSLEVPEDESIKTLYVGGLDARVT 240
HEMP TGELSQQNIKDRYYGVNDPVAMKLLGKAGEM +LE P+DESIKTLYVGGL++R+
Sbjct: 181 HEMPETGELSQQNIKDRYYGVNDPVAMKLLGKAGEMGTLESPDDESIKTLYVGGLNSRIL 240
Query: 241 EQDLRDNFYAHGEIESVKMVLQRACAFVTYTTREGAEKAAEELSNKLVIKGLRLKLMWGR 300
EQD+RD FYAHGEIES++++ +ACAFVTYT+REGAEKAA+ELSN+LVI G RLKL WGR
Sbjct: 241 EQDIRDQFYAHGEIESIRILADKACAFVTYTSREGAEKAAQELSNRLVINGQRLKLTWGR 300
Query: 301 PQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGMLYYNNPPPPQQERNYYP 360
P KP+ DG A QQ VAHSGL PRAVISQQ NQ YY +PPP Q++ YYP
Sbjct: 301 P---KPDQDG---ANQQGGVAHSGLLPRAVISQQHNQPPPMQQ-YYMHPPPANQDKPYYP 353
Query: 361 SMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQY---HHQYYP 417
SMDPQRMGA++ +Q+ GG S SS H P PPP G + Q YP
Sbjct: 354 SMDPQRMGAVISTQEA--GGSSTENNGASSSSYMMPPHQSYP--PPPYGYMPSPYQQQYP 409
Query: 418 P---YQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQPGSSQTGSGQ 474
P +Q P Q+ A P PY Q P + P P +A P P G+ S Q
Sbjct: 410 PNHHHQPSPMQHYAPPPAAYPYPQQPGPGSRPAPSPTAVSAISPDSAPAGSGAPSGSSQQ 469
Query: 475 AGSAPAETGTS 485
A TG+S
Sbjct: 470 APDVSTATGSS 480
>At2g29580 putative RNA-binding protein
Length = 483
Score = 673 bits (1737), Expect = 0.0
Identities = 342/499 (68%), Positives = 383/499 (76%), Gaps = 35/499 (7%)
Query: 1 MAHRLLRDHEADGWERSDFPIICESCLGDSPYVRMTKAEYDKECKICTRPFTVFRWRPGR 60
MAHR+LRDHEADGWERSDFPIICESCLGD+PYVRMTKA YDKECKICTRPFTVFRWRPGR
Sbjct: 1 MAHRILRDHEADGWERSDFPIICESCLGDNPYVRMTKANYDKECKICTRPFTVFRWRPGR 60
Query: 61 DARYKKSEICQTCSKLKNVCQVCLLDLEYGLPVQVRDTALSISSNDAIPKSDVNREYFAE 120
DARYKK+E+CQTC KLKNVCQVCLLDLEYGLPVQVRDTAL+IS++D+IPKSDVNRE+FAE
Sbjct: 61 DARYKKTEVCQTCCKLKNVCQVCLLDLEYGLPVQVRDTALNISTHDSIPKSDVNREFFAE 120
Query: 121 EHDRKARAGIDYESSFGKARPNDTILKLQRTTPYYKRNRAHICSFYTRGECTRGAECPYR 180
EHDRK RAG+DYESSFGK RPNDTI LQRTTPYYKRNRAHICSF+ RGECTRG ECPYR
Sbjct: 121 EHDRKTRAGLDYESSFGKIRPNDTIRMLQRTTPYYKRNRAHICSFFIRGECTRGDECPYR 180
Query: 181 HEMPVTGELSQQNIKDRYYGVNDPVAMKLLGKAGEMSSLEVPEDESIKTLYVGGLDARVT 240
HEMP TGELSQQNIKDRYYGVNDPVA+KLLGKAGEM +LE PED+SI+TLYVGGL++RV
Sbjct: 181 HEMPETGELSQQNIKDRYYGVNDPVALKLLGKAGEMGTLESPEDQSIRTLYVGGLNSRVL 240
Query: 241 EQDLRDNFYAHGEIESVKMVLQRACAFVTYTTREGAEKAAEELSNKLVIKGLRLKLMWGR 300
EQD+RD FYAHGEIES++++ ++ACAFVTYTTREGAEKAAEELSN+LV+ G RLKL WGR
Sbjct: 241 EQDIRDQFYAHGEIESIRILAEKACAFVTYTTREGAEKAAEELSNRLVVNGQRLKLTWGR 300
Query: 301 PQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGMLYYNNPPPPQ---QERN 357
PQ KP+ DG + QQ SVAHSGL PRAVISQQQNQ + YY +PPPPQ Q+R
Sbjct: 301 PQVPKPDQDG---SNQQGSVAHSGLLPRAVISQQQNQPPPM-LQYYMHPPPPQPPHQDRP 356
Query: 358 YYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQY-HHQYY 416
+YPSMDPQRMGA+ S++ G S S SS M PP G Y HQ Y
Sbjct: 357 FYPSMDPQRMGAVSSSKE---SGSSTSDNRGASSSSYTM---------PPHGHYPQHQPY 404
Query: 417 PPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQPGSSQT---GSG 473
P PP Y M PPYQQYPP ++ A+H Y PGS S
Sbjct: 405 P-----PPSYGGYMQ--PPYQQYPPYHHG-----HSQQADHDYPQQPGPGSRPNPPHPSS 452
Query: 474 QAGSAPAETGTSTSGSQEQ 492
+ P + SGS +Q
Sbjct: 453 VSAPPPDSVSAAPSGSSQQ 471
>At5g07060 RNA-binding protein-like
Length = 363
Score = 463 bits (1191), Expect = e-130
Identities = 243/420 (57%), Positives = 291/420 (68%), Gaps = 67/420 (15%)
Query: 7 RDHEADGWERSDFPIICESCLGDSPYVRMTKAEYDKECKICTRPFTVFRWRPGRDARYKK 66
RDH ADGWE +DFPI CESC GD+PY+RMT+A+YDKECKIC+RPFT FRWRPGR+AR+KK
Sbjct: 4 RDHGADGWESADFPITCESCFGDNPYMRMTRADYDKECKICSRPFTAFRWRPGRNARFKK 63
Query: 67 SEICQTCSKLKNVCQVCLLDLEYGLPVQVRDTALSISSNDAIPKSDVNREYFAEEHDRKA 126
+EICQTCSKLKNVCQVCLLDL +GLPVQVRD+AL+I+S+ ++P S VNREYFA+EHD K
Sbjct: 64 TEICQTCSKLKNVCQVCLLDLGFGLPVQVRDSALNINSHYSVPMSHVNREYFADEHDPKT 123
Query: 127 RAGIDYESSFGKARPNDTILKLQRTTPYYKRNRAHICSFYTRGECTRGAECPYRHEMPVT 186
RAG+DYESSFGK +PNDTILKLQR TP Y++NR ICSFYT G+C RGAEC +RHEMP T
Sbjct: 124 RAGLDYESSFGKMQPNDTILKLQRRTPSYEKNRPKICSFYTIGQCKRGAECSFRHEMPET 183
Query: 187 GELSQQNIKDRYYGVNDPVAMKLLGKAGEMSSLEVPEDESIKTLYVGGLDARVTEQDLRD 246
GELS QNI+DRYY VNDPVAMKLL KAGEM +LE PEDESIKTLYVGGL++R+ EQD+ D
Sbjct: 184 GELSHQNIRDRYYSVNDPVAMKLLRKAGEMGTLEPPEDESIKTLYVGGLNSRIFEQDIHD 243
