

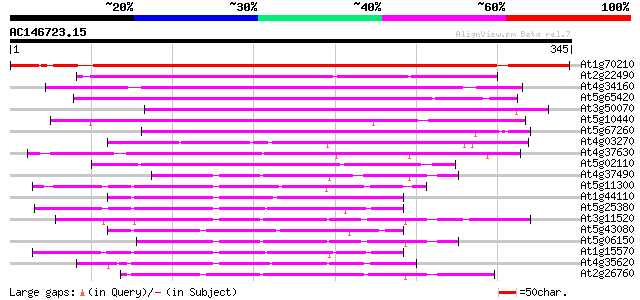
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146723.15 + phase: 0
(345 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g70210 unknown protein 363 e-101
At2g22490 putative cyclin D 182 2e-46
At4g34160 cyclin delta-3 171 5e-43
At5g65420 D-type cyclin (emb|CAB41347.1) 169 2e-42
At3g50070 cyclin D3-like protein 163 1e-40
At5g10440 cyclin protein - like 161 6e-40
At5g67260 cyclin D3-like protein 150 8e-37
At4g03270 putative D-type cyclin 140 1e-33
At4g37630 putative protein 119 2e-27
At5g02110 putative protein 101 5e-22
At4g37490 cyclin cyc1 78 8e-15
At5g11300 cyclin 3b 77 2e-14
At1g44110 mitotic cyclin a2-type, putative 75 5e-14
At5g25380 cyclin 3a 74 9e-14
At3g11520 cyclin box 74 2e-13
At5g43080 cyclin A-type 72 4e-13
At5g06150 mitosis-specific cyclin 1b 67 2e-11
At1g15570 putative cyclin (At1g15570) 67 2e-11
At4g35620 cyclin 2b protein 65 5e-11
At2g26760 putative cyclin 63 3e-10
>At1g70210 unknown protein
Length = 339
Score = 363 bits (933), Expect = e-101
Identities = 198/346 (57%), Positives = 247/346 (71%), Gaps = 21/346 (6%)
Query: 1 MPLSSTSDCELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEH 60
M +S ++D +L CGEDS V +G E + D SS P +SIA FIE
Sbjct: 12 MSVSFSNDMDLFCGEDSG-VFSG---ESTVDFSSSEVDSWPG---------DSIACFIED 58
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E FVPG DY+SRFQ+RSL++S RE+++AWILKV YY FQPLTAYL+VNYMDRFL +R
Sbjct: 59 ERHFVPGHDYLSRFQTRSLDASAREDSVAWILKVQAYYNFQPLTAYLAVNYMDRFLYARR 118
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
LPE++GWP+QLL+VACLSLAAKMEE LVPSL DFQ+ G KY+F+ +TI RMELLVL++LD
Sbjct: 119 LPETSGWPMQLLAVACLSLAAKMEEILVPSLFDFQVAGVKYLFEAKTIKRMELLVLSVLD 178
Query: 181 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
WRLRS+TP F+SFFA K+D +GTF F IS ATEIILSNI++ASFL Y PS IAAAAIL
Sbjct: 179 WRLRSVTPFDFISFFAYKIDPSGTFLGFFISHATEIILSNIKEASFLEYWPSSIAAAAIL 238
Query: 241 SAANEIPNW-SFVNP-EHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVT 298
ANE+P+ S VNP E E+WC+GLSKEKI+ CY L++ + NN+ N PKV+ +LRV+
Sbjct: 239 CVANELPSLSSVVNPHESPETWCDGLSKEKIVRCYRLMKAMAIENNRLNTPKVIAKLRVS 298
Query: 299 ARTRRWSTVSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVDKGNS 344
R SS+ + PS SS S S K+RKL+ WV + S
Sbjct: 299 VR------ASSTLTRPSDESSFSSSSPCKRRKLSGYSWVGDETSTS 338
>At2g22490 putative cyclin D
Length = 361
Score = 182 bits (462), Expect = 2e-46
Identities = 106/259 (40%), Positives = 153/259 (58%), Gaps = 7/259 (2%)
Query: 42 SSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQ 101
SSSL E+ I + E +F PG DYV R S L+ S R +A+ WILKV +Y F
Sbjct: 59 SSSL----SEDRIKEMLVREIEFCPGTDYVKRLLSGDLDLSVRNQALDWILKVCAHYHFG 114
Query: 102 PLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKY 161
