

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.12 + phase: 0
(139 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
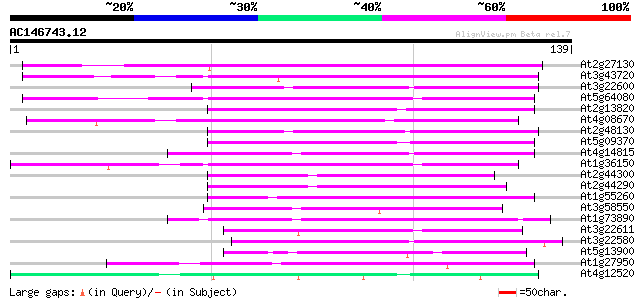
At2g27130 predicted GPI-anchored protein 101 1e-22
At3g43720 lipid-transfer protein-like, predicted GPI-anchored pr... 100 2e-22
At3g22600 predicted GPI-anchored protein 71 2e-13
At5g64080 predicted GPI-anchored protein 69 8e-13
At2g13820 predicted GPI-anchored protein 69 8e-13
At4g08670 lipid transfer protein - like predicted GPI-anchored p... 67 4e-12
At2g48130 predicted GPI-anchored protein 65 1e-11
At5g09370 putative lipid transfer protein, predicted GPI-anchore... 64 3e-11
At4g14815 Expressed protein 62 7e-11
At1g36150 hypothetical protein 60 3e-10
At2g44300 predicted GPI-anchored protein 54 3e-08
At2g44290 non-specific lipid transfer protein (nLTP) like protein 54 3e-08
At1g55260 unknown protein 52 1e-07
At3g58550 predicted GPI-anchored protein 50 3e-07
At1g73890 predicted GPI-anchored protein 46 5e-06
At3g22611 predicted GPI-anchored protein 43 5e-05
At3g22580 unknown protein 42 1e-04
At5g13900 predicted GPI-anchored protein 41 2e-04
At1g27950 GPI-anchored protein (LTPL) 41 2e-04
At4g12520 pEARLI 1-like protein 40 3e-04
>At2g27130 predicted GPI-anchored protein
Length = 159
Score = 101 bits (251), Expect = 1e-22
Identities = 50/130 (38%), Positives = 74/130 (56%), Gaps = 11/130 (8%)
Query: 4 TTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADD-GCLMALTNMSDCLT 62
TT +A++ L+F SL QSP + A P+ CL+++ N+SDC +
Sbjct: 3 TTNTLAVLLLLFLSL----------CSGQSPPAPEPIAADGPSSPVNCLVSMLNVSDCFS 52
Query: 63 FVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGV 122
+V+ GS KP+ CCPELAG++ +P C+C L G + FG+K++ +A +L TICGV
Sbjct: 53 YVQVGSNEIKPEAACCPELAGMVQSSPECVCNLYGGGASPRFGVKLDKQRAEQLSTICGV 112
Query: 123 TTPPVSACSG 132
P S CSG
Sbjct: 113 KAPSPSLCSG 122
>At3g43720 lipid-transfer protein-like, predicted GPI-anchored
protein
Length = 193
Score = 100 bits (249), Expect = 2e-22
Identities = 60/130 (46%), Positives = 77/130 (59%), Gaps = 12/130 (9%)
Query: 4 TTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLTF 63
+ +VV V LI ASL+ H +A + SP+ PS A D C+ L NM+ CL++
Sbjct: 2 SNVVVIAVVLIVASLTGH----VSAQMDMSPSS-----GPSGAPD-CMANLMNMTGCLSY 51
Query: 64 VE--DGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICG 121
V +G KPDK CCP LAGL++ +P CLC LL + A GIKI+ KALKLP +CG
Sbjct: 52 VTVGEGGGAAKPDKTCCPALAGLVESSPQCLCYLLSGDMAAQLGIKIDKAKALKLPGVCG 111
Query: 122 VTTPPVSACS 131
V TP S CS
Sbjct: 112 VITPDPSLCS 121
>At3g22600 predicted GPI-anchored protein
Length = 170
Score = 71.2 bits (173), Expect = 2e-13
Identities = 34/86 (39%), Positives = 49/86 (56%), Gaps = 3/86 (3%)
Query: 46 ADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFG 105
A C AL +MS CL ++ S T P++ CC +L+ ++ +P CLC++L G
Sbjct: 24 AQSSCTNALISMSPCLNYITGNS--TSPNQQCCNQLSRVVQSSPDCLCQVLNGG-GSQLG 80
Query: 106 IKINVNKALKLPTICGVTTPPVSACS 131
I +N +AL LP C V TPPVS C+
