

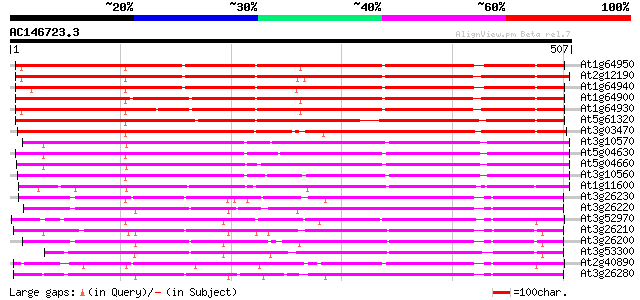
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146723.3 - phase: 0
(507 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g64950 unknown protein 509 e-144
At2g12190 pseudogene 503 e-142
At1g64940 hypothetical protein 491 e-139
At1g64900 unknown protein 491 e-139
At1g64930 hypothetical protein 478 e-135
At5g61320 cytochrome P450 - like protein 449 e-126
At3g03470 cytochrome P450 nlike protein 448 e-126
At3g10570 putative cytochrome P450 321 7e-88
At5g04630 cytochrome P450 - like protein 316 2e-86
At5g04660 cytochrom P450 - like protein 312 2e-85
At3g10560 putative cytochrome P450 305 4e-83
At1g11600 cytochrome P450 like protein 295 3e-80
At3g26230 cytochrome P450, putative 197 9e-51
At3g26220 cytochrome P450 monooxygenase (CYP71B3) 197 1e-50
At3g52970 cytochrome P450 - like protein 192 3e-49
At3g26210 cytochrome P450 like protein 192 5e-49
At3g26200 cytochrome P450 like protein 190 1e-48
At3g53300 CYTOCHROME P450-like protein 190 2e-48
At2g40890 cytochrome P450 like protein 188 7e-48
At3g26280 cytochrome P450 monooxygenase (CYP71B4) 186 3e-47
>At1g64950 unknown protein
Length = 510
Score = 509 bits (1311), Expect = e-144
Identities = 263/512 (51%), Positives = 353/512 (68%), Gaps = 29/512 (5%)
Query: 6 WFII--TALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
W +I + L LLL L+ RLR S S+ LPP P Y P I I L+ L +R +H
Sbjct: 4 WLLILGSLFLSLLLNLLFFRLRDSSSLPLPPDPNYFPFIGTIQWLRQGLGGLNNYLRSVH 63
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSC 117
+ GPIIT S P IF++DR LAHQAL+ N VFADRP K + ++Q NI+S
Sbjct: 64 HRLGPIITLRITSRPSIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSCL 123
Query: 118 YGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHA 177
YG TW +R NL S +L PSR +S+S AR+ VL+ L + F N + +++++ +A
Sbjct: 124 YGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDR--FGKNRGEEPIVVVDHLHYA 181
Query: 178 IFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQ 237
+F LL MCFG+ +D ++I++++ +QRR +L + LN +PK ++++ RKRW+EF Q
Sbjct: 182 MFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKF-TKLILRKRWEEFFQ 240
Query: 238 LRKLQNDLLVELIQARKKVIEKNN--------DDDEYVVCYVDTLLNLRLPEEKRKLNEG 289
+R+ Q+D+L+ LI+AR+K++E+ D+ EYV YVDTLL L LP+EKRKLNE
Sbjct: 241 MRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKEYVQSYVDTLLELELPDEKRKLNED 300
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
E+V+LCSEFL GTDTT+T L+W+MANLVK+ +Q+ L EEI+ VVGE E EV EED +
Sbjct: 301 EIVSLCSEFLNGGTDTTATALQWIMANLVKNPDIQKRLYEEIKSVVGE-EANEVEEEDAQ 359
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
K+PYL+AV++EGLRRHPP H+ +P VTED VL GY VPKNGT+NFMVA+IG DP VWE+
Sbjct: 360 KMPYLEAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKNGTINFMVAEIGRDPKVWEE 419
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF++ +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V
Sbjct: 420 PMAFKPERFME--------EAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 471
Query: 470 LNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
F+WK + G VDLTEK EFT+VMK+PL+A
Sbjct: 472 REFDWK-EVQGHEVDLTEKLEFTVVMKHPLKA 502
>At2g12190 pseudogene
Length = 512
Score = 503 bits (1295), Expect = e-142
Identities = 263/517 (50%), Positives = 355/517 (67%), Gaps = 27/517 (5%)
Query: 6 WFII--TALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
W +I + L LLL L++ R R S S+ LPP P + P + + L+ L +R +H
Sbjct: 4 WLLILGSLFLSLLLNLLLFRRRDSSSLPLPPDPNFFPFLGTLQWLRQGLGGLNNYLRSVH 63
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSC 117
+ GPIIT S P IF++DR LAHQAL+ N VFADRP K + ++Q NI+SS
Sbjct: 64 HRLGPIITLRITSRPAIFVADRSLAHQALVLNGAVFADRPPAAPISKIISSNQHNISSSL 123
Query: 118 YGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHA 177
YG TW +R NL S +L PSR +S+S AR+ VL+ L + F + + +++++ +A
Sbjct: 124 YGATWRLLRRNLTSEILHPSRVRSYSHARRWVLEILFDR--FGKSRGEEPIVVVDHLHYA 181
Query: 178 IFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQ 237
+F LL MCFG+ +D ++I++++ +QRR +L + LN +PK ++++ RKRW+EF Q
Sbjct: 182 MFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILNLWPKF-TKLILRKRWEEFFQ 240
Query: 238 LRKLQNDLLVELIQARKKVIE--KNNDDDE------YVVCYVDTLLNLRLPEEKRKLNEG 289
+R+ Q+D+L+ LI+AR+K++E KN +E YV YVDTLL L LP+EKRKLNE
Sbjct: 241 MRREQHDVLLPLIRARRKIVEERKNRSSEEEEDNKVYVQSYVDTLLELELPDEKRKLNED 300
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
E+V+LCSEFL GTDTT+T L+W+MANLVK+ +Q+ L EEI+ VVGE E +EV EED +
Sbjct: 301 EIVSLCSEFLNGGTDTTATALQWIMANLVKNPEIQKRLYEEIKSVVGE-EAKEVEEEDAQ 359
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
K+PYLKAV++EGLRRHPP H+ +P VTED VL GY VPK GT+NFMVA+IG DP VWE+
Sbjct: 360 KMPYLKAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKKGTINFMVAEIGRDPMVWEE 419
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF+ E +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V
Sbjct: 420 PMAFKPERFM------GEEEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 473
Query: 470 LNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
FEWK + G VDLTEK EFT+VMK+ L+A LR
Sbjct: 474 REFEWK-EVQGHEVDLTEKFEFTVVMKHSLKALAVLR 509
