

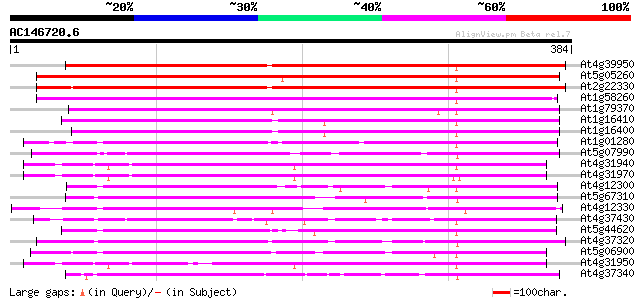
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146720.6 - phase: 0 /pseudo
(384 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g39950 cytochrome P450 like protein 391 e-109
At5g05260 cytochrome P450 CYP79A2 (CYP79A2) 388 e-108
At2g22330 putative cytochrome P450 386 e-108
At1g58260 hypothetical protein 286 1e-77
At1g79370 hypothetical protein 273 1e-73
At1g16410 putative cytochrome P450 protein 251 6e-67
At1g16400 cytochrome P450-like protein (At1g16400) 233 2e-61
At1g01280 cytochrome P450, putative 166 2e-41
At5g07990 flavonoid 3'-hydroxylase - like protein 159 2e-39
At4g31940 Cytochrome P450-like protein 141 5e-34
At4g31970 Cytochrome P450-like protein 139 2e-33
At4g12300 flavonoid 3',5'-hydroxylase -like protein 135 4e-32
At5g67310 cytochrome P450 131 5e-31
At4g12330 flavonoid 3',5'-hydroxylase like protein 130 2e-30
At4g37430 cytochrome P450 monooxygenase (CYP91A2) 126 2e-29
At5g44620 flavonoid 3',5'-hydroxylase-like; cytochrome P450 125 4e-29
At4g37320 cytochrome P450-like protein 124 9e-29
At5g06900 cytochrome P450 123 1e-28
At4g31950 cytochrome P450-like protein 123 1e-28
At4g37340 cytochrome P450-like protein 122 3e-28
>At4g39950 cytochrome P450 like protein
Length = 541
Score = 391 bits (1005), Expect = e-109
Identities = 184/365 (50%), Positives = 258/365 (70%), Gaps = 26/365 (7%)
Query: 39 KPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPE 98
K KP LPPGPT WPI+G +P ML +RP FRW+ +M LNT+IAC++LGN HVIT++ P+
Sbjct: 51 KKKPYLPPGPTGWPIIGMIPTMLKSRPVFRWLHSIMKQLNTEIACVKLGNTHVITVTCPK 110
Query: 99 IARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKW 158
IARE+ +QDA+FASRP +++ + ++NGY T +TPFG+Q+KK++KV+ ELV P RH+W
Sbjct: 111 IAREILKQQDALFASRPLTYAQKILSNGYKTCVITPFGDQFKKMRKVVMTELVCPARHRW 170
Query: 159 LHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFG-NGSED 217
LH KR EE D++ +VYN + G V+ ++Y G+ I++L+ R F N + D
Sbjct: 171 LHQKRSEENDHLTAWVYN---MVKNSGSVDFRFMTRHYCGNAIKKLMFGTRTFSKNTAPD 227
Query: 218 YGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDPII 277
GP +E++E++EA+F L + FAF +SD++P L GLDL+GHE+I++++ IM KYHDPII
Sbjct: 228 GGPTVEDVEHMEAMFEALGFTFAFCISDYLPMLTGLDLNGHEKIMRESSAIMDKYHDPII 287
Query: 278 EDRIQQWKNGKKIEKEDLLDVLISLKD----------------------AVDNPSNAVEW 315
++RI+ W+ GK+ + ED LD+ IS+KD A DNPSNAVEW
Sbjct: 288 DERIKMWREGKRTQIEDFLDIFISIKDEQGNPLLTADEIKPTIKELVMAAPDNPSNAVEW 347
Query: 316 GLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPH 375
+AE++N+PE+L+KA EE+D VVGK RLVQE D PKLNYVKA +EAFR HP+ FNLPH
Sbjct: 348 AMAEMVNKPEILRKAMEEIDRVVGKERLVQESDIPKLNYVKAILREAFRLHPVAAFNLPH 407
Query: 376 VRCNE 380
V ++
Sbjct: 408 VALSD 412
>At5g05260 cytochrome P450 CYP79A2 (CYP79A2)
Length = 523
Score = 388 bits (997), Expect = e-108
Identities = 189/385 (49%), Positives = 263/385 (68%), Gaps = 27/385 (7%)