L LS+NY+DRFL S LP+ W QLL+V+CLSLA+KMEE VP ++D Q+E K+
Sbjct: 115 HLCICLSMNYLDRFLTSYELPKDKDWAAQLLAVSCLSLASKMEETDVPHIVDLQVEDPKF 174
Query: 162 IFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNI 221
+F+ +TI RMELLV+T L+WRL+++TP SF+ +F K+ +G + +I R++ IL+
Sbjct: 175 VFEAKTIKRMELLVVTTLNWRLQALTPFSFIDYFVDKI--SGHVSENLIYRSSRFILNTT 232
Query: 222 QDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVS 281
+ FL +RPS IAAAA +S + ++ E A S + +E++ C L++ +
Sbjct: 233 KAIEFLDFRPSEIAAAAAVSVSIS-GETECIDEEKALSSLIYVKQERVKRCLNLMRSLTG 291
Query: 282 SNNQRNAPKVLPQLRVTAR 300
N R Q RV R
Sbjct: 292 EENVRGTSLSQEQARVAVR 310
>At4g34160 cyclin delta-3
Length = 376
Score = 171 bits (433), Expect = 5e-43
Identities = 105/294 (35%), Positives = 169/294 (56%), Gaps = 15/294 (5%)
Query: 23 GDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESS 82
G+ E +S L SSSS + E+E+ + +F + E + + D V S+
Sbjct: 33 GEEVEENSSLSSSSSPFVVLQQDLFWEDEDLVTLFSKEEEQGLSCLDDVYL-------ST 85
Query: 83 TREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAK 142
R+EA+ WIL+V+ +YGF L A L++ Y+D+F+ S L W LQL+SVACLSLAAK
Sbjct: 86 DRKEAVGWILRVNAHYGFSTLAAVLAITYLDKFICSYSLQRDKPWMLQLVSVACLSLAAK 145
Query: 143 MEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDST 202
+EE VP LLDFQ+E KY+F+ +TI RMELL+L+ L+W++ ITP+SF+ +L
Sbjct: 146 VEETQVPLLLDFQVEETKYVFEAKTIQRMELLILSTLEWKMHLITPISFVDHIIRRLGLK 205
Query: 203 GTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCE 262
+++ ++LS I D+ F+ Y PS +AAA ++ ++ + ++ +
Sbjct: 206 NNAHWDFLNKCHRLLLSVISDSRFVGYLPSVVAAATMMRIIEQVDPFDPLSYQTNLLGVL 265
Query: 263 GLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTARTRRWSTVSSSS-SSPS 315
L+KEK+ CY+LI ++ ++ Q+++ + +R S SSSS +SPS
Sbjct: 266 NLTKEKVKTCYDLILQL-------PVDRIGLQIQIQSSKKRKSHDSSSSLNSPS 312
>At5g65420 D-type cyclin (emb|CAB41347.1)
Length = 308
Score = 169 bits (428), Expect = 2e-42
Identities = 105/275 (38%), Positives = 155/275 (56%), Gaps = 6/275 (2%)
Query: 40 LPSSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESST-REEAIAWILKVHEYY 98
+P E EE I +E E + +P DY+ R +S L+ + R +A+ WI K E +
Sbjct: 33 IPQMGFSQSESEEIIMEMVEKEKQHLPSDDYIKRLRSGDLDLNVGRRDALNWIWKACEVH 92
Query: 99 GFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEG 158
F PL L++NY+DRFL LP GW LQLL+VACLSLAAK+EE VP L+D Q+
Sbjct: 93 QFGPLCFCLAMNYLDRFLSVHDLPSGKGWILQLLAVACLSLAAKIEETEVPMLIDLQVGD 152
Query: 159 AKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTF-THFIISRATEII 217
+++F+ +++ RMELLVL L WRLR+ITP S++ +F K+ ++ +ISR+ ++I
Sbjct: 153 PQFVFEAKSVQRMELLVLNKLKWRLRAITPCSYIRYFLRKMSKCDQEPSNTLISRSLQVI 212
Query: 218 LSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQ 277
S + FL +RPS +AAA LS + E+ F N + + L KE++ E+I+