>At1g64940 hypothetical protein
Length = 511
Score = 491 bits (1265), Expect = e-139
Identities = 254/513 (49%), Positives = 348/513 (67%), Gaps = 30/513 (5%)
Query: 6 WFIITALLVLLLC---LIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKL 62
W +I L L L L+ R S S+ LPP P + P I + L+ L +R +
Sbjct: 4 WLLILGSLFLSLLVNHLLFRRRDSFSSLPLPPDPNFFPFIGTLKWLRKGLGGLDNYLRSV 63
Query: 63 HAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASS 116
H GPIIT S P IF++DR LAHQAL+ N VFADRP K + ++Q NI+S
Sbjct: 64 HHHLGPIITLRITSRPAIFVTDRSLAHQALVLNGAVFADRPPAESISKIISSNQHNISSC 123
Query: 117 CYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRH 176
YG TW +R NL S +L PSR +S+S AR+ VL+ L + F N + +++++ +
Sbjct: 124 LYGATWRLLRRNLTSEILHPSRLRSYSHARRWVLEILFGR--FGKNRGEEPIVVVDHLHY 181
Query: 177 AIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFT 236
A+F LL MCFG+ +D ++I++++ +QRR +L + L +PK +++++RKRW+EF
Sbjct: 182 AMFALLVLMCFGDKLDEKQIKQVEYVQRRQLLGFSRFNILTLWPKF-TKLIYRKRWEEFF 240
Query: 237 QLRKLQNDLLVELIQARKKVI--------EKNNDDDEYVVCYVDTLLNLRLPEEKRKLNE 288
Q++ Q D+L+ LI+AR+K++ E+ D+ EYV YVDTLL++ LP+EKRKLNE
Sbjct: 241 QMQSEQQDVLLPLIRARRKIVDERKKRSSEEEKDNKEYVQSYVDTLLDVELPDEKRKLNE 300
Query: 289 GEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDL 348
E+V+LCSEFL AGTDTT+T L+W+MANLVK+ +Q+ L EEI+ +VGE E +EV E+D
Sbjct: 301 DEIVSLCSEFLNAGTDTTATALQWIMANLVKNPEIQRRLYEEIKSIVGE-EAKEVEEQDA 359
Query: 349 EKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWE 408
+K+PYLKAV++EGLRRHPP H+ +P VTED VL GY VPK GT+NFMVA+IG DP VWE
Sbjct: 360 QKMPYLKAVVMEGLRRHPPGHFVLPHSVTEDTVLGGYKVPKKGTINFMVAEIGRDPKVWE 419
Query: 409 DPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANL 468
+P FKPERF++ +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+
Sbjct: 420 EPMAFKPERFME--------EAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANM 471
Query: 469 VLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
V FEW+ + G VDLTEK EFT+VMK+PL+A
Sbjct: 472 VREFEWQ-EVQGHEVDLTEKLEFTVVMKHPLKA 503
>At1g64900 unknown protein
Length = 506
Score = 491 bits (1264), Expect = e-139
Identities = 258/508 (50%), Positives = 347/508 (67%), Gaps = 25/508 (4%)
Query: 6 WFIITALLV-LLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHA 64
W +I A L LL ++LR R+S S LPP P +LP + + L+ L+ +R +H
Sbjct: 4 WLLILASLSGSLLLHLLLRRRNSSSPPLPPDPNFLPFLGTLQWLREGLGGLESYLRSVHH 63
Query: 65 KHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCY 118
+ GPI+T S P IF++DR L H+AL+ N V+ADRP K + H NI+S Y
Sbjct: 64 RLGPIVTLRITSRPAIFVADRSLTHEALVLNGAVYADRPPPAVISKIVDEH--NISSGSY 121
Query: 119 GPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAI 178
G TW +R N+ S +L PSR +S+S AR VL+ L + + + LI+++ +A+
Sbjct: 122 GATWRLLRRNITSEILHPSRVRSYSHARHWVLEILFERFRNHGG--EEPIVLIHHLHYAM 179
Query: 179 FCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQL 238
F LL MCFG+ +D ++I++++ IQR +LS + N +PK ++++ RKRWQEF Q+
Sbjct: 180 FALLVLMCFGDKLDEKQIKEVEFIQRLQLLSLTKFNIFNIWPKF-TKLILRKRWQEFLQI 238
Query: 239 RKLQNDLLVELIQARKKVIEKNN-----DDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVA 293
R+ Q D+L+ LI+AR+K++E+ D +YV YVDTLL+L LPEE RKLNE +++
Sbjct: 239 RRQQRDVLLPLIRARRKIVEERKRSEQEDKKDYVQSYVDTLLDLELPEENRKLNEEDIMN 298
Query: 294 LCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPY 353
LCSEFLTAGTDTT+T L+W+MANLVK +Q+ L EEI+ VVGE E +EV EED+EK+PY
Sbjct: 299 LCSEFLTAGTDTTATALQWIMANLVKYPEIQERLHEEIKSVVGE-EAKEVEEEDVEKMPY 357
Query: 354 LKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVF 413
LKAV+LEGLRRHPP H+ +P VTED VL GY VPKNGT+NFMVA+IG DP WE+P F
Sbjct: 358 LKAVVLEGLRRHPPGHFLLPHSVTEDTVLGGYKVPKNGTINFMVAEIGRDPVEWEEPMAF 417
Query: 414 KPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFE 473
KPERF+ E +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V F+
Sbjct: 418 KPERFM------GEEEAVDLTGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMVREFQ 471
Query: 474 WKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
WK + G VDLTEK EFT+VMK+PL+A
Sbjct: 472 WK-EVQGHEVDLTEKLEFTVVMKHPLKA 498
>At1g64930 hypothetical protein
Length = 511
Score = 478 bits (1229), Expect = e-135
Identities = 248/512 (48%), Positives = 342/512 (66%), Gaps = 28/512 (5%)
Query: 6 WFII--TALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
W +I + L LLL L+ RLR S S+ LPP P + P + + L+ +R +H
Sbjct: 4 WLLILGSLSLSLLLNLLFFRLRDSSSLPLPPAPNFFPFLGTLQWLRQGLGGFNNYVRSVH 63
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSC 117
+ GPIIT S P IF++D LAHQAL+ N VFADRP K L N+Q I S
Sbjct: 64 HRLGPIITLRITSRPAIFVADGSLAHQALVLNGAVFADRPPAAPISKILSNNQHTITSCL 123
Query: 118 YGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHA 177
YG TW +R N+ I L PSR KS+S R VL+ L +L+ + + +++ +A
Sbjct: 124 YGVTWRLLRRNITEI-LHPSRMKSYSHVRHWVLEILFDRLRKSGG--EEPIVVFDHLHYA 180
Query: 178 IFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQ 237
+F +L MCFG+ +D ++I++++ +QR+++L + LN PK ++++ RKRW+EF Q
Sbjct: 181 MFAVLVLMCFGDKLDEKQIKQVEYVQRQMLLGFARYSILNLCPKF-TKLILRKRWEEFFQ 239
Query: 238 LRKLQNDLLVELIQARKKVIEKNN--------DDDEYVVCYVDTLLNLRLPEEKRKLNEG 289