Query: 247 NFYAHGEIESVKMVLQRACAFVTYTTREGAEKAAEELSNKLVIKGLRLKLMWGRPQSAKP 306
+FYA+GE+ES++++ + K
Sbjct: 244 HFYAYGEMESIRVM----------------------------------------AEDGKY 263
Query: 307 ESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGMLYYNNPPPPQQERNYYPSMDPQR 366
+ G +Q +QQ S+AH+GL ISQQQNQ +Q YY PPPP E ++YPSMD QR
Sbjct: 264 DQSGSNQ-QQQGSIAHTGL-----ISQQQNQ-HSQMQQYYMQPPPP-NEYSHYPSMDTQR 315
Query: 367 MGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQY 426
MGA +Q+ G S S N+ S Y PM P HQ YP PP Y
Sbjct: 316 MGAAFSTQES--DGSSTSENNRAYSS------YSYPMPP-------HQPYPT----PPPY 356
>At5g04210 RNA binding protein - like
Length = 193
Score = 107 bits (266), Expect = 2e-23
Identities = 63/143 (44%), Positives = 89/143 (62%), Gaps = 11/143 (7%)
Query: 162 ICSFYTRG-ECTRGAECPYRHEMPVTGELSQQNIKDRYYGVNDPVAMKLLGKAGEMSSLE 220
IC + G +CTRG C YRHE+ + ++ G+ +P +++L +A L+
Sbjct: 16 ICPLFDAGRQCTRGTMCLYRHEINRGLDCHGRS------GLENPFVLEVLARACNTGPLK 69
Query: 221 VPEDESIKTLYVGGL-DARVTEQDLRDNFYAHGEIESVKMVLQRA---CAFVTYTTREGA 276
PED+SIKTLY+ L ++ V EQD+RD+F +GEIES+ + R CAF+TYTTR A
Sbjct: 70 PPEDQSIKTLYIRRLINSSVLEQDIRDHFCPYGEIESIVIFPHRGGGTCAFLTYTTRLAA 129
Query: 277 EKAAEELSNKLVIKGLRLKLMWG 299
EKA ELS+ IKG R+KL+WG
Sbjct: 130 EKAMLELSSWTDIKGQRVKLLWG 152
>At1g49600 DNA binding protein ACBF, putative
Length = 445
Score = 68.6 bits (166), Expect = 8e-12
Identities = 41/112 (36%), Positives = 62/112 (54%), Gaps = 2/112 (1%)
Query: 202 NDPVAMKLLGKAGEMSSLEVPEDESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMVL 261
N A+ L G G S+ E + T++VGGLDA VTE+DL F GE+ SVK+ +
Sbjct: 302 NGSQALTLAGGHGGNGSMSDGESNN-STIFVGGLDADVTEEDLMQPFSDFGEVVSVKIPV 360
Query: 262 QRACAFVTYTTREGAEKAAEELSNKLVIKGLRLKLMWGRPQSAKPESDGLDQ 313
+ C FV + R+ AE+A L+ ++ K ++L WGR + + SD +Q
Sbjct: 361 GKGCGFVQFANRQSAEEAIGNLNGTVIGKN-TVRLSWGRSPNKQWRSDSGNQ 411
Score = 30.8 bits (68), Expect = 1.8
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Query: 225 ESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQR------ACAFVTYTTREGAEK 278
+ +KTL+VG L + E L F E+ SVK++ + FV + +R AE+
Sbjct: 116 DDVKTLWVGDLLHWMDETYLHTCFSHTNEVSSVKVIRNKQTCQSEGYGFVEFLSRSAAEE 175
Query: 279 AAEELS 284
A + S
Sbjct: 176 ALQSFS 181
>At1g47490 unknown protein
Length = 432
Score = 66.2 bits (160), Expect = 4e-11
Identities = 38/93 (40%), Positives = 56/93 (59%), Gaps = 2/93 (2%)
Query: 218 SLEVPEDESIKT-LYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRACAFVTYTTREGA 276
+L PE + + T ++VGGLD+ VT++DL+ F GEI SVK+ + + C FV + R A
Sbjct: 293 TLTRPEGDIMNTTIFVGGLDSSVTDEDLKQPFNEFGEIVSVKIPVGKGCGFVQFVNRPNA 352
Query: 277 EKAAEELSNKLVIKGLRLKLMWGRPQSAKPESD 309
E+A E+L N VI ++L WGR + K D
Sbjct: 353 EEALEKL-NGTVIGKQTVRLSWGRNPANKQPRD 384
Score = 42.0 bits (97), Expect = 8e-04
Identities = 25/73 (34%), Positives = 30/73 (40%), Gaps = 14/73 (19%)
Query: 372 PSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMP 431
P PP P+ EN+P + PPP H YPP P Q A P
Sbjct: 24 PPPPPPPQQPAKEEENQPKTSPT----------PPP----HWMRYPPTVIIPHQMMYAPP 69
Query: 432 PVPPYQQYPPQYN 444
P PPY QYP ++
Sbjct: 70 PFPPYHQYPNHHH 82
Score = 31.2 bits (69), Expect = 1.4
Identities = 21/81 (25%), Positives = 31/81 (37%), Gaps = 13/81 (16%)
Query: 386 ENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPP---- 441
E++ S + + P PPPQ P ++ Q + P P + +YPP
Sbjct: 9 ESESSDSHPVVDNQPPPPPPPPQ--------QPAKEEENQPKTSPTPPPHWMRYPPTVII 60
Query: 442 -QYNAAMPPPQPPAANHPYQH 461
PPP PP +P H
Sbjct: 61 PHQMMYAPPPFPPYHQYPNHH 81
>At1g47500 unknown protein
Length = 434
Score = 65.9 bits (159), Expect = 5e-11
Identities = 35/81 (43%), Positives = 50/81 (61%), Gaps = 1/81 (1%)
Query: 229 TLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRACAFVTYTTREGAEKAAEELSNKLV 288
T++VGGLD+ VT++DL+ F GEI SVK+ + + C FV + R AE+A E+L N V
Sbjct: 307 TIFVGGLDSSVTDEDLKQPFSEFGEIVSVKIPVGKGCGFVQFVNRPNAEEALEKL-NGTV 365
Query: 289 IKGLRLKLMWGRPQSAKPESD 309
I ++L WGR + K D
Sbjct: 366 IGKQTVRLSWGRNPANKQPRD 386
Score = 33.9 bits (76), Expect = 0.21
Identities = 23/79 (29%), Positives = 33/79 (41%), Gaps = 13/79 (16%)