Sbjct: 213 ASTTKGIDFLEFRPSEVAAAVALSVSGELQRVHFDNSSFSPLF-SLLQKERVKKIGEMIE 271
Query: 278 EIVSSNNQRNAPKVLPQLRVTARTRRWSTVSSSSS 312
S + V L V+A + T SSSS
Sbjct: 272 SDGSDLCSQTPNGV---LEVSACCFSFKTHDSSSS 303
>At3g50070 cyclin D3-like protein
Length = 361
Score = 163 bits (413), Expect = 1e-40
Identities = 97/253 (38%), Positives = 140/253 (54%), Gaps = 5/253 (1%)
Query: 84 REEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKM 143
RE+A+ WI KV +YGF LTA L+VNY DRF+ SR W QL ++ACLSLAAK+
Sbjct: 86 REKALDWIFKVKSHYGFNSLTALLAVNYFDRFITSRKFQTDKPWMSQLTALACLSLAAKV 145
Query: 144 EEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTG 203
EE VP LLDFQ+E A+Y+F+ +TI RMELLVL+ LDWR+ +TP+SF +
Sbjct: 146 EEIRVPFLLDFQVEEARYVFEAKTIQRMELLVLSTLDWRMHPVTPISFFDHIIRRYSFKS 205
Query: 204 TFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEG 263
+SR ++LS I D+ FL++ PS +A A ++S ++ +
Sbjct: 206 HHQLEFLSRCESLLLSIIPDSRFLSFSPSVLATAIMVSVIRDLKMCDEAVYQSQLMTLLK 265
Query: 264 LSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTARTRRWSTVSSS-----SSSPSSSS 318
+ EK+ CYEL+ + S + P + +S+ SS+ S+S S SS
Sbjct: 266 VDSEKVNKCYELVLDHSPSKKRMMNWMQQPASPIGVFDASFSSDSSNESWVVSASASVSS 325
Query: 319 SPSFSLSYKKRKL 331
SPS K+R++
Sbjct: 326 SPSSEPLLKRRRV 338
>At5g10440 cyclin protein - like
Length = 317
Score = 161 bits (407), Expect = 6e-40
Identities = 106/317 (33%), Positives = 162/317 (50%), Gaps = 31/317 (9%)
Query: 26 PECSSDLDSSSSSQLPSSSLFAE----EEEESIAVFIEHEFKFVPGFDYVSRFQSRSLES 81
P S+ D S+ + + S+F E EE + IE E + P DY+ R ++ L+
Sbjct: 7 PNLVSNFDDEKSNSVDTRSIFQMGFPLESEEIVREMIEKERQHSPRDDYLKRLRNGDLDF 66
Query: 82 STREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAA 141
+ R +A+ WI K E F PL L++NY+DRFL LP W +QLL+VACLSLAA
Sbjct: 67 NVRIQALGWIWKACEELQFGPLCICLAMNYLDRFLSVHDLPSGKAWTVQLLAVACLSLAA 126
Query: 142 KMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDS 201
K+EE VP L+ Q+ ++F+ +++ RMELLVL +L WRLR++TP S++ +F K++
Sbjct: 127 KIEETNVPELMQLQVGAPMFVFEAKSVQRMELLVLNVLRWRLRAVTPCSYVRYFLSKING 186
Query: 202 TGTFTHF-IISRATEIILSNIQ-------------------DASFLTYRPSCIAAAAILS 241
H +++R+ ++I S + FL +R S IAAA LS
Sbjct: 187 YDQEPHSRLVTRSLQVIASTTKGDRLGLFFFKGVLIVDVWAGIDFLEFRASEIAAAVALS 246
Query: 242 AANE-IPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTAR 300
+ E +SF S L KE++ E+I+ SS++ + + Q +
Sbjct: 247 VSGEHFDKFSF------SSSFSSLEKERVKKIGEMIERDGSSSSSQTPNNTVLQFKSRRY 300
Query: 301 TRRWSTVSSSSSSPSSS 317
+ ST S SSS S S
Sbjct: 301 SHSLSTASVSSSLTSLS 317
>At5g67260 cyclin D3-like protein
Length = 367
Score = 150 bits (380), Expect = 8e-37
Identities = 91/245 (37%), Positives = 140/245 (57%), Gaps = 8/245 (3%)
Query: 82 STREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAA 141
S R+EA+ W+L+V +YGF LTA L+VNY DRF+ S L W QL++VA LSLAA