+R+ Q D+L+ LI AR+K++E+ ++ EYV YVDTLL++ LP+EKRKLNE
Sbjct: 240 MRREQQDVLLRLIYARRKIVEERKKRSSEEEEENKEYVQSYVDTLLDVELPDEKRKLNED 299
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
E+V+LCSEFL AG+DTT+T L+W+MANLVK++ +Q+ L EEI VVGE E + V E+D +
Sbjct: 300 EIVSLCSEFLIAGSDTTATVLQWIMANLVKNQEIQERLYEEITNVVGE-EAKVVEEKDTQ 358
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
K+PYLKAV++E LRRHPP + +P VTED VL GY VPK GT+NF+VA+IG DP VWE+
Sbjct: 359 KMPYLKAVVMEALRRHPPGNTVLPHSVTEDTVLGGYKVPKKGTINFLVAEIGRDPKVWEE 418
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF+ E +A D+ G + IKMMPFGAGRRICP LAMLHLEY+VAN+V
Sbjct: 419 PMAFKPERFM------GEEEAVDITGSRGIKMMPFGAGRRICPGIGLAMLHLEYYVANMV 472
Query: 470 LNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEA 501
F+WK + G VDLTEK EFT++MK+PL+A
Sbjct: 473 REFQWK-EVEGHEVDLTEKVEFTVIMKHPLKA 503
>At5g61320 cytochrome P450 - like protein
Length = 497
Score = 449 bits (1155), Expect = e-126
Identities = 239/504 (47%), Positives = 328/504 (64%), Gaps = 33/504 (6%)
Query: 6 WFIITALLVLLLCLIILRLR-SSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHA 64
WF I + + L+ L R ++ ++ LPP P + P+ L+ F +R +H
Sbjct: 4 WFHIFCVSFTVNFLLYLFFRRTNNNLPLPPNPNFFPMPGPFQWLRQGFDDFYSYLRSIHH 63
Query: 65 KHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCY 118
+ GPII+ +S P IF+SDR LAH+AL+ N VF+DRP K + ++Q I+S Y
Sbjct: 64 RLGPIISLRIFSVPAIFVSDRSLAHKALVLNGAVFSDRPPALPTGKIITSNQHTISSGSY 123
Query: 119 GPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAI 178
G TW +R NL S +L PSR KS+S AR+ VL+ L S+++ V +++++R+A+
Sbjct: 124 GATWRLLRRNLTSEILHPSRVKSYSNARRSVLENLCSRIRNHGEEAKPIV-VVDHLRYAM 182
Query: 179 FCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQL 238
F LL MCFG+ +D +I++++ +QRR +++ + LN FP +++ RKRW+EF
Sbjct: 183 FSLLVLMCFGDKLDEEQIKQVEFVQRRELITLPRFNILNVFPSF-TKLFLRKRWEEFLTF 241
Query: 239 RKLQNDLLVELIQARKKV-IEKNNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALCSE 297
R+ ++L+ LI++R+K+ IE + EY+ YVDTLL+L LP+EKRKLNE E+V+LCSE
Sbjct: 242 RREHKNVLLPLIRSRRKIMIESKDSGKEYIQSYVDTLLDLELPDEKRKLNEDEIVSLCSE 301
Query: 298 FLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAV 357
FL AGTDTT+TTL+W+MANL V+GE EE+E+ EE+++K+PYLKAV
Sbjct: 302 FLNAGTDTTATTLQWIMANL----------------VIGEEEEKEIEEEEMKKMPYLKAV 345
Query: 358 ILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPER 417
+LEGLR HPP H +P V+ED L GY VPK GT N VA IG DPTVWE+P FKPER
Sbjct: 346 VLEGLRLHPPGHLLLPHRVSEDTELGGYRVPKKGTFNINVAMIGRDPTVWEEPMEFKPER 405
Query: 418 FLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTS 477
F+ + K DV G + IKMMPFGAGRRICP AMLHLEYFV NLV FEWK
Sbjct: 406 FIGEDKE------VDVTGSRGIKMMPFGAGRRICPGIGSAMLHLEYFVVNLVKEFEWK-E 458
Query: 478 LPGSNVDLTEKQEFTMVMKYPLEA 501
+ G VDL+EK EFT+VMKYPL+A
Sbjct: 459 VEGYEVDLSEKWEFTVVMKYPLKA 482
>At3g03470 cytochrome P450 nlike protein
Length = 511
Score = 448 bits (1153), Expect = e-126
Identities = 247/513 (48%), Positives = 341/513 (66%), Gaps = 29/513 (5%)
Query: 8 IITALLVLLLCLIILRLRSSQSIH-LPPGPLYLPIITDI-FLLQISFSKLKPIIRKLHAK 65
II ++ L I L+L S H LPPGP P+I +I +L + +FS + ++R L ++
Sbjct: 6 IIFLIISSLTFSIFLKLIFFFSTHKLPPGPPRFPVIGNIIWLKKNNFSDFQGVLRDLASR 65
Query: 66 HGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCYG 119
HGPIIT H S P I+++DR LAHQAL+QN VF+DR K + ++Q +I SS YG
Sbjct: 66 HGPIITLHVGSKPSIWVTDRSLAHQALVQNGAVFSDRSLALPTTKVITSNQHDIHSSVYG 125
Query: 120 PTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIF 179
W +R NL S +LQPSR K+ + +RK L+ L+ + E + + ++++RHA+F
Sbjct: 126 SLWRTLRRNLTSEILQPSRVKAHAPSRKWSLEILVDLFETEQREKGHISDALDHLRHAMF 185
Query: 180 CLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLR 239
LL MCFGE + +I +I++ Q ++++S + LN FP V++ L R++W+EF +LR
Sbjct: 186 YLLALMCFGEKLRKEEIREIEEAQYQMLISYTKFSVLNIFPS-VTKFLLRRKWKEFLELR 244
Query: 240 KLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEE------KRKLNEGEVVA 293
K Q +++ + AR K E D V+CYVDTLLNL +P E KRKL++ E+V+
Sbjct: 245 KSQESVILRYVNARSK--ETTGD----VLCYVDTLLNLEIPTEEKEGGKKRKLSDSEIVS 298
Query: 294 LCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEV-VGEREERE-VREEDLEKL 351
LCSEFL A TD T+T+++W+MA +VK +Q+ + EE++ V GE EERE +REEDL KL
Sbjct: 299 LCSEFLNAATDPTATSMQWIMAIMVKYPEIQRKVYEEMKTVFAGEEEEREEIREEDLGKL 358
Query: 352 PYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPT 411
YLKAVILE LRRHPP HY VT D VL G+L+P+ GT+NFMV ++G DP +WEDP
Sbjct: 359 SYLKAVILECLRRHPPGHYLSYHKVTHDTVLGGFLIPRQGTINFMVGEMGRDPKIWEDPL 418
Query: 412 VFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLN 471
FKPERFL++ E FD+ G +EIKMMPFGAGRR+CP Y L++LHLEY+VANLV
Sbjct: 419 TFKPERFLEN----GEACDFDMTGTREIKMMPFGAGRRMCPGYALSLLHLEYYVANLVWK 474
Query: 472 FEWKTSLPGSNVDLTEKQEF-TMVMKYPLEAQI 503
FEWK + G VDL+EKQ+F TMVMK P +A I
Sbjct: 475 FEWK-CVEGEEVDLSEKQQFITMVMKNPFKANI 506
>At3g10570 putative cytochrome P450
Length = 513
Score = 321 bits (822), Expect = 7e-88
Identities = 179/504 (35%), Positives = 277/504 (54%), Gaps = 21/504 (4%)
Query: 12 LLVLLLCLIILRLRSSQS--IHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAKHGPI 69