Sbjct: 81 INVNQTQALGLPRACNVQTPPVSRCN 106
>At5g64080 predicted GPI-anchored protein
Length = 173
Score = 68.9 bits (167), Expect = 8e-13
Identities = 41/127 (32%), Positives = 62/127 (48%), Gaps = 15/127 (11%)
Query: 4 TTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLTF 63
TT + IV L +S+S G A AP+P+ D C + NM+DCL+F
Sbjct: 10 TTPLFLIVLLSLSSVSVLG------------ASHHHATAPAPSVD-CSTLILNMADCLSF 56
Query: 64 VEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGVT 123
V G + KP+ CC L ++ + CLC+ S+ S G+ +N+ KA LP C +
Sbjct: 57 VSSGGTVAKPEGTCCSGLKTVLKADSQCLCEAFKSSA--SLGVTLNITKASTLPAACKLH 114
Query: 124 TPPVSAC 130
P ++ C
Sbjct: 115 APSIATC 121
>At2g13820 predicted GPI-anchored protein
Length = 169
Score = 68.9 bits (167), Expect = 8e-13
Identities = 30/81 (37%), Positives = 47/81 (57%), Gaps = 2/81 (2%)
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C + NM+DCL+FV GS + KP+ CC L ++ P CLC+ + S G+ ++
Sbjct: 27 CSSLILNMADCLSFVTSGSTVVKPEGTCCSGLKTVVRTGPECLCEAF--KNSGSLGLTLD 84
Query: 110 VNKALKLPTICGVTTPPVSAC 130
++KA LP++C V PP + C
Sbjct: 85 LSKAASLPSVCKVAAPPSARC 105
>At4g08670 lipid transfer protein - like predicted GPI-anchored
protein
Length = 194
Score = 66.6 bits (161), Expect = 4e-12
Identities = 40/126 (31%), Positives = 63/126 (49%), Gaps = 11/126 (8%)
Query: 5 TLVVAIVSLIFASLSF----HGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDC 60
+L+++ V L+ +S S H S A + AP AP P+ C + +M DC
Sbjct: 4 SLLLSFVLLLLSSSSLVTPIHARNKSNPAKSPVGAP-----APGPSSSDCSTVIYSMMDC 58
Query: 61 LTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTIC 120
L ++ GS TKP+K CC + ++ NP C+C G +A GI++N +AL P C
Sbjct: 59 LGYLGVGSNETKPEKSCCTGIETVLQYNPQCIC--AGLVSAGEMGIELNSTRALATPKAC 116
Query: 121 GVTTPP 126
++ P
Sbjct: 117 KLSIAP 122
>At2g48130 predicted GPI-anchored protein
Length = 183
Score = 64.7 bits (156), Expect = 1e-11
Identities = 29/82 (35%), Positives = 47/82 (56%), Gaps = 3/82 (3%)
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C+ LT +S CL+++ S T P + CC L +I +P C+C + S + G+ IN
Sbjct: 30 CVSTLTTLSPCLSYITGNS--TTPSQPCCSRLDSVIKSSPQCICSAVNS-PIPNIGLNIN 86
Query: 110 VNKALKLPTICGVTTPPVSACS 131
+AL+LP C + TPP++ C+
Sbjct: 87 RTQALQLPNACNIQTPPLTQCN 108
>At5g09370 putative lipid transfer protein, predicted GPI-anchored
protein
Length = 129
Score = 63.9 bits (154), Expect = 3e-11
Identities = 31/81 (38%), Positives = 40/81 (49%), Gaps = 3/81 (3%)
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C + + CL F+ G P CC L ++D PICLC+ L GIK+N
Sbjct: 28 CDTLVITLFPCLPFISIGGTADTPTASCCSSLKNILDTKPICLCEGL---KKAPLGIKLN 84
Query: 110 VNKALKLPTICGVTTPPVSAC 130
V K+ LP C + PPVSAC
Sbjct: 85 VTKSATLPVACKLNAPPVSAC 105
>At4g14815 Expressed protein
Length = 156
Score = 62.4 bits (150), Expect = 7e-11
Identities = 32/91 (35%), Positives = 48/91 (52%), Gaps = 3/91 (3%)
Query: 40 VLAPSPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSN 99
V++ A C L +M+ CL+F+ + L P + CC +LA ++ + CLC++L
Sbjct: 16 VMSIVSAQSSCTNVLISMAPCLSFITQNTSL--PSQQCCNQLAHVVRYSSECLCQVLDGG 73