Query: 386 ENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPY-QQYP---- 440
E++ S + + P PPP +PP + Q + P PP+ +YP
Sbjct: 9 ESESSDSHPLVDYQSLPPYPPP--------HPPVEVEENQPKTSPTPPPPHWMRYPPVLM 60
Query: 441 PQYNAAMPPPQPPAANHPY 459
PQ A PPP P + H Y
Sbjct: 61 PQMMYAPPPPMPFSPYHQY 79
Score = 33.9 bits (76), Expect = 0.21
Identities = 24/70 (34%), Positives = 29/70 (41%), Gaps = 16/70 (22%)
Query: 377 PPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVP-- 434
PP P EN+P + P PPP H YPP P A PP+P
Sbjct: 29 PPHPPVEVEENQPKTS---------PTPPPP----HWMRYPPVLM-PQMMYAPPPPMPFS 74
Query: 435 PYQQYPPQYN 444
PY QYP ++
Sbjct: 75 PYHQYPNHHH 84
>At4g27000 putative DNA binding protein
Length = 415
Score = 65.5 bits (158), Expect = 7e-11
Identities = 46/136 (33%), Positives = 60/136 (43%), Gaps = 16/136 (11%)
Query: 224 DESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRACAFVTYTTREGAEKAAEEL 283
D + T++VG +D VTE DL+ F GE+ VK+ + C FV Y R AE+A L
Sbjct: 274 DPTNTTIFVGAVDQSVTEDDLKSVFGQFGELVHVKIPAGKRCGFVQYANRACAEQALSVL 333
Query: 284 SNKLVIKGLRLKLMWGRPQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGM 343
N + G ++L WGR S K DQA+ + G P QG
Sbjct: 334 -NGTQLGGQSIRLSWGRSPSNKQTQP--DQAQYGGGGGYYGYPP-------------QGY 377
Query: 344 LYYNNPPPPQQERNYY 359
Y PPPQ YY
Sbjct: 378 EAYGYAPPPQDPNAYY 393
>At2g18510 putative spliceosome associated protein
Length = 363
Score = 62.8 bits (151), Expect = 4e-10
Identities = 78/314 (24%), Positives = 128/314 (39%), Gaps = 56/314 (17%)
Query: 187 GELSQQNIKDRYYGVNDPVAMKLLGK-------AGEMSSLEVPEDESIKTLYVGGLDARV 239
G + ++ +D Y + +KL GK + + SL+V + L++G LD V
Sbjct: 69 GFIEYRSEEDADYAIKVLNMIKLHGKPIRVNKASQDKKSLDVGAN-----LFIGNLDPDV 123
Query: 240 TEQDLRDNFYAHGEIESVKMVLQ-------RACAFVTYTTREGAEKAAEELSNKLVIKGL 292
E+ L D F A G I S +++ R F++Y + E ++ A E ++ + +
Sbjct: 124 DEKLLYDTFSAFGVIASNPKIMRDPDTGNSRGFGFISYDSFEASDAAIESMTGQY-LSNR 182
Query: 293 RLKLMWGRPQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGMLYYNNPPPP 352
++ + + + K E G R A+ + Q+++ T L+ PP
Sbjct: 183 QITVSYAYKKDTKGERHGTPAERLLAATNPTA---------QKSRPHT---LFAMGPPSS 230
Query: 353 QQERNYYPSMDPQRMGAL----LPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQ 408
+ N P P G++ +P+ PP P + +P S Q Q + M+PPP
Sbjct: 231 APQVNGLPR--PFANGSMQPVPIPAPRQPPPPPPQVYQTQPPSWPSQPQQHS--MVPPPM 286
Query: 409 GQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQH---PMQP 465
Q+ PP Q PP PP P + + + PPP PP A +QH P Q
Sbjct: 287 -----QFRPP-QGMPP------PPPPQFLNHQQGFGGPRPPP-PPQAMGMHQHGGWPPQH 333
Query: 466 GSSQTGSGQAGSAP 479
Q G Q P
Sbjct: 334 MQQQGGPPQQQQPP 347
Score = 33.1 bits (74), Expect = 0.36
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 6/62 (9%)
Query: 229 TLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRAC------AFVTYTTREGAEKAAEE 282
T+YVGGLDA+++E+ L + F G + +V + R F+ Y + E A+ A +
Sbjct: 26 TVYVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAIKV 85
Query: 283 LS 284
L+
Sbjct: 86 LN 87
>At4g16290 FCA delta protein
Length = 533
Score = 62.4 bits (150), Expect = 6e-10
Identities = 68/264 (25%), Positives = 105/264 (39%), Gaps = 45/264 (17%)
Query: 230 LYVGGLDARVTEQDLRDNFYAHGEIESVKMV-----LQRACAFVTYTTREGAEKAAEELS 284
L+VG L+ + TE+++ + F G +E V ++ R C FV Y+++E A A + L+
Sbjct: 213 LFVGSLNKQATEKEVEEIFLQFGHVEDVYLMRDEYRQSRGCGFVKYSSKETAMAAIDGLN 272
Query: 285 NKLVIKGLRLKLM--WGRPQSAKPESDGLDQARQQASVAHSGLFPRAVIS---QQQNQDQ 339
++G L+ + P+ KP ++R+ A G PR S N
Sbjct: 273 GTYTMRGCNQPLIVRFAEPKRPKP-----GESREMAPPVGLGSGPRFQASGPRPTSNFGD 327
Query: 340 TQGMLYYNNPPPPQQERNYYP-----------SMDPQRMGALLPSQDGPPGGPSG----- 383
+ G + + NP P RN P P+ A LPS G P G G
Sbjct: 328 SSGDVSHTNPWRPATSRNVGPPSNTGIRGAGSDFPPKPGQATLPSNQGGPLGGYGVPPLN 387
Query: 384 ----SGENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYP---------PYQQYPPQYNAAM 430
G + ++ +QQ + G+ + P + + Q P P Q P Q A
Sbjct: 388 PLPVPGVSSSATLQQQNRAAGQHITPLKKPLHSPQGLPLPLRPQTNFPGAQAPLQNPYAY 447
Query: 431 PPVPPYQQYPPQYN-AAMPPPQPP 453
P Q PPQ N + PQ P
Sbjct: 448 SSQLPTSQLPPQQNISRATAPQTP 471
Score = 48.5 bits (114), Expect = 8e-06