+L+ L L+IL RS++S + LPPGP P++ ++F S + + + K+GPI
Sbjct: 21 ILISSLVLLILTRRSAKSKIVKLPPGPPGWPVVGNLFQFARSGKQFYEYVDDVRKKYGPI 80
Query: 70 ITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPK------FLKNHQFNIASSCYGPTWH 123
T S I ISD L H LIQ +FA RP ++ F + +S YGP W
Sbjct: 81 YTLRMGSRTMIIISDSALVHDVLIQRGPMFATRPTENPTRTIFSSNTFTVNASAYGPVWR 140
Query: 124 AIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIFCLLF 183
++R N+ ML RF+ F R+ +D L+ ++K EA + V ++ R A FC+L
Sbjct: 141 SLRKNMVQNMLSSIRFREFGSLRQSAMDKLVERIKSEAKDNDGLVWVLRNARFAAFCILL 200
Query: 184 SMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLRKLQN 243
MCFG +D I + + ++++++ L+ + I++ ++R + ++R Q
Sbjct: 201 EMCFGIEMDEESILNMDQVMKKVLITLNP--RLDDYLPILAPFYSKERARAL-EVRCEQV 257
Query: 244 DLLVELIQARKKVIEK-NNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALCSEFLTAG 302
D +V+LI+ R++ I+K D Y+DTL +L+ + E+V+LCSEFL G
Sbjct: 258 DFIVKLIERRRRAIQKPGTDKTASSFSYLDTLFDLKTEGRITTPSNEELVSLCSEFLNGG 317
Query: 303 TDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAVILEGL 362
TDTT T +EW +A L+ + +Q L +EI+ VG+RE V E+D++K+ +L+AV+ E L
Sbjct: 318 TDTTGTAIEWGIAQLIVNPEIQSRLYDEIKSTVGDRE---VEEKDVDKMVFLQAVVKEIL 374
Query: 363 RRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPERFLKDA 422
R+HPPT++T+ VTE + GY VP V F + I DP +W DP F P+RF+
Sbjct: 375 RKHPPTYFTLTHSVTEPTTVAGYDVPVGINVEFYLPGINEDPKLWSDPKKFNPDRFISGK 434
Query: 423 KNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTSLPGSN 482
+ D+ G+ +KMMPFG GRRICP +A +H+ +A +V FEW P S
Sbjct: 435 EEA------DITGVTGVKMMPFGIGRRICPGLAMATVHVHLMLAKMVQEFEWSAYPPESE 488
Query: 483 VDLTEKQEFTMVMKYPLEAQISLR 506
+D K EFT+VMK PL A + R
Sbjct: 489 IDFAGKLEFTVVMKKPLRAMVKPR 512
>At5g04630 cytochrome P450 - like protein
Length = 509
Score = 316 bits (810), Expect = 2e-86
Identities = 179/511 (35%), Positives = 274/511 (53%), Gaps = 21/511 (4%)
Query: 6 WFIITALLVLLLCLIILRLRSSQS---IHLPPGPLYLPIITDIFLLQISFSKLKPIIRKL 62
+F T + +LL CL+ + R S + +LPPGP P++ ++ S + + ++
Sbjct: 9 FFFFTLVTILLSCLVYILTRHSHNPKCANLPPGPKGWPVVGNLLQFARSGKQFFEYVDEM 68
Query: 63 HAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASS 116
+GPI T I ISD LAHQALI+ FA RP K + + S+
Sbjct: 69 RNIYGPIFTLKMGIRTMIIISDANLAHQALIERGAQFATRPAETPTRKIFSSSDITVHSA 128
Query: 117 CYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRH 176
YGP W ++R N+ ML +R K F RK +D L+ K+K EA + V ++ R
Sbjct: 129 MYGPVWRSLRRNMVQNMLCSNRLKEFGSIRKSAIDKLVEKIKSEAKENDGLVWVLRNARF 188
Query: 177 AIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFT 236
A FC+L MCFG ++ IEK+ + ++ + ++ + I++ F++R
Sbjct: 189 AAFCILLDMCFGVKMEEESIEKMDQMMTEILTAVDP--RIHDYLPILTPFYFKERKNSLE 246
Query: 237 QLRKLQNDLLVELIQARKKVIEK-NNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALC 295
RKL +V I+ R+ I +D Y+DTL +LR+ + ++ ++V LC
Sbjct: 247 LRRKLVQ-FVVGFIEKRRLAIRNLGSDKTASSFAYLDTLFDLRVDGRETSPSDEDLVTLC 305
Query: 296 SEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLK 355
SEFL AGTDTT T +EW +A L+ + +Q L +EI+ VG+ +R V E+DL K+ +L+
Sbjct: 306 SEFLNAGTDTTGTAIEWGIAELISNPKIQSRLYDEIKSTVGD--DRTVEEKDLNKMVFLQ 363
Query: 356 AVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKP 415
A + E LRRHPPT++T+ VTE L GY +P V F + I DP +W P F P
Sbjct: 364 AFVKELLRRHPPTYFTLTHGVTEPTNLAGYDIPVGANVEFYLPGISEDPKIWSKPEKFDP 423
Query: 416 ERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWK 475
+RF+ ++ D+ G+ +KMMPFG GRRICP +A++H+E ++ +V FEW
Sbjct: 424 DRFITGGEDA------DLTGVAGVKMMPFGIGRRICPGLGMAVVHVELMLSRMVQEFEWS 477
Query: 476 TSLPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
+ P S VD T K F +VMK PL A++ R
Sbjct: 478 SYPPESQVDFTGKLVFAVVMKNPLRARVKAR 508
>At5g04660 cytochrom P450 - like protein
Length = 512
Score = 312 bits (800), Expect = 2e-85
Identities = 173/509 (33%), Positives = 274/509 (52%), Gaps = 20/509 (3%)
Query: 7 FIITALLVLLLCLIILRLRSSQS--IHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHA 64
F A+++ II R S+ ++LPPGP P++ ++F S L
Sbjct: 14 FTFFAIIISGFVFIITRWNSNSKKRLNLPPGPPGWPVVGNLFQFARSGKPFFEYAEDLKK 73
Query: 65 KHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPK------FLKNHQFNIASSCY 118
+GPI T + I +SD L H+ALIQ +FA RP ++F + ++ Y
Sbjct: 74 TYGPIFTLRMGTRTMIILSDATLVHEALIQRGALFASRPAENPTRTIFSCNKFTVNAAKY 133
Query: 119 GPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAI 178
GP W ++R N+ ML +R K F K R+ +D LI ++K EA + + ++ R A
Sbjct: 134 GPVWRSLRRNMVQNMLSSTRLKEFGKLRQSAMDKLIERIKSEARDNDGLIWVLKNARFAA 193
Query: 179 FCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQL 238
FC+L MCFG +D IEK+ +I + ++++ ++ P + F K + ++
Sbjct: 194 FCILLEMCFGIEMDEETIEKMDEILKTVLMTVDPR-IDDYLPILAP--FFSKERKRALEV 250
Query: 239 RKLQNDLLVELIQARKKVIEK-NNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALCSE 297
R+ Q D +V +I+ R++ I+ +D Y+DTL +L++ K + E+V LCSE
Sbjct: 251 RREQVDYVVGVIERRRRAIQNPGSDKTASSFSYLDTLFDLKIEGRKTTPSNEELVTLCSE 310
Query: 298 FLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAV 357
FL GTDTT T +EW +A L+ + +Q L +EI+ VG+ +R V E+D++K+ +L+A
Sbjct: 311 FLNGGTDTTGTAIEWGIAQLIANPEIQSRLYDEIKSTVGD--DRRVDEKDVDKMVFLQAF 368