Query: 19 LMLLVYKFLIKHQRSTQK-SEKPKPKLPPGPTPWPIVGNLPEMLA-NRPTFRWIQKMMND 76
L+LL K ++ T S LPPGP WP++GNLPE+L N+P FRWI +M +
Sbjct: 8 LLLLALTMKRKEKKKTMLISPTRNLSLPPGPKSWPLIGNLPEILGRNKPVFRWIHSLMKE 67
Query: 77 LNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFG 136
LNTDIACIRL N HVI ++ P IARE+ KQD++FA+RP + EY + GYLT A+ P G
Sbjct: 68 LNTDIACIRLANTHVIPVTSPRIAREILKKQDSVFATRPLTMGTEYCSRGYLTVAVEPQG 127
Query: 137 EQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDG--IVNVSVAAQ 194
EQWKK+++V+++ + S + + KR EEADN+VRY+ N+ K G+ ++++ +A +
Sbjct: 128 EQWKKMRRVVASHVTSKKSFQMMLQKRTEEADNLVRYINNRSVKNRGNAFVVIDLRLAVR 187
Query: 195 YYSGDVIRRLLLNKRYFGNGSED-YGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGL 253
YSG+V R+++ R+FG GSED GPGLEEIE+VE++FTVL +L+AF++SD++P LR L
Sbjct: 188 QYSGNVARKMMFGIRHFGKGSEDGSGPGLEEIEHVESLFTVLTHLYAFALSDYVPWLRFL 247
Query: 254 DLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKD--------- 304
DL+GHE+++ A + + KY+DP +++R+ QW+NGK E +D LD+ I KD
Sbjct: 248 DLEGHEKVVSNAMRNVSKYNDPFVDERLMQWRNGKMKEPQDFLDMFIIAKDTDGKPTLSD 307
Query: 305 -------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPK 351
VDNPSNA EWG+AE+IN+P +++KA EE+D VVGK RLV E D P
Sbjct: 308 EEIKAQVTELMLATVDNPSNAAEWGMAEMINEPSIMQKAVEEIDRVVGKDRLVIESDLPN 367
Query: 352 LNYVKACAKEAFRRHPICDFNLPHV 376
LNYVKAC KEAFR HP+ FNLPH+
Sbjct: 368 LNYVKACVKEAFRLHPVAPFNLPHM 392
>At2g22330 putative cytochrome P450
Length = 566
Score = 386 bits (992), Expect = e-108
Identities = 188/385 (48%), Positives = 266/385 (68%), Gaps = 27/385 (7%)
Query: 19 LMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLN 78
L L + ++ +S+ +++K P LPPGPT +PIVG +P ML NRP FRW+ +M +LN
Sbjct: 57 LAALCFLMILNKIKSSSRNKKLHP-LPPGPTGFPIVGMIPAMLKNRPVFRWLHSLMKELN 115
Query: 79 TDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQ 138
T+IAC+RLGN HVI ++ P+IARE+ +QDA+FASRP +++ + ++NGY T +TPFGEQ
Sbjct: 116 TEIACVRLGNTHVIPVTCPKIAREIFKQQDALFASRPLTYAQKILSNGYKTCVITPFGEQ 175
Query: 139 WKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSG 198
+KK++KVI E+V P RH+WLHD R EE D++ ++YN + V++ ++Y G
Sbjct: 176 FKKMRKVIMTEIVCPARHRWLHDNRAEETDHLTAWLYN---MVKNSEPVDLRFVTRHYCG 232
Query: 199 DVIRRLLLNKRYFGNGSE-DYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDG 257
+ I+RL+ R F +E D GP LE+IE+++A+F L + FAF +SD++P L GLDL+G
Sbjct: 233 NAIKRLMFGTRTFSEKTEADGGPTLEDIEHMDAMFEGLGFTFAFCISDYLPMLTGLDLNG 292
Query: 258 HERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKD------------- 304
HE+I++++ IM KYHDPII++RI+ W+ GK+ + ED LD+ IS+KD
Sbjct: 293 HEKIMRESSAIMDKYHDPIIDERIKMWREGKRTQIEDFLDIFISIKDEAGQPLLTADEIK 352
Query: 305 ---------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYV 355
A DNPSNAVEW +AE+IN+PE+L KA EE+D VVGK R VQE D PKLNYV
Sbjct: 353 PTIKELVMAAPDNPSNAVEWAIAEMINKPEILHKAMEEIDRVVGKERFVQESDIPKLNYV 412