Sbjct: 94 SCRKEALDWVLRVKSHYGFTSLTAILAVNYFDRFMTSIKLQTDKPWMSQLVAVASLSLAA 153
Query: 142 KMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDS 201
K+EE VP LLD Q+E A+Y+F+ +TI RMELL+L+ L WR+ +TP+SF + S
Sbjct: 154 KVEEIQVPLLLDLQVEEARYLFEAKTIQRMELLILSTLQWRMHPVTPISFFDHIIRRFGS 213
Query: 202 TGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWC 261
+ +++S I D F+ Y PS +A A ++ E+ V + +
Sbjct: 214 KWHQQLDFCRKCERLLISVIADTRFMRYFPSVLATAIMILVFEELKPCDEVEYQSQITTL 273
Query: 262 EGLSKEKIIGCYELIQEIVSSNNQ------RNAPKVLPQLRVTARTRRWSTVSSSSSSPS 315
+++EK+ CYEL+ E S + +++P + ++ + W+ VS+++S S
Sbjct: 274 LKVNQEKVNECYELLLEHNPSKKRMMNLVDQDSPSGVLDFDDSSNS-SWN-VSTTASVSS 331
Query: 316 SSSSP 320
SSSSP
Sbjct: 332 SSSSP 336
>At4g03270 putative D-type cyclin
Length = 302
Score = 140 bits (353), Expect = 1e-33
Identities = 94/268 (35%), Positives = 150/268 (55%), Gaps = 13/268 (4%)
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
EF+ +P Y +S + S R +AI+ I + + LT YL+VNY+DRFL S
Sbjct: 35 EFQHMPSSHYFHSLKSSAFLLSNRNQAISSITQYSRKFDDPSLT-YLAVNYLDRFLSSED 93
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
+P+S W L+L+S++C+SL+AKM +P + S+ D +EG F + I RME ++L L
Sbjct: 94 MPQSKPWILKLISLSCVSLSAKMRKPDM-SVSDLPVEGE--FFDAQMIERMENVILGALK 150
Query: 181 WRLRSITPLSFLSF----FACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAA 236
WR+RS+TP SFL+F F K + H + S+ +++ S D SFL ++PS IA
Sbjct: 151 WRMRSVTPFSFLAFFISLFELKEEDPLLLKHSLKSQTSDLTFSLQHDISFLEFKPSVIAG 210
Query: 237 AAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQE---IVSSN--NQRNAPKV 291
AA+L A+ E+ F + + C ++K++++ CY+ IQE IV N + A V
Sbjct: 211 AALLFASFELCPLQFPCFSNRINQCTYVNKDELMECYKAIQERDIIVGENEGSTETAVNV 270
Query: 292 LPQLRVTARTRRWSTVSSSSSSPSSSSS 319
L Q + + + T+++SSS +S
Sbjct: 271 LDQQFSSCESDKSITITASSSPKRRKTS 298
>At4g37630 putative protein
Length = 321
Score = 119 bits (299), Expect = 2e-27
Identities = 88/315 (27%), Positives = 144/315 (44%), Gaps = 25/315 (7%)
Query: 12 LCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFVPGFDYV 71
LC E S L E + S Q P + ++E+ + ++ +F
Sbjct: 12 LCHESESS-----LNEDDDETIERSDKQEPHFTTTIDDEDYVADLVLKENLRF------- 59
Query: 72 SRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQL 131
S++ SS R AI WIL VH+ + P + L N + R + + + W ++L
Sbjct: 60 ETLPSKTTSSSDRLIAIDWILTVHKNKIWVPTSNSLHCNLILRSVSPQKIHRYETWAMRL 119
Query: 132 LSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSF 191
LSVACLSLAAKMEE +VP L + + ++F+P I + ELL+L+ LDW++ ITP +
Sbjct: 120 LSVACLSLAAKMEERIVPGLSQYP-QDHDFVFKPDVIRKTELLILSTLDWKMNLITPFHY 178
Query: 192 LSFFACKL--DSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAAN----- 244
++F K+ D+ ++ R+++ +L+ ++ SF YR +AA L A++
Sbjct: 179 FNYFLAKISQDNHSVSKDLVLLRSSDSLLALTKEISFTEYRQFVVAAVTTLLASSSTSSD 238