Query: 358 ILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPER 417
+ E LR+HPPT++++ V E L GY +P V + I DP +W +P F P+R
Sbjct: 369 VKELLRKHPPTYFSLTHAVMETTTLAGYDIPAGVNVEVYLPGISEDPRIWNNPKKFDPDR 428
Query: 418 FLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTS 477
F+ ++ D+ GI +KM+PFG GRRICP +A +H+ +A +V FEW
Sbjct: 429 FMLGKEDA------DITGISGVKMIPFGVGRRICPGLAMATIHVHLMLARMVQEFEWCAH 482
Query: 478 LPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
PGS +D K EFT+VMK PL A + R
Sbjct: 483 PPGSEIDFAGKLEFTVVMKNPLRAMVKPR 511
>At3g10560 putative cytochrome P450
Length = 514
Score = 305 bits (781), Expect = 4e-83
Identities = 176/506 (34%), Positives = 270/506 (52%), Gaps = 22/506 (4%)
Query: 8 IITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAKHG 67
I+ ++ LL I R +S ++LPPGP P+I ++F S + + L +G
Sbjct: 22 ILAIIISGLLKTITYRKHNSNHLNLPPGPPGWPVIGNLFQFTRSGKQFFEYVEDLVKIYG 81
Query: 68 PIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCYGPT 121
PI+T + I ISD LAH+ALI+ FA RP K + + + S+ YGP
Sbjct: 82 PILTLRLGTRTMIIISDASLAHEALIERGAQFATRPVETPTRKIFSSSEITVHSAMYGPV 141
Query: 122 WHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIFCL 181
W ++R N+ ML +R K F RK +D LI ++K EA + V ++ R+A FC+
Sbjct: 142 WRSLRRNMVQNMLSSNRLKEFGSVRKSAMDKLIERIKSEARDNDGLVWVLQNSRYAAFCV 201
Query: 182 LFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLRKL 241
L +CFG ++ IEK+ + ++ + L+ + I++ + +R +LR+
Sbjct: 202 LLDICFGVEMEEESIEKMDQLMTAILNAVDP--KLHDYLPILTPFNYNER-NRALKLRRE 258
Query: 242 QNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEEKRKL-NEGEVVALCSEFLT 300
D +V I+ R+K I Y+DTL +LR+ E ++ ++V LCSEFL
Sbjct: 259 LVDFVVGFIEKRRKAIRTATVSS---FSYLDTLFDLRIIEGSETTPSDEDLVTLCSEFLN 315
Query: 301 AGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAVILE 360
AGTDTT +EW +A L+ + +Q L +EI+ VG+R V E D++K+ L+AV+ E
Sbjct: 316 AGTDTTGAAIEWGIAELIANPEIQSRLYDEIKSTVGDRA---VDERDVDKMVLLQAVVKE 372
Query: 361 GLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPERFLK 420
LRRHPPT++T+ VTE L+GY +P + F + I DP +W +P F P+RFL
Sbjct: 373 ILRRHPPTYFTLSHGVTEPTTLSGYNIPVGVNIEFYLPGISEDPKIWSEPKKFDPDRFLS 432
Query: 421 DAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTSLPG 480
++ D+ G+ +KMMPFG GRRICP +A +H+ +A +V FEW P
Sbjct: 433 GREDA------DITGVAGVKMMPFGVGRRICPGMGMATVHVHLMIARMVQEFEWLAYPPQ 486
Query: 481 SNVDLTEKQEFTMVMKYPLEAQISLR 506
S +D K F +VMK PL A + R
Sbjct: 487 SEMDFAGKLVFAVVMKKPLRAMVRPR 512
>At1g11600 cytochrome P450 like protein
Length = 510
Score = 295 bits (756), Expect = 3e-80
Identities = 185/520 (35%), Positives = 285/520 (54%), Gaps = 35/520 (6%)
Query: 9 ITALLVLLLCLIILRL---------RSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPI- 58
+T +++ L L + L S S+++PPGP P++ + LLQ+ F + +
Sbjct: 3 LTDVIIFLFALYFINLWWRRYFSAGSSQCSLNIPPGPKGWPLVGN--LLQVIFQRRHFVF 60
Query: 59 -IRKLHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPK------FLKNHQF 111
+R L K+GPI T I I+D L H+AL+Q FA RP +
Sbjct: 61 LMRDLRKKYGPIFTMQMGQRTMIIITDEKLIHEALVQRGPTFASRPPDSPIRLMFSVGKC 120
Query: 112 NIASSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELI 171
I S+ YG W +R N + ++ R K S R + + ++K E N VE++
Sbjct: 121 AINSAEYGSLWRTLRRNFVTELVTAPRVKQCSWIRSWAMQNHMKRIKTE-NVEKGFVEVM 179
Query: 172 NYIRHAIFCLLFSMCFGENVDSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKR 231
+ R I +L +CFG + KI+ I+++ + ++L + +F P V LFR++
Sbjct: 180 SQCRLTICSILICLCFGAKISEEKIKNIENVLKDVMLITSPT-LPDFLP--VFTPLFRRQ 236
Query: 232 WQEFTQLRKLQNDLLVELIQARKKVIE-KNNDDDEYV----VCYVDTLLNLRLPEEKRKL 286
+E +LRK Q + LV LI+ R+K ++ K N ++E V YVD+L L L E +L
Sbjct: 237 VREARELRKTQLECLVPLIRNRRKFVDAKENPNEEMVSPIGAAYVDSLFRLNLIERGGEL 296
Query: 287 NEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREE 346
+ E+V LCSE ++AGTDT++TTLEW + +LV D+++Q+ L EE+ VVG+ V E+
Sbjct: 297 GDEEIVTLCSEIVSAGTDTSATTLEWALFHLVTDQNIQEKLYEEVVGVVGKNGV--VEED 354
Query: 347 DLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTV 406
D+ K+PYL+A++ E LRRHPP H+ + +D L GY +P V A + +P +
Sbjct: 355 DVAKMPYLEAIVKETLRRHPPGHFLLSHAAVKDTELGGYDIPAGAYVEIYTAWVTENPDI 414
Query: 407 WEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVA 466
W DP F+PERFL + DA D G + + M+PFGAGRRICPA+ L +LH+ +A
Sbjct: 415 WSDPGKFRPERFLTGG---DGVDA-DWTGTRGVTMLPFGAGRRICPAWSLGILHINLMLA 470
Query: 467 NLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLEAQISLR 506
++ +F+W +P S D TE FT+VMK L+AQI R
Sbjct: 471 RMIHSFKW-IPVPDSPPDPTETYAFTVVMKNSLKAQIRSR 509
>At3g26230 cytochrome P450, putative
Length = 498
Score = 197 bits (502), Expect = 9e-51
Identities = 146/510 (28%), Positives = 248/510 (48%), Gaps = 39/510 (7%)
Query: 9 ITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAKHGP 68
I + LL LII++ + +LPP PL LP+I +++ L+ F K + L KHGP
Sbjct: 3 ILLYFIALLSLIIIKKIKDSNRNLPPSPLKLPVIGNLYQLRGLFHKC---LHDLSKKHGP 59
Query: 69 IITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP------KFLKNHQFNIASSCYGPTW 122
++ + IS A +AL + + RP K ++ Q +I + YG +
Sbjct: 60 VLLLRLGFLDMVVISSTEAAEEALKVHDLECCTRPITNVTSKLWRDGQ-DIGLAPYGESL 118
Query: 123 HAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIFCLL 182