Query: 356 KACAKEAFRRHPICDFNLPHVRCNE 380
KA +EAFR HP+ FNLPHV ++
Sbjct: 413 KAIIREAFRLHPVAAFNLPHVALSD 437
>At1g58260 hypothetical protein
Length = 670
Score = 286 bits (733), Expect = 1e-77
Identities = 154/380 (40%), Positives = 229/380 (59%), Gaps = 24/380 (6%)
Query: 19 LMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLN 78
L+L+ ++ R + KPK +LPPGP WPIVGN+ +M+ NRP WI ++M +L
Sbjct: 12 LVLISITLVLALARRFSRFMKPKGQLPPGPRGWPIVGNMLQMIINRPAHLWIHRVMEELQ 71
Query: 79 TDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQ 138
TDIAC R HVIT++ +IARE+ ++D + A R S+++ +++GY + + +GE
Sbjct: 72 TDIACYRFARFHVITVTSSKIAREVLREKDEVLADRSESYASHLISHGYKNISFSSYGEN 131
Query: 139 WKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSG 198
WK VKKV++ +L+SP R EADNIV YVYN C +NV Y
Sbjct: 132 WKLVKKVMTTKLMSPTTLSKTLGYRNIEADNIVTYVYNLCQLGSVRKPINVRDTILTYCH 191
Query: 199 DVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGH 258
V+ R++ +R+F E+ G G +E E+++AI+ L F+F++++++P LRG ++D
Sbjct: 192 AVMMRMMFGQRHFDEVVENGGLGPKEKEHMDAIYLALDCFFSFNLTNYIPFLRGWNVDKA 251
Query: 259 ERIIKKACKIMKKYHDPIIEDRIQQW-KNGKKIEKEDLLDVLISLKD------------- 304
E +++A I+ +DPII++RI W K G K +ED LD+LI+LKD
Sbjct: 252 ETEVREAVHIINICNDPIIQERIHLWRKKGGKQMEEDWLDILITLKDDQGMHLFTFDEIR 311
Query: 305 ---------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYV 355
+DN N VEW +AE++N PE+L+KAT ELD +VGK RLVQE D +LNY+
Sbjct: 312 AQCKEINLATIDNTMNNVEWTIAEMLNHPEILEKATNELDIIVGKDRLVQESDISQLNYI 371
Query: 356 KACAKEAFRRHPICDFNLPH 375
KAC+KE+FR HP F +PH
Sbjct: 372 KACSKESFRLHPANVF-MPH 390
>At1g79370 hypothetical protein
Length = 437
Score = 273 bits (698), Expect = 1e-73
Identities = 153/373 (41%), Positives = 217/373 (58%), Gaps = 37/373 (9%)
Query: 41 KPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEIA 100
+ KLPP P +PI+GNL ML NRPT +WI ++MND+ TDIAC R G VHVI I+ IA
Sbjct: 37 RKKLPPCPRGFPIIGNLVGMLKNRPTSKWIVRVMNDMKTDIACFRFGRVHVIVITSDVIA 96
Query: 101 RELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKWLH 160
RE+ ++DA+FA RP S+S EY++ GY +GE+ K+KKV+S+EL+S L
Sbjct: 97 REVVREKDAVFADRPDSYSAEYISGGYNGVVFDEYGERQMKMKKVMSSELMSTKALNLLL 156
Query: 161 DKRVEEADNIVRYVYNKC----TKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNGSE 216
R E+DN++ YV+N +K +VNV ++ +V RLL +R+F +
Sbjct: 157 KVRNLESDNLLAYVHNLYNKDESKTKHGAVVNVRDIVCTHTHNVKMRLLFGRRHFKETTM 216
Query: 217 DYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDPI 276
D GL E E+ +AIF L F+F V+D+ P LRG +L G E +++A ++ +Y+ I
Sbjct: 217 DGSLGLMEKEHFDAIFAALDCFFSFYVADYYPFLRGWNLQGEEAELREAVDVIARYNKMI 276
Query: 277 IEDRIQQWKNGKKIEK-----------EDLLDVLISLKD--------------------- 304
I+++I+ W+ K +D LD+L +LKD
Sbjct: 277 IDEKIELWRGQNKDYNRAETKNDVPMIKDWLDILFTLKDENGKPLLTPQEITHLSVDLDV 336
Query: 305 -AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAF 363