Identities = 24/84 (28%), Positives = 42/84 (49%), Gaps = 6/84 (7%)
Query: 214 GEMSSLEVPEDESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMV------LQRACAF 267
G + +V + S L+VG + TE+++R F HG + V ++ Q+ C F
Sbjct: 106 GSFTGTDVSDRSSTVKLFVGSVPRTATEEEIRPYFEQHGNVLEVALIKDKRTGQQQGCCF 165
Query: 268 VTYTTREGAEKAAEELSNKLVIKG 291
V Y T + A++A L N++ + G
Sbjct: 166 VKYATSKDADRAIRALHNQITLPG 189
>At4g16280 FCA gamma protein
Length = 747
Score = 62.4 bits (150), Expect = 6e-10
Identities = 68/264 (25%), Positives = 105/264 (39%), Gaps = 45/264 (17%)
Query: 230 LYVGGLDARVTEQDLRDNFYAHGEIESVKMV-----LQRACAFVTYTTREGAEKAAEELS 284
L+VG L+ + TE+++ + F G +E V ++ R C FV Y+++E A A + L+
Sbjct: 213 LFVGSLNKQATEKEVEEIFLQFGHVEDVYLMRDEYRQSRGCGFVKYSSKETAMAAIDGLN 272
Query: 285 NKLVIKGLRLKLM--WGRPQSAKPESDGLDQARQQASVAHSGLFPRAVIS---QQQNQDQ 339
++G L+ + P+ KP ++R+ A G PR S N
Sbjct: 273 GTYTMRGCNQPLIVRFAEPKRPKP-----GESREMAPPVGLGSGPRFQASGPRPTSNFGD 327
Query: 340 TQGMLYYNNPPPPQQERNYYP-----------SMDPQRMGALLPSQDGPPGGPSG----- 383
+ G + + NP P RN P P+ A LPS G P G G
Sbjct: 328 SSGDVSHTNPWRPATSRNVGPPSNTGIRGAGSDFPPKPGQATLPSNQGGPLGGYGVPPLN 387
Query: 384 ----SGENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYP---------PYQQYPPQYNAAM 430
G + ++ +QQ + G+ + P + + Q P P Q P Q A
Sbjct: 388 PLPVPGVSSSATLQQQNRAAGQHITPLKKPLHSPQGLPLPLRPQTNFPGAQAPLQNPYAY 447
Query: 431 PPVPPYQQYPPQYN-AAMPPPQPP 453
P Q PPQ N + PQ P
Sbjct: 448 SSQLPTSQLPPQQNISRATAPQTP 471
Score = 48.5 bits (114), Expect = 8e-06
Identities = 24/84 (28%), Positives = 42/84 (49%), Gaps = 6/84 (7%)
Query: 214 GEMSSLEVPEDESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMV------LQRACAF 267
G + +V + S L+VG + TE+++R F HG + V ++ Q+ C F
Sbjct: 106 GSFTGTDVSDRSSTVKLFVGSVPRTATEEEIRPYFEQHGNVLEVALIKDKRTGQQQGCCF 165
Query: 268 VTYTTREGAEKAAEELSNKLVIKG 291
V Y T + A++A L N++ + G
Sbjct: 166 VKYATSKDADRAIRALHNQITLPG 189
>At1g11650 putative DNA binding protein
Length = 405
Score = 62.4 bits (150), Expect = 6e-10
Identities = 53/192 (27%), Positives = 75/192 (38%), Gaps = 43/192 (22%)
Query: 224 DESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRACAFVTYTTREGAEKAAEEL 283
D + T++VGGLDA VT+ L++ F +GEI VK+ + C FV ++ + AE+A L
Sbjct: 257 DPNNTTVFVGGLDASVTDDHLKNVFSQYGEIVHVKIPAGKRCGFVQFSEKSCAEEALRML 316
Query: 284 SNKLVIKGLRLKLMWGRPQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGM 343
N + + G ++L WGR S K D
Sbjct: 317 -NGVQLGGTTVRLSWGRSPSNKQSGD-------------------------------PSQ 344
Query: 344 LYYNNPPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPM 403
YY Q++ Y DP A G GG SG + P + +Q P
Sbjct: 345 FYYGGYGQGQEQYGYTMPQDP---NAYYGGYSG--GGYSGGYQQTPQAGQQP------PQ 393
Query: 404 MPPPQGQYHHQY 415
PP Q Q Y
Sbjct: 394 QPPQQQQVGFSY 405
Score = 29.3 bits (64), Expect = 5.2
Identities = 29/142 (20%), Positives = 50/142 (34%), Gaps = 10/142 (7%)
Query: 181 HEMPVTGELSQQNIKDRY--YGVNDPVAMKLLGKAGEMSSLEVPEDESIKTLYVGGLDAR 238
H P Q + Y G P +A S++ + I+TL++G L
Sbjct: 13 HHAPPPSAQQQYGYQQPYGIAGAAPPPPQMWNPQAAAPPSVQPTTADEIRTLWIGDLQYW 72
Query: 239 VTEQDLRDNFYAHGEIESVKMVLQR------ACAFVTYTTREGAEKAAEELSNKLV--IK 290
+ E L F GE+ S K++ + F+ + + AE+ + +N +
Sbjct: 73 MDENFLYGCFAHTGEMVSAKVIRNKQTGQVEGYGFIEFASHAAAERVLQTFNNAPIPSFP 132
Query: 291 GLRLKLMWGRPQSAKPESDGLD 312
+L W S D D
Sbjct: 133 DQLFRLNWASLSSGDKRDDSPD 154
Score = 28.5 bits (62), Expect = 8.8
Identities = 19/52 (36%), Positives = 22/52 (41%), Gaps = 8/52 (15%)
Query: 405 PPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAAN 456
PPP Q + Y P Y A PP Q + PQ AA P QP A+
Sbjct: 16 PPPSAQQQYGYQQP-------YGIAGAAPPPPQMWNPQ-AAAPPSVQPTTAD 59
>At5g07750 putative protein
Length = 1289
Score = 62.0 bits (149), Expect = 7e-10
Identities = 41/128 (32%), Positives = 47/128 (36%), Gaps = 6/128 (4%)
Query: 345 YYNNPPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMM 404
Y + PPPP +Y P S PP P G G P YG P
Sbjct: 940 YGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYG--SPPPPPPPPPSYGSPPP 997
Query: 405 PPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQ 464