Query: 100 TADSFGIKINVNKALKLPTICGVTTPPVSAC 130
GI +N +AL LP C V TPP S C
Sbjct: 74 -GSQLGINVNETQALALPKACHVETPPASRC 103
>At1g36150 hypothetical protein
Length = 256
Score = 60.5 bits (145), Expect = 3e-10
Identities = 41/127 (32%), Positives = 66/127 (51%), Gaps = 9/127 (7%)
Query: 1 MATTTLVVAIVSLIFASLSFHGD-GVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSD 59
M + ++++IV L+ +SLS D G + + +P P+ PS + D C + +M D
Sbjct: 1 MKPSFVLLSIVLLLSSSLSDAADFGSPSQPPSMAPTPQ-----PSNSTD-CSSVIYSMVD 54
Query: 60 CLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTI 119
CL+F+ GS P K CC + +++ +P CLC L S+ G ++ KAL +P I
Sbjct: 55 CLSFLTVGSTDPSPTKTCCVGVKTVLNYSPKCLCSALESSR--EMGFVLDDTKALAMPKI 112
Query: 120 CGVTTPP 126
C V P
Sbjct: 113 CNVPIDP 119
>At2g44300 predicted GPI-anchored protein
Length = 204
Score = 53.9 bits (128), Expect = 3e-08
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C L M+ CL +V+ +K PD CC L +++ N CLC ++ G++IN
Sbjct: 35 CTEQLVGMATCLPYVQGQAKSPTPD--CCSGLKQVLNSNKKCLCVIIQDRNDPDLGLQIN 92
Query: 110 VNKALKLPTIC 120
V+ AL LP++C
Sbjct: 93 VSLALALPSVC 103
>At2g44290 non-specific lipid transfer protein (nLTP) like protein
Length = 205
Score = 53.9 bits (128), Expect = 3e-08
Identities = 25/74 (33%), Positives = 40/74 (53%), Gaps = 2/74 (2%)
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C L M+ CL +V+ +K PD CC L +I+ + CLC ++ G+++N
Sbjct: 36 CTAQLVGMATCLPYVQGKAKSPTPD--CCSGLKQVINSDMKCLCMIIQERNDPDLGLQVN 93
Query: 110 VNKALKLPTICGVT 123
V+ AL LP++C T
Sbjct: 94 VSLALALPSVCHAT 107
>At1g55260 unknown protein
Length = 184
Score = 51.6 bits (122), Expect = 1e-07
Identities = 26/81 (32%), Positives = 37/81 (45%), Gaps = 2/81 (2%)
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKIN 109
C L +S C+ +V G P K CC +I + C+C L+ GIKIN
Sbjct: 33 CTNQLIELSTCIPYV--GGDAKAPTKDCCAGFGQVIRKSEKCVCILVRDKDDPQLGIKIN 90
Query: 110 VNKALKLPTICGVTTPPVSAC 130
A LP+ C +T P ++ C
Sbjct: 91 ATLAAHLPSACHITAPNITDC 111
>At3g58550 predicted GPI-anchored protein
Length = 177
Score = 50.4 bits (119), Expect = 3e-07
Identities = 27/76 (35%), Positives = 39/76 (50%), Gaps = 4/76 (5%)
Query: 49 GCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPI--CLCKLLGSNTADSFGI 106
GC A++++ CL FV + +K PD CC L ID CLC L+ G
Sbjct: 37 GCQDAMSDLYSCLPFVTNKAKA--PDSTCCSTLKVKIDKGQTRKCLCTLVKDRDDPGLGF 94
Query: 107 KINVNKALKLPTICGV 122
K++ N+A+ LP+ C V
Sbjct: 95 KVDANRAMSLPSACHV 110
>At1g73890 predicted GPI-anchored protein
Length = 193
Score = 46.2 bits (108), Expect = 5e-06
Identities = 31/95 (32%), Positives = 44/95 (45%), Gaps = 5/95 (5%)
Query: 40 VLAPSPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSN 99
VLA PA C L +++ C FV+ ++L P + CC L + CLC L +
Sbjct: 22 VLAQVPAT--CASRLLSLAPCGPFVQGFAQL--PAQPCCDSLNQIYSQEATCLCLFLNNT 77
Query: 100 TADSFGIKINVNKALKLPTICGVTTPPVSACSGKF 134
+ S IN AL+LP +C + S CS F
Sbjct: 78 STLSPAFPINQTLALQLPPLCNIPANS-STCSSSF 111
>At3g22611 predicted GPI-anchored protein
Length = 203