+DN N +EW LAE++NQ E+L+KA EE+D VVGK RLVQE D P LNYVKAC +E
Sbjct: 337 VGIDNAVNVIEWTLAEMLNQREILEKAVEEIDMVVGKERLVQESDVPNLNYVKACCRETL 396
Query: 364 RRHPICDFNLPHV 376
R HP F +PH+
Sbjct: 397 RLHPTNPFLVPHM 409
>At1g16410 putative cytochrome P450 protein
Length = 538
Score = 251 bits (640), Expect = 6e-67
Identities = 140/368 (38%), Positives = 211/368 (57%), Gaps = 30/368 (8%)
Query: 36 KSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITIS 95
K++ +LPPGP WPI+GNLPE+ RP ++ + M +L TDIAC + ITI+
Sbjct: 37 KTKDRSCQLPPGPPGWPILGNLPELFMTRPRSKYFRLAMKELKTDIACFNFAGIRAITIN 96
Query: 96 DPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLR 155
EIARE ++DA A RP + E + + Y + ++P+GEQ+ K+K+VI+ E++S
Sbjct: 97 SDEIAREAFRERDADLADRPQLFIMETIGDNYKSMGISPYGEQFMKMKRVITTEIMSVKT 156
Query: 156 HKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNG- 214
K L R EADN++ YV++ + V+V ++ Y V R+L +R+
Sbjct: 157 LKMLEAARTIEADNLIAYVHSMYQR---SETVDVRELSRVYGYAVTMRMLFGRRHVTKEN 213
Query: 215 --SEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMP-CLRGLDLDGHERIIKKACKIMKK 271
S+D G E ++E IF L L +FS +D++ LRG ++DG E+ + + C I++
Sbjct: 214 VFSDDGRLGNAEKHHLEVIFNTLNCLPSFSPADYVERWLRGWNVDGQEKRVTENCNIVRS 273
Query: 272 YHDPIIEDRIQQWK-NGKKIEKEDLLDVLISLKD----------------------AVDN 308
Y++PII++R+Q W+ G K ED LD I+LKD A+DN
Sbjct: 274 YNNPIIDERVQLWREEGGKAAVEDWLDTFITLKDQNGKYLVTPDEIKAQCVEFCIAAIDN 333
Query: 309 PSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPI 368
P+N +EW L E++ PE+L+KA +ELD VVG+ RLVQE D P LNY+KAC +E FR HP
Sbjct: 334 PANNMEWTLGEMLKNPEILRKALKELDEVVGRDRLVQESDIPNLNYLKACCRETFRIHPS 393
Query: 369 CDFNLPHV 376
+ H+
Sbjct: 394 AHYVPSHL 401
>At1g16400 cytochrome P450-like protein (At1g16400)
Length = 537
Score = 233 bits (593), Expect = 2e-61
Identities = 136/361 (37%), Positives = 196/361 (53%), Gaps = 30/361 (8%)
Query: 43 KLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARE 102
+LPPG WPI+GNLPE++ RP ++ M +L TDIAC H ITI+ EIARE
Sbjct: 43 QLPPGRPGWPILGNLPELIMTRPRSKYFHLAMKELKTDIACFNFAGTHTITINSDEIARE 102
Query: 103 LCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDK 162
++DA A RP E + + Y T + +GE + K+KKVI+ E++S L
Sbjct: 103 AFRERDADLADRPQLSIVESIGDNYKTMGTSSYGEHFMKMKKVITTEIMSVKTLNMLEAA 162
Query: 163 RVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNG---SEDYG 219
R EADN++ Y+++ + V+V ++ Y V R+L +R+ S+D
Sbjct: 163 RTIEADNLIAYIHSMYQR---SETVDVRELSRVYGYAVTMRMLFGRRHVTKENMFSDDGR 219
Query: 220 PGLEEIEYVEAIFTVLQYLFAFSVSDFMP-CLRGLDLDGHERIIKKACKIMKKYHDPIIE 278
G E ++E IF L L FS D++ L G ++DG E K +++ Y++PII+
Sbjct: 220 LGKAEKHHLEVIFNTLNCLPGFSPVDYVDRWLGGWNIDGEEERAKVNVNLVRSYNNPIID 279
Query: 279 DRIQQWK-NGKKIEKEDLLDVLISLKD----------------------AVDNPSNAVEW 315
+R++ W+ G K ED LD I+LKD A+DNP+N +EW
Sbjct: 280 ERVEIWREKGGKAAVEDWLDTFITLKDQNGNYLVTPDEIKAQCVEFCIAAIDNPANNMEW 339