PPP H PP PP + A PP PP PP + A PPP PP + P
Sbjct: 998 PPPPPFSHVSSIPPPPPPPPMHGGAPPPPPP----PPMHGGAPPPPPPPPMHGGAPPPPP 1053
Query: 465 PGSSQTGS 472
P G+
Sbjct: 1054 PPPMHGGA 1061
Score = 60.8 bits (146), Expect = 2e-09
Identities = 45/139 (32%), Positives = 48/139 (34%), Gaps = 8/139 (5%)
Query: 346 YNNPPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGS-GENKPSSEKQQMQHYGRPMM 404
Y +PPPP Y S P PS PP P G P YG P
Sbjct: 940 YGSPPPPPPPPPSYGSPPPPPPPP--PSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPP 997
Query: 405 PPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQ 464
PPP H PP PP + A PP PP PP + A PPP PP H P
Sbjct: 998 PPPPPFSHVSSIPPPPPPPPMHGGAPPPPPP----PPMHGGA-PPPPPPPPMHGGAPPPP 1052
Query: 465 PGSSQTGSGQAGSAPAETG 483
P G P G
Sbjct: 1053 PPPPMHGGAPPPPPPPMFG 1071
Score = 58.5 bits (140), Expect = 8e-09
Identities = 41/126 (32%), Positives = 47/126 (36%), Gaps = 9/126 (7%)
Query: 345 YYNNPPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMM 404
Y + PPPP Y P S PP P + P H G P
Sbjct: 966 YGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAPPP 1025
Query: 405 PPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPP---AANHPYQH 461
PPP H PP PP + A PP PP PP + A PPP PP A P
Sbjct: 1026 PPPPPM--HGGAPPPPPPPPMHGGAPPPPPP----PPMHGGAPPPPPPPMFGGAQPPPPP 1079
Query: 462 PMQPGS 467
PM+ G+
Sbjct: 1080 PMRGGA 1085
Score = 51.2 bits (121), Expect = 1e-06
Identities = 40/134 (29%), Positives = 48/134 (34%), Gaps = 12/134 (8%)
Query: 346 YNNPPPPQQERNYYPSMDPQRM-------GALLPSQDGPPGGPSGSGENKPSSEKQQMQH 398
Y PPPP + PS + + P + PP P N S
Sbjct: 880 YILPPPPLPYTSIAPSPSVKILPLHGISSAPSPPVKTAPPPPPPPPFSNAHSVLSPPPPS 939
Query: 399 YGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHP 458
YG P PPP + PP PP Y + PP PP PP Y + PPP PP +
Sbjct: 940 YGSPPPPPPPPPSYGS-PPPPPPPPPSYGSPPPPPPP----PPGYGSPPPPPPPPPSYGS 994
Query: 459 YQHPMQPGSSQTGS 472
P P S S
Sbjct: 995 PPPPPPPPFSHVSS 1008
Score = 50.1 bits (118), Expect = 3e-06
Identities = 40/131 (30%), Positives = 47/131 (35%), Gaps = 22/131 (16%)
Query: 349 PPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQ 408
PPPP N + + P PS PP P P S YG P PPP
Sbjct: 920 PPPPPPFSNAHSVLSPPP-----PSYGSPPPPPP-----PPPS-------YGSPPPPPPP 962
Query: 409 GQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQPGSS 468
+ PP PP Y + PP PP PP Y + PPP PP ++ P P
Sbjct: 963 PPSYGSPPPPPPP-PPGYGSPPPPPPP----PPSYGSPPPPPPPPFSHVSSIPPPPPPPP 1017
Query: 469 QTGSGQAGSAP 479
G P
Sbjct: 1018 MHGGAPPPPPP 1028
Score = 47.8 bits (112), Expect = 1e-05
Identities = 45/163 (27%), Positives = 50/163 (30%), Gaps = 27/163 (16%)
Query: 349 PPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQ 408
PPPP P P M P PP G+ P H G P PPP
Sbjct: 1013 PPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGA----PPPPPPPPMHGGAPPPPPPP 1068
Query: 409 GQYHHQYYPPYQQY--------PPQYNAAMPPVPPYQQY-------PPQYNAAMPPPQPP 453
Q PP PP A PP PP + PP + A PPP PP
Sbjct: 1069 MFGGAQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAPPPPPPP 1128
Query: 454 AANHPYQHPMQPGSSQTGS--------GQAGSAPAETGTSTSG 488
P PG G+ G+A P G G
Sbjct: 1129 MRGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPG 1171
Score = 46.2 bits (108), Expect = 4e-05
Identities = 40/125 (32%), Positives = 43/125 (34%), Gaps = 16/125 (12%)
Query: 349 PPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQ 408
PPPP P P R GA PP P P M+ P PPP
Sbjct: 1065 PPPPMFGGAQPPPPPPMRGGA-------PPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPM 1117
Query: 409 GQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQPGSS 468
H PP P + A PP PP + P A PPP PP P P PG
Sbjct: 1118 ----HGGAPPPPPPPMRGGAPPPPPPPGGRGP----GAPPPPPPPGGRAP-GPPPPPGPR 1168
Query: 469 QTGSG 473
G G
Sbjct: 1169 PPGGG 1173
Score = 42.0 bits (97), Expect = 8e-04
Identities = 31/121 (25%), Positives = 40/121 (32%), Gaps = 18/121 (14%)
Query: 360 PSMDPQRMGALLPS---QDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQ----GQYH 412
P P+ + LP PP PS + E K Q P PPP Y
Sbjct: 604 PDSSPKETPSSLPPASPHQAPPPLPSLTSEAKTVLHSSQAVASPPPPPPPPPLPTYSHYQ 663