Query: 245 -EIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVL----PQLRVTA 299
+ N + SW E + CY+ EI + P++ P +
Sbjct: 239 IRLTREEIANKFGSISWWTSNENENVYLCYQRTLEIEERKHMTPPPEIAVSREPPASGSG 298
Query: 300 RTRRWSTVSSSSSSP 314
RR S S SSP
Sbjct: 299 AKRRLSFDDSDQSSP 313
>At5g02110 putative protein
Length = 341
Score = 101 bits (252), Expect = 5e-22
Identities = 69/226 (30%), Positives = 113/226 (49%), Gaps = 11/226 (4%)
Query: 51 EESIAVFIEHEFKFVP-GFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSV 109
EE+IA+ +E E F G +V F S+ L + R A W+++ T + +
Sbjct: 45 EEAIAMDLEKELCFNNHGDKFVEFFVSKKL-TDYRFHAFQWLIQTRSRLNLSYETVFSAA 103
Query: 110 NYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTIL 169
N DRF+ E W ++L++V LS+A+K E P L + ++EG ++F T+
Sbjct: 104 NCFDRFVYMTCCDEWTNWMVELVAVTSLSIASKFNEVTTPLLEELEMEGLTHMFHVNTVA 163
Query: 170 RMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTY 229
+MEL++L L+WR+ ++T +F K+ G H I++R T +L I D L Y
Sbjct: 164 QMELIILKALEWRVNAVTSYTFSQTLVSKIGMVG--DHMIMNRITNHLLDVICDLKMLQY 221
Query: 230 RPSCIAAAAI-LSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYE 274
PS +A AAI + +++ S +N E KEKI+ C +
Sbjct: 222 PPSVVATAAIWILMEDKVCRESIMN------LFEQNHKEKIVKCVD 261
>At4g37490 cyclin cyc1
Length = 428
Score = 77.8 bits (190), Expect = 8e-15
Identities = 54/195 (27%), Positives = 98/195 (49%), Gaps = 21/195 (10%)
Query: 88 IAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPL 147
+ W++ VH + P T YL+VN +DRFL +P+P LQL+ ++ L ++AK EE
Sbjct: 201 VEWLIDVHVRFELNPETFYLTVNILDRFLSVKPVPRKE---LQLVGLSALLMSAKYEEIW 257
Query: 148 VPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFF---ACKLDSTGT 204
P + D ++ A + + + IL ME +L+ L+W L T FL+ F + +
Sbjct: 258 PPQVEDL-VDIADHAYSHKQILVMEKTILSTLEWYLTVPTHYVFLARFIKASIADEKMEN 316
Query: 205 FTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAAN---EIPNWSFVNPEHAESWC 261
H++ L + + + + PS +AA+AI +A + ++P W+ H
Sbjct: 317 MVHYLAE------LGVMHYDTMIMFSPSMVAASAIYAARSSLRQVPIWTSTLKHHT---- 366
Query: 262 EGLSKEKIIGCYELI 276
G S+ +++ C +L+
Sbjct: 367 -GYSETQLMDCAKLL 380
>At5g11300 cyclin 3b
Length = 434
Score = 76.6 bits (187), Expect = 2e-14
Identities = 73/245 (29%), Positives = 118/245 (47%), Gaps = 24/245 (9%)
Query: 15 EDSSEVLTGDLPECSSDLDSSSSSQLPSS-SLFAEEEEESIAVFIEHEFKFVPGFDYVSR 73
ED S V+ E +D S+ + P SL+A + ++I V E + P +Y+
Sbjct: 145 EDGSGVM-----ELLQVVDIDSNVEDPQCCSLYAADIYDNIHVA---ELQQRPLANYMEL 196
Query: 74 FQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLS 133
Q R ++ R+ I W+++V + Y P T YL+VN +DRFL + + LQLL
Sbjct: 197 VQ-RDIDPDMRKILIDWLVEVSDDYKLVPDTLYLTVNLIDRFLSNSYIERQR---LQLLG 252
Query: 134 VACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLS 193
V+C+ +A+K EE P + +F A +P +L ME+ +L + +RL T +FLS