Query: 316 GLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPH 375
L E++ PE+L+KA +ELD VVGK RLVQE D LNY+KAC +E FR HP + PH
Sbjct: 340 TLGEMLKNPEILRKALKELDEVVGKDRLVQESDIRNLNYLKACCRETFRIHPSAHYVPPH 399
Query: 376 V 376
V
Sbjct: 400 V 400
>At1g01280 cytochrome P450, putative
Length = 510
Score = 166 bits (421), Expect = 2e-41
Identities = 116/390 (29%), Positives = 202/390 (51%), Gaps = 40/390 (10%)
Query: 10 LSTTLWYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRW 69
L++ LIL +L++++L + S K+++ LPPGP PI+GNL ++ P
Sbjct: 5 LASLFAVLILNVLLWRWL---KASACKAQR----LPPGPPRLPILGNLLQL---GPLPHR 54
Query: 70 IQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLT 129
+ D + +RLGNV IT +DP+ RE+ ++QD +F+SRP + + ++ G
Sbjct: 55 DLASLCDKYGPLVYLRLGNVDAITTNDPDTIREILLRQDDVFSSRPKTLAAVHLAYGCGD 114
Query: 130 TALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNV 189
AL P G WK+++++ L++ R + +R EEA ++R V+ + ++ G +N+
Sbjct: 115 VALAPMGPHWKRMRRICMEHLLTTKRLESFTTQRAEEARYLIRDVFKR-SETGKP--INL 171
Query: 190 SVAAQYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYV-EAIFTVLQYLFAFSVSDFMP 248
+S + + R+LL K++FG GS +E ++ +F +L ++ + D++P
Sbjct: 172 KEVLGAFSMNNVTRMLLGKQFFGPGSLVSPKEAQEFLHITHKLFWLLGVIY---LGDYLP 228
Query: 249 CLRGLDLDGHERIIKKACKIMKKYHDPIIED-RIQQWKNGKKIEKEDLLDVLISLKD--- 304
R +D G E+ ++ K + ++H II++ R + ++ K D +DVL+SL
Sbjct: 229 FWRWVDPSGCEKEMRDVEKRVDEFHTKIIDEHRRAKLEDEDKNGDMDFVDVLLSLPGENG 288
Query: 305 -------------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQ 345
A D + EW +AE I QP +++K EELD+VVG R+V
Sbjct: 289 KAHMEDVEIKALIQDMIAAATDTSAVTNEWAMAEAIKQPRVMRKIQEELDNVVGSNRMVD 348
Query: 346 EYDFPKLNYVKACAKEAFRRHPICDFNLPH 375
E D LNY++ +E FR HP F +PH
Sbjct: 349 ESDLVHLNYLRCVVRETFRMHPAGPFLIPH 378
>At5g07990 flavonoid 3'-hydroxylase - like protein
Length = 513
Score = 159 bits (402), Expect = 2e-39
Identities = 114/387 (29%), Positives = 181/387 (46%), Gaps = 43/387 (11%)
Query: 16 YLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMN 75
+L ++L FLI S +++ +LPPGP PWPI+GNLP M +P R + M+
Sbjct: 5 FLTILLATVLFLILRIFSHRRNRSHNNRLPPGPNPWPIIGNLPHM-GTKP-HRTLSAMVT 62
Query: 76 DLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPF 135
I +RLG V V+ + +A + DA FASRP + +++ Y P+
Sbjct: 63 TYGP-ILHLRLGFVDVVVAASKSVAEQFLKIHDANFASRPPNSGAKHMAYNYQDLVFAPY 121
Query: 136 GEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDG-IVNVSVAAQ 194
G +W+ ++K+ S L S + R EE + R + TK G +VN+ V
Sbjct: 122 GHRWRLLRKISSVHLFSAKALEDFKHVRQEEVGTLTRELVRVGTKPVNLGQLVNMCVV-- 179
Query: 195 YYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLD 254
+ + R ++ +R FG ++ E+ + ++ F++ DF+P L LD
Sbjct: 180 ----NALGREMIGRRLFGADADHKAD-----EFRSMVTEMMALAGVFNIGDFVPSLDWLD 230
Query: 255 LDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA--------- 305
L G +K+ K + I+++ NG+ + D+L LISLK