+R ++ +SF R+ D ++ KLK EA +SV+L + + ++
Sbjct: 119 RELRKLSFLKFFSTTKVRSFRYIREEENDLMVKKLK-EAALKKSSVDLSQTLFGLVGSII 177
Query: 183 FSMCFGENVDSRK---IEKIKD----IQRRLILSSGQL--GALNFFPKIVSRVLFRKRWQ 233
F FG+ D EKI+D +Q+ LS+ L G L +F VS +K +
Sbjct: 178 FRSAFGQRFDEGNHVNAEKIEDLMFEVQKLGALSNSDLFPGGLGWFVDFVSGHN-KKLHK 236
Query: 234 EFTQLRKLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEEKR---KLNEGE 290
F ++ L N ++ + ++ + I + D +D+LL++ +E+ KL
Sbjct: 237 VFVEVDTLLNHIIDDHLKNSIEEITHDRPD------IIDSLLDMIRKQEQGDSFKLTIDN 290
Query: 291 VVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEK 350
+ + + AG DT++ T+ W MA LVK+ V + + +EI +G ++ ++ E+D++K
Sbjct: 291 LKGIIQDIYLAGVDTSAITMIWAMAELVKNPRVMKKVQDEIRTCIGIKQNEKIEEDDVDK 350
Query: 351 LPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDP 410
L YLK V+ E LR HP +PR I + GY +P + V IG DP W++P
Sbjct: 351 LQYLKLVVKETLRLHPAAPLLLPRETMSQIKIQGYNIPSKTILLVNVWSIGRDPKHWKNP 410
Query: 411 TVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVL 470
F PERF+ D +G +M+PFG+GRRICP A+ +E + NL+
Sbjct: 411 EEFNPERFID--------CPIDYKG-NSFEMLPFGSGRRICPGIAFAIATVELGLLNLLY 461
Query: 471 NFEWKTSLPGSNVDLTEKQEFTMVMKYPLE 500
+F+W+ ++D+ E + T++ K PL+
Sbjct: 462 HFDWRLPEEDKDLDMEEAGDVTIIKKVPLK 491
>At3g26220 cytochrome P450 monooxygenase (CYP71B3)
Length = 501
Score = 197 bits (501), Expect = 1e-50
Identities = 143/505 (28%), Positives = 237/505 (46%), Gaps = 37/505 (7%)
Query: 13 LVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAKHGPIITF 72
L ++L LI ++ +LPP P LPII ++ L+ F + + L KHGP++
Sbjct: 10 LPVILSLIFMKKFKDSKRNLPPSPPKLPIIGNLHQLRGLFHRC---LHDLSKKHGPVLLL 66
Query: 73 HFWSTPHIFISDRLLAHQALIQNAVVFADRPKFLKNHQFN-----IASSCYGPTWHAIRN 127
+ IS + A + L + + RPK + +F+ IA + YG +R
Sbjct: 67 RLGFIDMVVISSKEAAEEVLKVHDLECCTRPKTNASSKFSRDGKDIAFAPYGEVSRELRK 126
Query: 128 NLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIFCLLFSMCF 187
+ +SF R+ D ++ KLK A N+V+L + + + ++F F
Sbjct: 127 LSLINFFSTQKVRSFRYIREEENDLMVKKLKESAK-KKNTVDLSQTLFYLVGSIIFRATF 185
Query: 188 GENVDSRKI---EKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLR-KLQN 243
G+ +D K EKI+++ + G L + + FP V + +F R K +
Sbjct: 186 GQRLDQNKHVNKEKIEELMFE-VQKVGSLSSSDIFPAGVGWFM------DFVSGRHKTLH 238
Query: 244 DLLVELIQARKKVIE--------KNNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALC 295
+ VE+ VI+ K N D ++ + + + +E KL + +
Sbjct: 239 KVFVEVDTLLNHVIDGHLKNPEDKTNQDRPDIIDSILETIYKQEQDESFKLTIDHLKGII 298
Query: 296 SEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLK 355
AG DT++ T+ W MA LVK+ V + EEI +G +++ + EED++KL YLK
Sbjct: 299 QNIYLAGVDTSAITMIWAMAELVKNPRVMKKAQEEIRTCIGIKQKERIEEEDVDKLQYLK 358
Query: 356 AVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKP 415
VI E LR HPP +PR DI + GY +P+ + IG +P +WE+P F P
Sbjct: 359 LVIKETLRLHPPAPLLLPRETMADIKIQGYDIPRKTILLVNAWSIGRNPELWENPEEFNP 418
Query: 416 ERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWK 475
ERF+ D +G +M+PFG+GR+ICP + +E + NL+ F+W+
Sbjct: 419 ERFID--------CPMDYKG-NSFEMLPFGSGRKICPGIAFGIATVELGLLNLLYYFDWR 469
Query: 476 TSLPGSNVDLTEKQEFTMVMKYPLE 500
+ ++D+ E + T+V K PLE
Sbjct: 470 LAEEDKDIDMEEAGDATIVKKVPLE 494
>At3g52970 cytochrome P450 - like protein
Length = 516
Score = 192 bits (489), Expect = 3e-49
Identities = 146/516 (28%), Positives = 252/516 (48%), Gaps = 32/516 (6%)
Query: 2 EAMAWFIITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRK 61
E + F A+L+ + CL + ++ LPPGP P+I +IF Q++ + K
Sbjct: 9 ELIGLFTSIAVLIYVTCLFYTKRCRTR---LPPGPNPWPVIGNIF--QLAGLPPHDSLTK 63
Query: 62 LHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRP-----KFLKNHQFNIASS 116
L +HGPI+T S + IS +A + ++ A R K K+ ++ ++
Sbjct: 64 LSRRHGPIMTLRIGSMLTVVISSSEVAREIFKKHDAALAGRKIYEAMKGGKSSDGSLITA 123
Query: 117 CYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRH 176
YG W +R + R + S R +D ++ ++ ++++ Y
Sbjct: 124 QYGAYWRMLRRLCTTQFFVTRRLDAMSDVRSRCVDQMLRFVEEGGQNGTKTIDVGRYFFL 183
Query: 177 AIFCLLFSMCFGENV---DSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQ 233
F L+ ++ F ++ DS++ + +++ +G+ +FFP + R L + +
Sbjct: 184 MAFNLIGNLMFSRDLLDPDSKRGSEFFYHTGKVMEFAGKPNVADFFPLL--RFLDPQGIR 241
Query: 234 EFTQLR-KLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLR----LPEEKRKLNE 288
TQ + ++ E I+ R +V E+ D++ Y+D LL R + EE +
Sbjct: 242 RKTQFHVEKAFEIAGEFIRERTEVREREKSDEK-TKDYLDVLLEFRGGDGVDEEPSSFSS 300
Query: 289 GEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDL 348
++ + E TAGTDTT++TLEW +A L+ + L E+ + ++++EEDL
Sbjct: 301 RDINVIVFEMFTAGTDTTTSTLEWALAELLHNPRTLTKLQTELRTYF-KSSNQKLQEEDL 359
Query: 349 EKLPYLKAVILEGLRRHPPTHYTIP-RVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVW 407
LPYL AVI+E LR HPP + +P + ++ + + Y +PK V V IG DP W
Sbjct: 360 PNLPYLSAVIMETLRLHPPLPFLVPHKAMSTCHIFDQYTIPKETQVLVNVWAIGRDPKTW 419
Query: 408 EDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVAN 467
DP +FKPERF+ D DA D +G ++ + +PFG+GRR+CPA LA L + +
Sbjct: 420 IDPIMFKPERFISDP------DARDFKG-QDYEFLPFGSGRRMCPALPLASRVLPLAIGS 472
Query: 468 LVLNFEW--KTSLPGSNVDLTEKQEFTMVMKYPLEA 501