Sbjct: 253 VSCMLIASKYEELSAPGVEEFCFITANTYTRPE-VLSMEIQILNFVHFRLSVPTTKTFLS 311
Query: 194 --FFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSF 251
F L ++ + E+ L + SFL + PS IAA+A+ A W+
Sbjct: 312 ALFLIIILQVPFIELEYLANYLAELTL---VEYSFLRFLPSLIAASAVFLA-----RWTL 363
Query: 252 VNPEH 256
+H
Sbjct: 364 DQTDH 368
>At1g44110 mitotic cyclin a2-type, putative
Length = 460
Score = 75.1 bits (183), Expect = 5e-14
Identities = 60/184 (32%), Positives = 89/184 (47%), Gaps = 8/184 (4%)
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E K P DY+ R Q + + SS R + W+++V E Y P T YL+VNY+DR+L
Sbjct: 206 EAKKRPDVDYMERVQ-KDVNSSMRGILVDWLIEVSEEYRLVPETLYLTVNYIDRYLSGNV 264
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDF-QIEGAKYIFQPRTILRMELLVLTIL 179
+ LQLL VAC+ +AAK EE P + +F I Y+ +L ME VL L
Sbjct: 265 ISRQK---LQLLGVACMMIAAKYEEICAPQVEEFCYITDNTYL--KDEVLDMESDVLNYL 319
Query: 180 DWRLRSITPLSFLSFFACKLDSTGTFTHFIIS-RATEIILSNIQDASFLTYRPSCIAAAA 238
+ + + T FL F + A I ++ + + L++ PS +AA+A
Sbjct: 320 KFEMTAPTTKCFLRRFVRAAHGVHEAPLMQLECMANYIAELSLLEYTMLSHSPSLVAASA 379
Query: 239 ILSA 242
I A
Sbjct: 380 IFLA 383
>At5g25380 cyclin 3a
Length = 444
Score = 74.3 bits (181), Expect = 9e-14
Identities = 68/232 (29%), Positives = 110/232 (47%), Gaps = 16/232 (6%)
Query: 16 DSSEVLTGDLPECSSDLDSSSSSQLPS-SSLFAEEEEESIAVFIEHEFKFVPGFDYVSRF 74
D +E D+ +C +D S Q P SL+A +SI V E + P Y+ +
Sbjct: 149 DCAEEDRSDVTDCVQIVDIDSGVQDPQFCSLYAASIYDSINVA---ELEQRPSTSYMVQV 205
Query: 75 QSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSV 134
Q R ++ + R I W+++V E Y T YL+VN +DRF+ + + LQLL +
Sbjct: 206 Q-RDIDPTMRGILIDWLVEVSEEYKLVSDTLYLTVNLIDRFMSHNYIEKQK---LQLLGI 261
Query: 135 ACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSF 194
C+ +A+K EE P L +F + +L ME+ VL L +RL T +FL
Sbjct: 262 TCMLIASKYEEISAPRLEEFCFI-TDNTYTRLEVLSMEIKVLNSLHFRLSVPTTKTFLRR 320
Query: 195 FACKLDSTGTF----THFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA 242
F ++ ++ + E+ L+ + +FL + PS IAA+A+ A
Sbjct: 321 FIRAAQASDKVPLIEMEYLANYFAELTLT---EYTFLRFLPSLIAASAVFLA 369
>At3g11520 cyclin box
Length = 414
Score = 73.6 bits (179), Expect = 2e-13
Identities = 76/302 (25%), Positives = 134/302 (44%), Gaps = 26/302 (8%)
Query: 29 SSDLDSSSSSQLPSSSLFAEEEEESIAV--FIEHEFKFVPGFDYVSRFQ-----SRSLES 81
SS LD+ S + + + ++E +A ++E + F S+ Q ++
Sbjct: 129 SSVLDARSKAASKTLDIDYVDKENDLAAVEYVEDMYIFYKEVVNESKPQMYMHTQPEIDE 188
Query: 82 STREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAA 141
R I W+++VH + P T YL+VN +DRFL + +P LQL+ V+ L +A+
Sbjct: 189 KMRSILIDWLVEVHVKFDLSPETLYLTVNIIDRFLSLKTVPRRE---LQLVGVSALLIAS 245
Query: 142 KMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDS 201
K EE P + D + + R IL ME +L L+W L T FL F K