Sbjct: 231 LQGVAGKMKRLHKRFDAFLSSILKEHEM---NGQDQKHTDMLSTLISLKGTDLDGDGGSL 287
Query: 306 ----------------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDF 349
D ++ V+W +AELI P+++ KA EELD VVG+ R V E D
Sbjct: 288 TDTEIKALLLNMFTAGTDTSASTVDWAIAELIRHPDIMVKAQEELDIVVGRDRPVNESDI 347
Query: 350 PKLNYVKACAKEAFRRHPICDFNLPHV 376
+L Y++A KE FR HP +LPH+
Sbjct: 348 AQLPYLQAVIKENFRLHPPTPLSLPHI 374
>At4g31940 Cytochrome P450-like protein
Length = 524
Score = 141 bits (356), Expect = 5e-34
Identities = 109/390 (27%), Positives = 192/390 (48%), Gaps = 38/390 (9%)
Query: 10 LSTTLWYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPT-PWPIVGNLPEMLANRPT-- 66
+ T+L+ L + +LV+ F+ + KS+KPK P P+ WPI+G+L +L +
Sbjct: 1 MDTSLFSLFVPILVFVFIALFK----KSKKPKYVKAPAPSGAWPIIGHL-HLLGGKEQLL 55
Query: 67 FRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNG 126
+R + KM + ++ ++LG+ +S E+A++ D ASRP + + +++
Sbjct: 56 YRTLGKMADHYGPAMS-LQLGSNEAFVVSSFEVAKDCFTVNDKALASRPMTAAAKHMGYN 114
Query: 127 YLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGI 186
+ P+ W++++K+ + EL+S R + L RV E V+ +Y+ K GG
Sbjct: 115 FAVFGFAPYSAFWREMRKIATIELLSNRRLQMLKHVRVSEITMGVKDLYSLWFKNGGTKP 174
Query: 187 VNVSVAA--QYYSGDVIRRLLLNKRYFGNGSEDYGPGLEE-IEYVEAIFTVLQYLFAFSV 243
V V + + + + ++I R++ KRYFG G EE ++ +AI + F+V
Sbjct: 175 VMVDLKSWLEDMTLNMIVRMVAGKRYFGGGGSVSSEDTEEAMQCKKAIAKFFHLIGIFTV 234
Query: 244 SDFMPCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWK-NGKKIEKEDLLDVLISL 302
SD P L DL GHE+ +K+ + + IE+ QQ K +G K D +DV++SL
Sbjct: 235 SDAFPTLSFFDLQGHEKEMKQTGSELDVILERWIENHRQQRKFSGTKENDSDFIDVMMSL 294
Query: 303 KD-------------------------AVDNPSNAVEWGLAELINQPELLKKATEELDSV 337
+ D ++ + W ++ L+N E+LKKA +E+D
Sbjct: 295 AEQGKLSHLQYDANTSIKSTCLALILGGSDTSASTLTWAISLLLNNKEMLKKAQDEIDIH 354
Query: 338 VGKGRLVQEYDFPKLNYVKACAKEAFRRHP 367
VG+ R V++ D L Y++A KE R +P
Sbjct: 355 VGRDRNVEDSDIENLVYLQAIIKETLRLYP 384
>At4g31970 Cytochrome P450-like protein
Length = 523
Score = 139 bits (351), Expect = 2e-33
Identities = 106/389 (27%), Positives = 192/389 (49%), Gaps = 37/389 (9%)
Query: 10 LSTTLWYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPT-PWPIVGNLPEMLANRPT-- 66
+ T+L+ L + +LV+ F+ + KS+KPK P P+ WPI+G+L +L+ +
Sbjct: 1 MDTSLFSLFVPILVFVFIALFK----KSKKPKHVKAPAPSGAWPIIGHL-HLLSGKEQLL 55
Query: 67 FRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNG 126
+R + KM + ++ +RLG+ +S E+A++ D ASRP + + +++
Sbjct: 56 YRTLGKMADQYGPAMS-LRLGSSETFVVSSFEVAKDCFTVNDKALASRPITAAAKHMGYD 114
Query: 127 YLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGI 186
P+ W++++K+ + EL+S R + L RV E +++ +Y+ K GG
Sbjct: 115 CAVFGFAPYSAFWREMRKIATLELLSNRRLQMLKHVRVSEISMVMQDLYSLWVKKGGSEP 174
Query: 187 VNVSVAA--QYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVS 244
V V + + + S +++ R++ KRYFG GS E + + + + F+VS
Sbjct: 175 VMVDLKSWLEDMSLNMMVRMVAGKRYFGGGSLSPEDAEEARQCRKGVANFFHLVGIFTVS 234