+V +F+W + L +D+ E+ T+ PLEA
Sbjct: 473 MVRSFDWALENGLNAEEMDMGERIGITLKKAVPLEA 508
>At3g26210 cytochrome P450 like protein
Length = 501
Score = 192 bits (487), Expect = 5e-49
Identities = 148/515 (28%), Positives = 249/515 (47%), Gaps = 39/515 (7%)
Query: 4 MAWFIITALLVLLLCLIILRLRSSQS--IHLPPGPLYLPIITDIFLLQISFSKLKPIIRK 61
M+ F+ LL+LLL + I+ R SQS + LPPGP LPII ++ L K + K
Sbjct: 1 MSIFLCFLLLLLLLLVTIIFTRKSQSSKLKLPPGPPKLPIIGNLHYLNGLPHKCLLNLWK 60
Query: 62 LHAKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPKFL--KNHQFN---IASS 116
+H GP++ P + IS A + L + + RP+ + K +N I +
Sbjct: 61 IH---GPVMQLQLGYVPLVVISSNQAAEEVLKTHDLDCCSRPETIASKTISYNFKDIGFA 117
Query: 117 CYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRH 176
YG W A+R + +F SF R+ D L+ KL EA+ + V L +
Sbjct: 118 PYGEEWRALRKLAVIELFSLKKFNSFRYIREEENDLLVKKLS-EASEKQSPVNLKKALFT 176
Query: 177 AIFCLLFSMCFGENVDSRKI---EKIKDIQRRLILSSGQLGALNFFPK--IVSRVLFRKR 231
++ + FG+N+ + + ++D+ R + NFFP I+ ++ + +
Sbjct: 177 LSASIVCRLAFGQNLHESEFIDEDSMEDLASRSEKIQAKFAFSNFFPGGWILDKITGQSK 236
Query: 232 W--QEFTQLRKLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEEKRKLNEG 289
+ F L N +L + ++ +KV+E + VV + ++N + + KL
Sbjct: 237 SLNEIFADLDGFFNQVLDDHLKPGRKVLETPD-----VVDVMIDMMNKQSQDGSFKLTTD 291
Query: 290 EVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLE 349
+ + S+ AG +T++TT+ W M L+++ V + + +E+ V+GE+ +R + E+DL
Sbjct: 292 HIKGIISDIFLAGVNTSATTILWAMTELIRNPRVMKKVQDEVRTVLGEKRDR-ITEQDLN 350
Query: 350 KLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWED 409
+L Y K VI E R HP +PR I + GY +P+ + V IG DP +WE+
Sbjct: 351 QLNYFKLVIKETFRLHPAAPLLLPREAMAKIKIQGYDIPEKTQIMVNVYAIGRDPDLWEN 410
Query: 410 PTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLV 469
P FKPERF+ + D RG+ +++PFG+GRRICP + + +E + NL+
Sbjct: 411 PEEFKPERFVD--------SSVDYRGL-NFELLPFGSGRRICPGMTMGIATVELGLLNLL 461
Query: 470 LNFEWKTSLPG----SNVDLTEKQEFTMVMKYPLE 500
F+W LP ++DL E+ + K LE
Sbjct: 462 YFFDW--GLPEGRTVKDIDLEEEGAIIIGKKVSLE 494
>At3g26200 cytochrome P450 like protein
Length = 500
Score = 190 bits (483), Expect = 1e-48
Identities = 142/506 (28%), Positives = 233/506 (45%), Gaps = 38/506 (7%)
Query: 12 LLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAKHGPIIT 71
LL+L L LI + S LPPGPL LPII ++ L S + KL +GP++
Sbjct: 8 LLLLPLFLIFFKKLSPSKGKLPPGPLGLPIIGNLHQLGKSLHRS---FHKLSQNYGPVMF 64
Query: 72 FHFWSTPHIFISDRLLAHQALIQNAVVFADRPKFLKNHQFN-----IASSCYGPTWHAIR 126
HF P + +S R A + L + + RPK F+ I + YG W +R
Sbjct: 65 LHFGVVPVVVVSTREAAEEVLKTHDLETCTRPKLTATKLFSYNYKDIGFAQYGDDWREMR 124
Query: 127 NNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAIFCLLFSMC 186
+ + K+F R+ + L++KL A V+L + ++ +
Sbjct: 125 KLAMLELFSSKKLKAFRYIREEESEVLVNKLSKSAE-TRTMVDLRKALFSYTASIVCRLA 183
Query: 187 FGENV---DSRKIEKIKDIQRRLILSSGQLGALNFFPKIVSRVLFRKRWQEFTQLRKLQN 243
FG+N D ++K++D+ + G +FFP + V+ R Q ++L K
Sbjct: 184 FGQNFHECDFVDMDKVEDLVLESETNLGSFAFTDFFPAGLGWVIDRISGQH-SELHKA-- 240
Query: 244 DLLVELIQARKKVIEKN-----NDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALCSEF 298
L + VI+ + + D ++ + ++N ++ + + S+
Sbjct: 241 --FARLSNFFQHVIDDHLKPGQSQDHSDIIGVMLDMINKESKVGSFQVTYDHLKGVMSDV 298
Query: 299 LTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAVI 358
AG + + T+ W M L + V + L +EI E++G+ +E+ + E+DLEK+ YLK VI
Sbjct: 299 FLAGVNAGAITMIWAMTELARHPRVMKKLQQEIREILGDNKEK-ITEQDLEKVHYLKLVI 357
Query: 359 LEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPERF 418
E R HPP +PR D+ + GY +PKN + IG DP WE+P F PERF
Sbjct: 358 EETFRLHPPAPLLLPRETMSDLKIQGYNIPKNTMIEINTYSIGRDPNCWENPNDFNPERF 417
Query: 419 LKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTSL 478
+ + +G + +++PFGAGRRICP + +E + N++ F+W SL
Sbjct: 418 ID--------SPVEYKG-QHYELLPFGAGRRICPGMATGITIVELGLLNVLYFFDW--SL 466
Query: 479 PG----SNVDLTEKQEFTMVMKYPLE 500
P ++D+ E F + K PLE
Sbjct: 467 PDGMKIEDIDMEEAGAFVVAKKVPLE 492
>At3g53300 CYTOCHROME P450-like protein
Length = 498
Score = 190 bits (482), Expect = 2e-48
Identities = 139/487 (28%), Positives = 224/487 (45%), Gaps = 40/487 (8%)
Query: 32 LPPGPLYLPIITDIFLLQISFSKLKPIIRKLHAKHGPIITFHFWSTPHIFISDRLLAHQA 91
LPPGP LP+I ++ L L + KL +HGP++ + P S A +
Sbjct: 28 LPPGPTGLPLIGNLHQLG---RLLHSSLHKLSLEHGPVMLVRWGVVPMAVFSSNEAAKEV 84
Query: 92 LIQNAVVFADRPKFLKNHQF-----NIASSCYGPTWHAIRNNLASIMLQPSRFKSFSKAR 146
L + + +RPK + N F +I + YG W ++ + + P + KSF R
Sbjct: 85 LKTHDLETCNRPKLVANGLFTHGYKDIGFTQYGEEWREMKKFVGLELFSPKKHKSFRYIR 144
Query: 147 KCVLDALISKLKFEANFLNNSVELINYIRHAIFCLLFSMCFGENV---DSRKIEKIKDIQ 203
+ D L+ K+ A V+L + ++F FG+N D ++K++++
Sbjct: 145 EEEGDLLVKKISNYAQ-TQTLVDLRKSLFSYTASIIFREAFGQNFRECDYINMDKLEELV 203
Query: 204 RRLILSSGQLGALNFFPKIVSRVLFRKRWQE------FTQLRKLQNDLLVELIQARKKVI 257
+ + L +FFP+ + ++ R Q F++L D++ EL++
Sbjct: 204 QETETNVCSLAFTDFFPRGLGWLVDRISGQHSRMNIAFSKLTTFFEDVIDELLKT----- 258
Query: 258 EKNNDDDEYVVCYVDTLLNLRLPEEKRKLNEGEVVALCSEFLTAGTDTTSTTLEWVMANL 317
K DD +V + ++N K+ ++A+ S+ + AG + + T+ W M L
Sbjct: 259 -KQLDDHSDLVTAMLDVINRPRKFGSLKITYDHLIAMMSDVVLAGVNAGTVTMIWTMTEL 317
Query: 318 VKDKHVQQMLVEEIEEVVGEREEREVREEDLEKLPYLKAVILEGLRRHPPTHYTIPRVVT 377