Sbjct: 246 KYEEIWPPQVNDL-VYVTDNSYNSRQILVMEKTILGNLEWYLTVPTQYVFLVRF-IKASG 303
Query: 202 TGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA---ANEIPNWSFVNPEHAE 258
+ ++ E+ + L + PS +AA+A+ +A N+ P W+ H
Sbjct: 304 SDQKLENLVHFLAEL---GLMHHDSLMFCPSMLAASAVYTARCCLNKTPTWTDTLKFHT- 359
Query: 259 SWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTARTRRWSTVSSSSSSPSSSS 318
G S+ +++ C +L+ I +++ K+ L+ ++ R + S + SS
Sbjct: 360 ----GYSESQLMDCSKLLAFI---HSKAGESKLRGVLKKYSKLGRGAVALISPAKSLMSS 412
Query: 319 SP 320
+P
Sbjct: 413 AP 414
>At5g43080 cyclin A-type
Length = 355
Score = 72.0 bits (175), Expect = 4e-13
Identities = 61/187 (32%), Positives = 93/187 (49%), Gaps = 14/187 (7%)
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E K P DY+ + Q + + S+ R + W+++V E Y T YL+V+Y+DRFL
Sbjct: 99 EVKSRPLVDYIEKIQ-KDVTSNMRGVLVDWLVEVAEEYKLLSDTLYLAVSYIDRFLS--- 154
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDF-QIEGAKYIFQPRTILRMELLVLTIL 179
L N LQLL V + +A+K EE P++ DF I Y Q I++ME +L L
Sbjct: 155 LKTVNKQRLQLLGVTSMLIASKYEEITPPNVDDFCYITDNTYTKQ--EIVKMEADILLAL 212
Query: 180 DWRLRSITPLSFLSFFACKLDSTGTFTH----FIISRATEIILSNIQDASFLTYRPSCIA 235
+ L + T +FL F +H F+ S +E+ + + Q FL PS +A
Sbjct: 213 QFELGNPTSNTFLRRFTRVAQEDFEMSHLQMEFLCSYLSELSMLDYQSVKFL---PSTVA 269
Query: 236 AAAILSA 242
A+A+ A
Sbjct: 270 ASAVFLA 276
>At5g06150 mitosis-specific cyclin 1b
Length = 445
Score = 66.6 bits (161), Expect = 2e-11
Identities = 57/201 (28%), Positives = 94/201 (46%), Gaps = 16/201 (7%)
Query: 79 LESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLS 138
+ R I W+L+VH + T YL+VN +DRFL + +P+ LQL+ ++ L
Sbjct: 209 MNEKMRAILIDWLLEVHIKFELNLETLYLTVNIIDRFLSVKAVPKRE---LQLVGISALL 265
Query: 139 LAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACK 198
+A+K EE P + D + + R IL ME +L L+W L T FL F K
Sbjct: 266 IASKYEEIWPPQVNDL-VYVTDNAYSSRQILVMEKAILGNLEWYLTVPTQYVFLVRF-IK 323
Query: 199 LDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA---ANEIPNWSFVNPE 255
+ ++ E+ + LT+ PS +AA+A+ +A N+ P W+
Sbjct: 324 ASMSDPEMENMVHFLAEL---GMMHYDTLTFCPSMLAASAVYTARCSLNKSPAWTDTLQF 380
Query: 256 HAESWCEGLSKEKIIGCYELI 276
H G ++ +I+ C +L+
Sbjct: 381 HT-----GYTESEIMDCSKLL 396
>At1g15570 putative cyclin (At1g15570)
Length = 450
Score = 66.6 bits (161), Expect = 2e-11
Identities = 67/234 (28%), Positives = 108/234 (45%), Gaps = 18/234 (7%)
Query: 15 EDSSEVLTGDLPECSSDLDSSSSSQLPS-SSLFAEEEEESIAVFIEHEFKFVPGFDYVSR 73
E S+ + + +P+ +D S + P L+A E ++ V E K P D++ R
Sbjct: 156 EKSAVIGSSTVPDIPKFVDIDSDDKDPLLCCLYAPEIHYNLRV---SELKRRPLPDFMER 212
Query: 74 FQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLS 133
Q + + S R + W+++V E Y T YL+V +D FL + LQLL
Sbjct: 213 IQ-KDVTQSMRGILVDWLVEVSEEYTLASDTLYLTVYLIDWFLHGNYVQRQQ---LQLLG 268