Query: 245 DFMPCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWK-NGKKIEKEDLLDVLISL- 302
D P L D GHE+ +K+ + + + IE+ QQ K +G K D +DV++SL
Sbjct: 235 DAFPKLGWFDFQGHEKEMKQTGRELDVILERWIENHRQQRKVSGTKHNDSDFVDVMLSLA 294
Query: 303 ---------KDAV---------------DNPSNAVEWGLAELINQPELLKKATEELDSVV 338
DA+ + + + W ++ L+N ++LKKA +E+D V
Sbjct: 295 EQGKFSHLQHDAITSIKSTCLALILGGSETSPSTLTWAISLLLNNKDMLKKAQDEIDIHV 354
Query: 339 GKGRLVQEYDFPKLNYVKACAKEAFRRHP 367
G+ R V++ D L Y++A KE R +P
Sbjct: 355 GRDRNVEDSDIENLVYIQAIIKETLRLYP 383
>At4g12300 flavonoid 3',5'-hydroxylase -like protein
Length = 516
Score = 135 bits (340), Expect = 4e-32
Identities = 104/365 (28%), Positives = 166/365 (44%), Gaps = 46/365 (12%)
Query: 40 PKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEI 99
P+P LPPGP PIVGNLP + P + + I + LG+ I ++ P +
Sbjct: 38 PQPSLPPGPRGLPIVGNLPFL---DPDLHTYFANLAQSHGPIFKLNLGSKLTIVVNSPSL 94
Query: 100 ARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHKWL 159
ARE+ QD F++R + T G + TP+G +W++++K+ +L+S
Sbjct: 95 AREILKDQDINFSNRDVPLTGRAATYGGIDIVWTPYGAEWRQLRKICVLKLLSRKTLDSF 154
Query: 160 HDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNGSEDYG 219
++ R +E RY+Y + K V V Q + + L +N + G+ +
Sbjct: 155 YELRRKEVRERTRYLYEQGRKQSP-----VKVGDQLFL--TMMNLTMNMLWGGSVKAEE- 206
Query: 220 PGLEEI--EYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDPII 277
+E + E+ I + + L +VSDF P L DL G + K + + D ++
Sbjct: 207 --MESVGTEFKGVISEITRLLSEPNVSDFFPWLARFDLQG----LVKRMGVCARELDAVL 260
Query: 278 EDRIQQWK---NGKKIEKEDLLDVLISLKD------------------------AVDNPS 310
+ I+Q K E +D L L+ LKD D +
Sbjct: 261 DRAIEQMKPLRGRDDDEVKDFLQYLMKLKDQEGDSEVPITINHVKALLTDMVVGGTDTST 320
Query: 311 NAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICD 370
N +E+ +AEL++ PEL+K+A EELD VVGK +V+E +L Y+ A KE R HP
Sbjct: 321 NTIEFAMAELMSNPELIKRAQEELDEVVGKDNIVEESHITRLPYILAIMKETLRLHPTLP 380
Query: 371 FNLPH 375
+PH
Sbjct: 381 LLVPH 385
>At5g67310 cytochrome P450
Length = 507
Score = 131 bits (330), Expect = 5e-31
Identities = 87/363 (23%), Positives = 169/363 (45%), Gaps = 43/363 (11%)
Query: 39 KPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPE 98
K K LPP P +P++G+L L P R ++ + +L D+ +RLG+ + ++
Sbjct: 37 KQKKNLPPNPVGFPVIGHLH--LLKEPVHRSLRDLSRNLGIDVFILRLGSRRAVVVTSAS 94
Query: 99 IARELCIKQ-DAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLRHK 157
A E +Q D +FA+RP + EY+ + P+GE W+++++ + +++S R +
Sbjct: 95 AAEEFLSQQNDVVFANRPLATLTEYMGYNNTLVSTAPYGEHWRRLRRFCAVDILSTARLR 154
Query: 158 WLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNGSED 217
D R +E ++R + + GG + + + +++ ++ KR
Sbjct: 155 DFSDIRRDEVRAMIRKINVELVTSGGSVRLKLQPFLYGLTYNILMSMVAGKR-------- 206
Query: 218 YGPGLEEIEYVEAIFTVLQYLFAFS----VSDFMPCLRGLDLDGHERIIKKACKIMKKYH 273
EE E + + +++ +F F+ V DF+P L+ DLDG+ + KK + K+