Score = 43.1 bits (100), Expect = 5e-05
Identities = 24/75 (32%), Positives = 34/75 (45%), Gaps = 3/75 (4%)
Query: 54 LTNMSDCLTFVEDGSKL-TKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNK 112
+T +S C+ F+ + S T P CC L L G CLC ++ F I IN
Sbjct: 34 MTTVSPCMGFITNSSSNGTSPSSDCCNSLRSLTTGGMGCLCLIVTGTV--PFNIPINRTT 91
Query: 113 ALKLPTICGVTTPPV 127
A+ LP C + P+
Sbjct: 92 AVSLPRACNMPRVPL 106
>At3g22580 unknown protein
Length = 127
Score = 42.0 bits (97), Expect = 1e-04
Identities = 24/83 (28%), Positives = 37/83 (43%), Gaps = 2/83 (2%)
Query: 56 NMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALK 115
++ C +++ D CCP L + + C C+ L S I N +A +
Sbjct: 37 SLKPCFSYLTSSYPSLPDDSDCCPSLLDISKTSVDCFCQYLNSG-GSILDINANFIQARR 95
Query: 116 LPTICGVTTPPVSACS-GKFFMS 137
LP ICGV S C+ GK F++
Sbjct: 96 LPEICGVDPYLASVCNEGKNFLT 118
>At5g13900 predicted GPI-anchored protein
Length = 151
Score = 41.2 bits (95), Expect = 2e-04
Identities = 26/78 (33%), Positives = 43/78 (54%), Gaps = 8/78 (10%)
Query: 54 LTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLG---SNTADSFGIKINV 110
L ++ CL ++ +G+K + + CC L +I NP CLC+++ S+ A+ G I+V
Sbjct: 33 LNQLAPCLNYL-NGTK--EVPQVCCNPLKSVIRNNPECLCRMISNRWSSQAERAG--IDV 87
Query: 111 NKALKLPTICGVTTPPVS 128
N A LP CG P++
Sbjct: 88 NDAQMLPARCGEHVNPIA 105
>At1g27950 GPI-anchored protein (LTPL)
Length = 193
Score = 41.2 bits (95), Expect = 2e-04
Identities = 28/109 (25%), Positives = 47/109 (42%), Gaps = 10/109 (9%)
Query: 25 VSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGL 84
+ A+ A +PA LA D C ++ CL F K T P K CC + +
Sbjct: 15 IVASIAAAAPAAPGGALA-----DECNQDFQKVTLCLDFAT--GKATIPSKKCCDAVEDI 67
Query: 85 IDGNPICLCKLLGSNTADSFGIK---INVNKALKLPTICGVTTPPVSAC 130
+ +P CLC ++ +K + +K ++LPT C + ++ C
Sbjct: 68 KERDPKCLCFVIQQAKTGGQALKDLGVQEDKLIQLPTSCQLHNASITNC 116
>At4g12520 pEARLI 1-like protein
Length = 129
Score = 40.4 bits (93), Expect = 3e-04
Identities = 39/137 (28%), Positives = 54/137 (38%), Gaps = 14/137 (10%)
Query: 1 MATTTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDG-CLMALTNMSD 59
MA+ ++ L F L F + +P P+ P P G C +
Sbjct: 1 MASKISASLVIFLTFNILFFTLTTACGGGCSSTPKPK-----PKPKSTGSCPKDTLKLGV 55
Query: 60 CLTFVEDGSKL---TKPDKGCCPELAGLID-GNPICLCKLLGSNTADSFGIKINVNKALK 115
C ++D K+ T P K CC L GL+D CLC L A GI +NV +L
Sbjct: 56 CANVLKDLLKIQLGTPPVKPCCSLLNGLVDLEAAACLCTAL---KAKVLGINLNVPVSLS 112
Query: 116 -LPTICGVTTPPVSACS 131
L +CG P C+
Sbjct: 113 LLLNVCGKKVPSGFVCA 129
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,208,630
Number of Sequences: 26719
Number of extensions: 130096
Number of successful extensions: 374
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 332
Number of HSP's gapped (non-prelim): 53
length of query: 139
length of database: 11,318,596
effective HSP length: 89
effective length of query: 50
effective length of database: 8,940,605
effective search space: 447030250
effective search space used: 447030250
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146743.12