Query: 134 VACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLS 193
+ C+ +A+K EE P + +F + +L ME VL +++ + TP +FL
Sbjct: 269 ITCMLIASKYEEISAPRIEEFCFI-TDNTYTRDQVLEMENQVLKHFSFQIYTPTPKTFLR 327
Query: 194 FF-----ACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA 242
F A +L S F+ S TE+ L D FL + PS +AA+A+ A
Sbjct: 328 RFLRAAQASRL-SPSLEVEFLASYLTELTLI---DYHFLKFLPSVVAASAVFLA 377
>At4g35620 cyclin 2b protein
Length = 429
Score = 65.1 bits (157), Expect = 5e-11
Identities = 60/211 (28%), Positives = 96/211 (45%), Gaps = 13/211 (6%)
Query: 42 SSSLFAEEEEESIAVFIE--HEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYG 99
++SL A E + + F F VP DY++ Q + R I W+++VH+ +
Sbjct: 164 NNSLAAVEYVQDLYDFYRKTERFSCVP-LDYMA--QQFDISDKMRAILIDWLIEVHDKFE 220
Query: 100 FQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGA 159
T +L+VN +DRFL + + LQL+ + L LA K EE VP + D +
Sbjct: 221 LMNETLFLTVNLIDRFLSKQAVARKK---LQLVGLVALLLACKYEEVSVPIVEDLVVISD 277
Query: 160 KYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILS 219
K + +L ME ++L+ L + + T FL F K + + S E+ L
Sbjct: 278 K-AYTRTDVLEMEKIMLSTLQFNMSLPTQYPFLKRF-LKAAQSDKKLEILASFLIELAL- 334
Query: 220 NIQDASFLTYRPSCIAAAAILSAANEIPNWS 250
D + Y PS +AA A+ +A I +S
Sbjct: 335 --VDYEMVRYPPSLLAATAVYTAQCTIHGFS 363
>At2g26760 putative cyclin
Length = 387
Score = 62.8 bits (151), Expect = 3e-10
Identities = 56/233 (24%), Positives = 104/233 (44%), Gaps = 16/233 (6%)
Query: 69 DYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWP 128
DY+ + R I W++ VH + P T YL++N +DRFL L +
Sbjct: 149 DYIG--SQPEINEKMRSILIDWLVDVHRKFELMPETLYLTINLVDRFLS---LTMVHRRE 203
Query: 129 LQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITP 188
LQLL + + +A K EE P + DF + + + + +L ME +L ++W + TP
Sbjct: 204 LQLLGLGAMLIACKYEEIWAPEVNDF-VCISDNAYNRKQVLAMEKSILGQVEWYITVPTP 262
Query: 189 LSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA---ANE 245
FL+ + ++ E+ L +Q + RPS +AA+A+ +A +
Sbjct: 263 YVFLARYVKAAVPCDAEMEKLVFYLAELGL--MQYPIVVLNRPSMLAASAVYAARQILKK 320
Query: 246 IPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVT 298
P W+ H G S+++I+ +++ ++ S ++ V + V+
Sbjct: 321 TPFWTETLKHHT-----GYSEDEIMEHAKMLMKLRDSASESKLIAVFKKYSVS 368
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,971,159
Number of Sequences: 26719
Number of extensions: 343689
Number of successful extensions: 2359
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 2177
Number of HSP's gapped (non-prelim): 114
length of query: 345
length of database: 11,318,596
effective HSP length: 100
effective length of query: 245
effective length of database: 8,646,696
effective search space: 2118440520
effective search space used: 2118440520
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146723.15