Query: 413 HQYYPPYQQYPPQYNAAMP-----------PVPPYQQYPPQYNAAMPPPQPPAANHPYQH 461
PP PP +++ P P PP+ P +PPP PP +
Sbjct: 664 TSQLPPPPPPPPPFSSERPNSGTVLPPPPPPPPPFSSERPNSGTVLPPPPPPPLPFSSER 723
Query: 462 P 462
P
Sbjct: 724 P 724
Score = 40.8 bits (94), Expect = 0.002
Identities = 36/117 (30%), Positives = 37/117 (30%), Gaps = 15/117 (12%)
Query: 349 PPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQ 408
PPPP P P G P PP P G P H G P PPP
Sbjct: 1076 PPPPPMRGGAPPPPPPPMRGGAPP----PPPPPMRGGAPPPPPPPM---HGGAPPPPPPP 1128
Query: 409 GQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQP 465
+ PP PP P PP PP A PP PP P P P
Sbjct: 1129 MRGGAPPPPP----PPGGRGPGAPPPP----PPPGGRAPGPPPPPGPRPPGGGPPPP 1177
Score = 35.8 bits (81), Expect = 0.055
Identities = 29/110 (26%), Positives = 38/110 (34%), Gaps = 29/110 (26%)
Query: 349 PPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQ 408
PPPP ++Y + LP PP P +P+S G + PPP
Sbjct: 652 PPPPLPTYSHYQTSQ-------LPP---PPPPPPPFSSERPNS--------GTVLPPPPP 693
Query: 409 GQYHHQYYPPYQQYPPQYNAAMPPVPP-----YQQYPPQYNAAMPPPQPP 453
PP+ P +PP PP + P PPP PP
Sbjct: 694 PP------PPFSSERPNSGTVLPPPPPPPLPFSSERPNSGTVLPPPPSPP 737
Score = 33.5 bits (75), Expect = 0.27
Identities = 24/84 (28%), Positives = 32/84 (37%), Gaps = 18/84 (21%)
Query: 377 PPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPY 436
PP P+ + SS+ Q Q P PPP PP+ +PP PP
Sbjct: 765 PPPPPAYYSVGQKSSDLQTSQLPSPPPPPPP---------PPFASVRRNSETLLPPPPPP 815
Query: 437 QQYPPQYNAA-------MPPPQPP 453
PP + + +PPP PP
Sbjct: 816 P--PPPFASVRRNSETLLPPPPPP 837
Score = 32.3 bits (72), Expect = 0.61
Identities = 44/206 (21%), Positives = 66/206 (31%), Gaps = 21/206 (10%)
Query: 277 EKAAEELSNKLVIKGLRLKLMWGRPQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQN 336
+ +E SN L + +P S KP++ Q A A +G + QQ+
Sbjct: 456 QSKGDEESNDLESMSQKTNTSLNKPISEKPQATLRKQVGANAKPAAAGDSLKPKSKQQET 515
Query: 337 QDQTQGMLYYNNPPP--PQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSE-- 392
Q M N P + +Y SM + S G+ S +
Sbjct: 516 QGPNVRMAKPNAVSRWIPSNKGSYKDSMHVAYPPTRINSAPASITTSLKDGKRATSPDGV 575
Query: 393 --KQQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPP--------- 441
K Y R + P + P + +++PP P+Q PP
Sbjct: 576 IPKDAKTKYLRASVSSPDMRSRAPICSSPDSSPKETPSSLPPASPHQAPPPLPSLTSEAK 635
Query: 442 ------QYNAAMPPPQPPAANHPYQH 461
Q A+ PPP PP Y H
Sbjct: 636 TVLHSSQAVASPPPPPPPPPLPTYSH 661
Score = 30.0 bits (66), Expect = 3.0
Identities = 28/113 (24%), Positives = 38/113 (32%), Gaps = 4/113 (3%)
Query: 349 PPPPQQERNYYPSMDPQRMGALLPSQDGPPG-GPSGSGENKPSSEKQQMQHYGRPMMPPP 407
PPPP + S + G +LP PP S P+ P PPP
Sbjct: 710 PPPPPPPLPF--SSERPNSGTVLPPPPSPPWKSVYASALAIPAICSTSQAPTSSPTPPPP 767
Query: 408 QGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNA-AMPPPQPPAANHPY 459
Y+ Q + PP PP + N+ + PP PP P+
Sbjct: 768 PPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSETLLPPPPPPPPPPF 820
>At3g19130 nuclear acid binding protein, putative
Length = 435
Score = 62.0 bits (149), Expect = 7e-10
Identities = 36/104 (34%), Positives = 55/104 (52%), Gaps = 2/104 (1%)
Query: 211 GKAGEMSSLEVPEDESIK-TLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRACAFVT 269
G G M + ES T++VGG+D V ++DLR F GE+ SVK+ + + C FV
Sbjct: 303 GSNGSMGYGSQSDGESTNATIFVGGIDPDVIDEDLRQPFSQFGEVVSVKIPVGKGCGFVQ 362
Query: 270 YTTREGAEKAAEELSNKLVIKGLRLKLMWGRPQSAKPESDGLDQ 313
+ R+ AE A E L+ ++ K ++L WGR + + D Q
Sbjct: 363 FADRKSAEDAIESLNGTVIGKN-TVRLSWGRSPNKQWRGDSGQQ 405
Score = 32.7 bits (73), Expect = 0.47
Identities = 28/95 (29%), Positives = 38/95 (39%), Gaps = 25/95 (26%)
Query: 368 GALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQG-----------QYHHQYY 416
GA P+Q PP + ++QQ Q + M PP Y HQY
Sbjct: 15 GATPPNQQTPPP------PQQWQQQQQQQQQWMAAMQYPPAAAMMMMQQQQMLMYPHQYV 68
Query: 417 P----PYQQYPPQYNAAMPPVPPYQQYPPQYNAAM 447
P PYQQ+ PQ + YQQ+ Q + A+
Sbjct: 69 PYNQGPYQQHHPQ----LHQYGSYQQHQHQQHKAI 99
Score = 32.3 bits (72), Expect = 0.61
Identities = 20/70 (28%), Positives = 35/70 (49%), Gaps = 6/70 (8%)
Query: 225 ESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRACA------FVTYTTREGAEK 278