+ V + L EEI +G +ER + EEDLEK+ YL VI E R HPP +PR
Sbjct: 318 TRHPRVMKKLQEEIRATLGPNKER-ITEEDLEKVEYLNLVIKESFRLHPPAPLLLPRETM 376
Query: 378 EDIVLNGYLVPKNGTVNFMVADIGLDPTVWEDPTVFKPERFLKDAKNRNEFDAFDVRGIK 437
DI + GY +PKN V IG DP W +P F PERFL + N +
Sbjct: 377 SDIEIQGYHIPKNAHVKINTYAIGRDPKRWTNPEEFNPERFLNTSINYKG---------Q 427
Query: 438 EIKMMPFGAGRRICPAYKLAMLHLEYFVANLVLNFEWKTSLPG----SNVDLTEKQEFTM 493
+++PFGAGRR CP L + LE + N++ F+W SLP ++D+ E +
Sbjct: 428 HYELLPFGAGRRNCPGMTLGITILELGLLNILYYFDW--SLPSGMTIKDIDMEEDGALNI 485
Query: 494 VMKYPLE 500
K PL+
Sbjct: 486 AKKVPLQ 492
>At2g40890 cytochrome P450 like protein
Length = 508
Score = 188 bits (477), Expect = 7e-48
Identities = 136/517 (26%), Positives = 259/517 (49%), Gaps = 46/517 (8%)
Query: 4 MAWFIITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
M+WF+I + + ++ +L PPGP PI+ +++ +KP+ + +
Sbjct: 1 MSWFLIA--VATIAAVVSYKLIQRLRYKFPPGPSPKPIVGNLY-------DIKPVRFRCY 51
Query: 64 AK----HGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPK------FLKNHQFNI 113
+ +GPII+ S ++ +S LA + L ++ ADR + F +N Q ++
Sbjct: 52 YEWAQSYGPIISVWIGSILNVVVSSAELAKEVLKEHDQKLADRHRNRSTEAFSRNGQ-DL 110
Query: 114 ASSCYGPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNN---SVEL 170
+ YGP + +R + P R +S R+ + A++ + + N N ++L
Sbjct: 111 IWADYGPHYVKVRKVCTLELFTPKRLESLRPIREDEVTAMVESVFRDCNLPENRAKGLQL 170
Query: 171 INYIRHAIFCLLFSMCFGEN-VDSRKIEKIKDIQRRLILSSG-QLGALNFFPKIVS--RV 226
Y+ F + + FG+ +++ + + ++ + I+S+G +LGA + + R
Sbjct: 171 RKYLGAVAFNNITRLAFGKRFMNAEGVVDEQGLEFKAIVSNGLKLGASLSIAEHIPWLRW 230
Query: 227 LFRKRWQEFTQLRKLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEEKRKL 286
+F + F + ++ L +++ +K++ ++ +VD LL L+ ++ L
Sbjct: 231 MFPADEKAFAEHGARRDRLTRAIMEEHTLARQKSSGAKQH---FVDALLTLK---DQYDL 284
Query: 287 NEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVREE 346
+E ++ L + +TAG DTT+ T EW MA ++K+ VQQ + EE + VVG +R + E
Sbjct: 285 SEDTIIGLLWDMITAGMDTTAITAEWAMAEMIKNPRVQQKVQEEFDRVVGL--DRILTEA 342
Query: 347 DLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPTV 406
D +LPYL+ V+ E R HPPT +P D+ + GY +PK V+ V + DP V
Sbjct: 343 DFSRLPYLQCVVKESFRLHPPTPLMLPHRSNADVKIGGYDIPKGSNVHVNVWAVARDPAV 402
Query: 407 WEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFVA 466
W++P F+PERFL+ + D++G + +++PFGAGRR+CP +L + + ++
Sbjct: 403 WKNPFEFRPERFLE--------EDVDMKG-HDFRLLPFGAGRRVCPGAQLGINLVTSMMS 453
Query: 467 NLVLNFEW--KTSLPGSNVDLTEKQEFTMVMKYPLEA 501
+L+ +F W +D++E M+ P++A
Sbjct: 454 HLLHHFVWTPPQGTKPEEIDMSENPGLVTYMRTPVQA 490
>At3g26280 cytochrome P450 monooxygenase (CYP71B4)
Length = 504
Score = 186 bits (471), Expect = 3e-47
Identities = 142/515 (27%), Positives = 239/515 (45%), Gaps = 42/515 (8%)
Query: 4 MAWFIITALLVLLLCLIILRLRSSQSIHLPPGPLYLPIITDIFLLQISFSKLKPIIRKLH 63
+++F++ + + L + +++ S+ +LPPGP LPII ++ LQ L + L
Sbjct: 5 LSFFLLLLVPIFFLLIFTKKIKESKQ-NLPPGPAKLPIIGNLHQLQ---GLLHKCLHDLS 60
Query: 64 AKHGPIITFHFWSTPHIFISDRLLAHQALIQNAVVFADRPKFLKNHQFN-----IASSCY 118
KHGP++ P + IS A +AL + + RP + + F+ I Y
Sbjct: 61 KKHGPVMHLRLGFAPMVVISSSEAAEEALKTHDLECCSRPITMASRVFSRNGKDIGFGVY 120
Query: 119 GPTWHAIRNNLASIMLQPSRFKSFSKARKCVLDALISKLKFEANFLNNSVELINYIRHAI 178
G W +R + +SF R+ D +I KLK E + V+L +
Sbjct: 121 GDEWRELRKLSVREFFSVKKVQSFKYIREEENDLMIKKLK-ELASKQSPVDLSKILFGLT 179
Query: 179 FCLLFSMCFGENV-DSRKI--EKIKDIQRRLILSSGQLGALNFFPKIVSRVLF------- 228
++F FG++ D++ + E IK++ LS+ +FFP +
Sbjct: 180 ASIIFRTAFGQSFFDNKHVDQESIKELMFES-LSNMTFRFSDFFPTAGLKWFIGFVSGQH 238
Query: 229 RKRWQEFTQLRKLQNDLLVELIQARKKVIEKNNDDDEYVVCYVDTLLNLRLPEEKR---K 285
++ + F ++ N ++ + KK + D VD +L++ E++ K
Sbjct: 239 KRLYNVFNRVDTFFNHIVDD--HHSKKATQDRPD-------MVDAILDMIDNEQQYASFK 289
Query: 286 LNEGEVVALCSEFLTAGTDTTSTTLEWVMANLVKDKHVQQMLVEEIEEVVGEREEREVRE 345
L + + S AG DT++ TL W MA LV++ V + +EI +G ++E + E
Sbjct: 290 LTVDHLKGVLSNIYHAGIDTSAITLIWAMAELVRNPRVMKKAQDEIRTCIGIKQEGRIME 349
Query: 346 EDLEKLPYLKAVILEGLRRHPPTHYTIPRVVTEDIVLNGYLVPKNGTVNFMVADIGLDPT 405
EDL+KL YLK V+ E LR HP +PR DI + GY +P+ + IG DP
Sbjct: 350 EDLDKLQYLKLVVKETLRLHPAAPLLLPRETMADIKIQGYDIPQKRALLVNAWSIGRDPE 409
Query: 406 VWEDPTVFKPERFLKDAKNRNEFDAFDVRGIKEIKMMPFGAGRRICPAYKLAMLHLEYFV 465
W++P F PERF+ D +G +++PFG+GRRICP +A+ +E +
Sbjct: 410 SWKNPEEFNPERFID--------CPVDYKG-HSFELLPFGSGRRICPGIAMAIATIELGL 460
Query: 466 ANLVLNFEWKTSLPGSNVDLTEKQEFTMVMKYPLE 500
NL+ F+W ++D+ E + T+ K PLE
Sbjct: 461 LNLLYFFDWNMPEKKKDMDMEEAGDLTVDKKVPLE 495
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,312,611
Number of Sequences: 26719
Number of extensions: 490396
Number of successful extensions: 3038
Number of sequences better than 10.0: 264
Number of HSP's better than 10.0 without gapping: 224
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 1991
Number of HSP's gapped (non-prelim): 328
length of query: 507
length of database: 11,318,596
effective HSP length: 104
effective length of query: 403
effective length of database: 8,539,820
effective search space: 3441547460
effective search space used: 3441547460
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146723.3