+ +KTL+VG L + E L F GE+ SVK++ + + FV + +R AE+
Sbjct: 105 DDVKTLWVGDLLHWMDETYLHSCFSHTGEVSSVKVIRNKLTSQSEGYGFVEFLSRAAAEE 164
Query: 279 AAEELSNKLV 288
+ S ++
Sbjct: 165 VLQNYSGSVM 174
>At5g54900 unknown protein
Length = 387
Score = 60.8 bits (146), Expect = 2e-09
Identities = 44/144 (30%), Positives = 61/144 (41%), Gaps = 33/144 (22%)
Query: 224 DESIKTLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRACAFVTYTTREGAEKAAEEL 283
D + T++VGGLDA VT+ +L+ F GE+ VK+ + C FV Y + AE A L
Sbjct: 256 DPNNTTIFVGGLDANVTDDELKSIFGQFGELLHVKIPPGKRCGFVQYANKASAEHALSVL 315
Query: 284 SNKLVIKGLRLKLMWGRPQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGM 343
N + G ++L WGR ++Q +Q Q G
Sbjct: 316 -NGTQLGGQSIRLSWGRSP-----------------------------NKQSDQAQWNGG 345
Query: 344 LYYNNPPPPQQERNYY---PSMDP 364
YY PP PQ Y P+ DP
Sbjct: 346 GYYGYPPQPQGGYGYAAQPPTQDP 369
>At3g16510 unknown protein
Length = 360
Score = 59.7 bits (143), Expect = 4e-09
Identities = 46/138 (33%), Positives = 53/138 (38%), Gaps = 37/138 (26%)
Query: 347 NNPPPPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPP 406
N PPP E N+YP L S PP P QHY P P
Sbjct: 188 NYPPPQSSEANFYPP---------LSSIGYPPSSPP--------------QHYSSPPYPY 224
Query: 407 PQGQYHHQYYP--PYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQ---PPAANHPYQH 461
P +H +YP P YPP PP YPP Y + PP PP H + H
Sbjct: 225 PNPYQYHSHYPEQPVAVYPP------PPPSASNLYPPPYYSTSPPQHQSYPPPPGHSF-H 277
Query: 462 PMQPGSSQTGSGQAGSAP 479
QP SQ+ G A S+P
Sbjct: 278 QTQP--SQSFHGFAPSSP 293
Score = 32.3 bits (72), Expect = 0.61
Identities = 22/72 (30%), Positives = 28/72 (38%), Gaps = 10/72 (13%)
Query: 399 YGRPMMPPPQGQYHHQYYPPYQQ-----YPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPP 453
Y PM P YPP Q YPP + PP PPQ+ ++ P P P
Sbjct: 172 YTNPMDIPSDFSSATTNYPPPQSSEANFYPPLSSIGYPP-----SSPPQHYSSPPYPYPN 226
Query: 454 AANHPYQHPMQP 465
+ +P QP
Sbjct: 227 PYQYHSHYPEQP 238
>At2g23350 putative poly(A) binding protein
Length = 662
Score = 58.5 bits (140), Expect = 8e-09
Identities = 73/272 (26%), Positives = 103/272 (37%), Gaps = 42/272 (15%)
Query: 230 LYVGGLDARVTEQDLRDNFYAHGEIESVKMV-----LQRACAFVTYTTREGAEKAAEELS 284
LYV LD VT++ LR+ F G I S K++ + FV ++ A + E++
Sbjct: 330 LYVKNLDDTVTDEKLRELFAEFGTITSCKVMRDPSGTSKGSGFVAFSAASEASRVLNEMN 389
Query: 285 NKLV-IKGLRLKLMWGRPQ-SAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQG 342
K+V K L + L + + AK ++ Q Q G+ PR I Q
Sbjct: 390 GKMVGGKPLYVALAQRKEERRAKLQA----QFSQMRPAFIPGVGPRMPIFTGGAPGLGQQ 445
Query: 343 MLYYNNPPP--PQQ-----ERNYYPSMDPQRMGA--LLPSQDGPPGGPSGSGENKPSSEK 393
+ Y PPP P Q + P M P G + P Q GP G SG+ +
Sbjct: 446 IFYGQGPPPIIPHQPGFGYQPQLVPGMRPAFFGGPMMQPGQQGPRPGGRRSGDGPMRHQH 505
Query: 394 QQMQHYGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVP-PYQQYPPQYNAAMPPPQP 452
QQ Y +P M P Y + P MP P P P Y+ +
Sbjct: 506 QQPMPYMQPQMMPRGRGYRY----------PSGGRNMPDGPMPGGMVPVAYDMNV----- 550
Query: 453 PAANHPYQHPMQPGSSQTGSGQAGSAPAETGT 484
PY PM G Q + A + PA+ T
Sbjct: 551 ----MPYSQPMSAG--QLATSLANATPAQQRT 576
Score = 36.2 bits (82), Expect = 0.042
Identities = 25/86 (29%), Positives = 42/86 (48%), Gaps = 7/86 (8%)
Query: 229 TLYVGGLDARVTEQDLRDNFYAHGEIESVKMVLQRAC------AFVTYTTREGAEKAAEE 282
+LYVG LD VT+ L D F ++ SV++ A +V Y+ + AEKA ++
Sbjct: 47 SLYVGDLDFNVTDSQLYDYFTEVCQVVSVRVCRDAATNTSLGYGYVNYSNTDDAEKAMQK 106
Query: 283 LSNKLVIKGLRLKLMWGRPQSAKPES 308
L N + G +++ + S+ S
Sbjct: 107 L-NYSYLNGKMIRITYSSRDSSARRS 131
>At5g51300 unknown protein
Length = 804
Score = 58.2 bits (139), Expect = 1e-08
Identities = 76/290 (26%), Positives = 98/290 (33%), Gaps = 68/290 (23%)
Query: 230 LYVGGLDARVTEQDLRDNFYAHGEIESVKMV------LQRACAFVTYTTREGAEKAAEEL 283
LY+G L + + L + F + GEI K++ L + FV Y + A A + +
Sbjct: 482 LYIGFLPPMLEDDGLINLFSSFGEIVMAKVIKDRVTGLSKGYGFVKYADVQMANTAVQAM 541
Query: 284 SNKLVIKGLRLKLMWGRPQSAKPESDGLDQARQQASVAHSGLFPRAVISQQQNQDQTQGM 343
+ G R + GR + + +A P A Q TQG
Sbjct: 542 N------GYRFE---GRTLAVR--------------IAGKSPPPIAPPGPPAPQPPTQGY 578
Query: 344 LYYNNPP---PPQQERNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYG 400
N PP P QQ S P G +PS P G P Q M YG
Sbjct: 579 PPSNQPPGAYPSQQYATGGYSTAPVPWGPPVPSYSPYALPPPPPGSYHPV-HGQHMPPYG 637
Query: 401 RPMMPPPQGQYHHQYYPP------------YQQYPPQYNA-------------------A 429
PPP H PP Q +PP A A
Sbjct: 638 MQYPPPPP---HVTQAPPPGTTQNPSSSEPQQSFPPGVQADSGAATSSIPPNVYGSSVTA 694
Query: 430 MPPVPPYQQYPPQYNAAMPPPQPPAANHPYQHPMQPGSSQTGSGQAGSAP 479
MP PPY YP YN A+PPP PPA H G+ + + S P
Sbjct: 695 MPGQPPYMSYPSYYN-AVPPPTPPAPASSTDHSQNMGNMPWANNPSVSTP 743
Score = 36.6 bits (83), Expect = 0.032
Identities = 35/131 (26%), Positives = 43/131 (32%), Gaps = 30/131 (22%)
Query: 377 PPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQYHHQYY-------------PPYQQYP 423
PP P G +P ++ G P P G Y Q Y PP Y
Sbjct: 561 PPIAPPGPPAPQPPTQ-------GYPPSNQPPGAYPSQQYATGGYSTAPVPWGPPVPSYS 613
Query: 424 PQYNAAMPPVPPY-------QQYPPQYNAAMPPPQPPAANHPYQHPMQPGSSQTGSGQAG 476
P A+PP PP Q PP PPP P P SS+
Sbjct: 614 PY---ALPPPPPGSYHPVHGQHMPPYGMQYPPPPPHVTQAPPPGTTQNPSSSEPQQSFPP 670
Query: 477 SAPAETGTSTS 487
A++G +TS
Sbjct: 671 GVQADSGAATS 681
>At5g14540 unknown protein
Length = 530
Score = 57.4 bits (137), Expect = 2e-08
Identities = 46/147 (31%), Positives = 58/147 (39%), Gaps = 14/147 (9%)
Query: 350 PPPQQERNYYPSMD-PQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQH------YGRP 402
PP Q PS+ PQ P Q+ P PSG + P+ + + P
Sbjct: 252 PPASQHGLSPPSLQLPQLPNQFSPQQE-PYFPPSGQSQPPPTIQPPYQPPPPTQSLHQPP 310
Query: 403 MMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHP---- 458
PPPQ + Q PP Q+P YN PP P Q YPP PP PP + P
Sbjct: 311 YQPPPQQPQYPQQPPPQLQHPSGYNPEEPPY-PQQSYPPN-PPRQPPSHPPPGSAPSQQY 368
Query: 459 YQHPMQPGSSQTGSGQAGSAPAETGTS 485
Y P P S G G ++ +G S
Sbjct: 369 YNAPPTPPSMYDGPGGRSNSGFPSGYS 395
Score = 47.0 bits (110), Expect = 2e-05
Identities = 60/219 (27%), Positives = 78/219 (35%), Gaps = 46/219 (21%)
Query: 300 RPQSAKPESDGLDQARQQASVAHSGLFPRAVI----SQQQNQDQTQGMLYYNNPPPPQQE 355
+P ++ P+ A S+ GL P+ I SQ + + N PQQE
Sbjct: 221 QPPASLPQPPA--SAAAPPSLTQQGLPPQQFIQPPASQHGLSPPSLQLPQLPNQFSPQQE 278
Query: 356 RNYYPSMDPQRMGALLPSQDGPPGGPSGSGENKPSSEKQQMQHYGRPMMPPPQGQYHHQY 415
+ PS Q + P PP P+ S P Q Y P PPPQ Q+ Y
Sbjct: 279 PYFPPSGQSQPPPTIQPPYQPPP--PTQSLHQPPYQPPPQQPQY--PQQPPPQLQHPSGY 334
Query: 416 YP-----PYQQYPP------------------QYNAAMPPVPPYQQYPP--QYNAAMPPP 450
P P Q YPP QY A PP PP P + N+ P
Sbjct: 335 NPEEPPYPQQSYPPNPPRQPPSHPPPGSAPSQQYYNA-PPTPPSMYDGPGGRSNSGFPSG 393
Query: 451 QPPAANHPYQHPMQPGSSQTGSGQAGSAPAETGTSTSGS 489
P ++PY P Q G+ P+ T SGS
Sbjct: 394 YSP-ESYPYTGP---------PSQYGNTPSVKPTHQSGS 422
>At3g22800 hypothetical protein
Length = 470
Score = 57.0 bits (136), Expect = 2e-08
Identities = 49/187 (26%), Positives = 67/187 (35%), Gaps = 37/187 (19%)
Query: 306 PESDGLDQARQQASVAH---SGLFPRAVISQQQNQDQTQGMLYYNNPPPP-------QQE 355
PE+ G ++ +Q ++AH SG P ++ + ++ T +++ PP
Sbjct: 284 PETIGDMKSLEQLNIAHNKFSGYIPESICRLPRLENFTYSYNFFSGEPPACLRLQEFDDR 343
Query: 356 RNYYPSMDPQR-----------------MGALLPSQDGPPGGPSGSGENKPSSEKQQMQH 398
RN PS QR G PS PP P P
Sbjct: 344 RNCLPSRPMQRSLAECKSFSSYPIDCASFGCSPPSPPPPPPPPPPPPPPPPPPPP----- 398
Query: 399 YGRPMMPPPQGQYHHQYYPPYQQYPPQYNAAMPPVPPYQQYPPQYNAAMPPPQPPAANHP 458
P PPP Y + PP PP Y PP PP YPP + P PP + P
Sbjct: 399 ---PPPPPPPPPYVYPSPPPPPPSPPPY--VYPPPPPPYVYPPPPSPPYVYPPPPPSPQP 453
Query: 459 YQHPMQP 465
Y +P P
Sbjct: 454 YMYPSPP 460
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,578,653
Number of Sequences: 26719
Number of extensions: 628698
Number of successful extensions: 8173
Number of sequences better than 10.0: 501
Number of HSP's better than 10.0 without gapping: 189
Number of HSP's successfully gapped in prelim test: 320
Number of HSP's that attempted gapping in prelim test: 2253
Number of HSP's gapped (non-prelim): 1866
length of query: 492
length of database: 11,318,596
effective HSP length: 103
effective length of query: 389
effective length of database: 8,566,539
effective search space: 3332383671
effective search space used: 3332383671
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146743.13