Sbjct: 207 -----EEDEETKEVRKLIREVFDFAGVNYVGDFLPTLKLFDLDGYRKRAKKLASKLDKFM 261
Query: 274 DPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA---------------------VDNPSNA 312
++++ + GK ++ ++ L+SL+++ D +
Sbjct: 262 QKLVDEHRKN--RGKAELEKTMITRLLSLQESEPECYTDDIIKGLVQVMLLAGTDTTAVT 319
Query: 313 VEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFN 372
+EW +A L+N PE+L+K EL+ V +GR+ +E D K Y+ E R P
Sbjct: 320 LEWAMANLLNHPEVLRKLKTELNEVSKEGRVFEESDTGKCPYLNNVISETLRLFPAAPLL 379
Query: 373 LPH 375
+PH
Sbjct: 380 VPH 382
>At4g12330 flavonoid 3',5'-hydroxylase like protein
Length = 518
Score = 130 bits (326), Expect = 2e-30
Identities = 107/405 (26%), Positives = 180/405 (44%), Gaps = 61/405 (15%)
Query: 2 EYLVVSDQLSTTLWYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEML 61
++++ + +S+ WY+ + + KS++ P LPPGP PIVGNLP +
Sbjct: 18 DFVLATIVISSVFWYIWVYV--------------KSKRLFPPLPPGPRGLPIVGNLPFL- 62
Query: 62 ANRPTFRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNE 121
P + + + + LG I I+ E R++ D IFA+ +
Sbjct: 63 --HPELHTYFHSLAQKHGPVFKLWLGAKLTIVITSSEATRDILRTNDVIFANDDVPVAGS 120
Query: 122 YVTNGYLTTALTPFGEQWKKVKKVISNELVS--PLRHKWLHDKRVEEADNIVRYVYNKC- 178
T G + +P+G +W ++K+ N+++S L R +E VRY+ ++
Sbjct: 121 LSTYGGVDIVWSPYGPEWPMLRKICINKMLSNATLDSNSFSALRRQETRRTVRYLADRAR 180
Query: 179 ----TKIGGDGIVNV-SVAAQYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFT 233
+G V + +V Q G+ + ++D E++E I
Sbjct: 181 AGLAVNVGEQIFVTILNVVTQMLWGETV-------------ADDEEREKVGAEFLELITE 227
Query: 234 VLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKE 293
++ + +VSDF P L DL G + ++++ + M + D II R+ K G K
Sbjct: 228 IIDVVGKPNVSDFFPVLSRFDLQGLAKRVRRSAQRMDRMFDRIISQRMGMDK-GSKGNGG 286
Query: 294 DLLDVLISLKDAVDNPS--------------------NAVEWGLAELINQPELLKKATEE 333
D L VL++ KD +N S N +E+ +AELIN+ E++K+A +E
Sbjct: 287 DFLMVLLNAKDEDENMSMNHVKALLMDMVLGGTDTSLNTIEFAMAELINKLEIMKRAQQE 346
Query: 334 LDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLPHVRC 378
LD VVGK +V+E KL Y+ + KE R HP +P RC
Sbjct: 347 LDKVVGKNNIVEEKHITKLPYILSIMKETLRLHPALPLLIP--RC 389
>At4g37430 cytochrome P450 monooxygenase (CYP91A2)
Length = 500
Score = 126 bits (317), Expect = 2e-29
Identities = 100/387 (25%), Positives = 185/387 (46%), Gaps = 54/387 (13%)
Query: 17 LILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMND 76
L+ +++ YKFL S+ + LPPGP P VG+L L P R +Q+ N
Sbjct: 9 LLFLVISYKFLY--------SKTQRFNLPPGPPSRPFVGHLH--LMKPPIHRLLQRYSNQ 58
Query: 77 LNTDIACIRLGNVHVITISDPEIARELCIKQ-DAIFASRPSSWSNEYVTNGYLTTALTPF 135
I +R G+ V+ I+ P +A+E Q D + +SRP + +YV + T P+
Sbjct: 59 YGP-IFSLRFGSRRVVVITSPSLAQESFTGQNDIVLSSRPLQLTAKYVAYNHTTVGTAPY 117
Query: 136 GEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYV--YNKCTKIGGDGIVNVSVAA 193
G+ W+ ++++ S E++S H+ ++ + + + D I+R + ++ T+ + +
Sbjct: 118 GDHWRNLRRICSQEILSS--HRLINFQHIRK-DEILRMLTRLSRYTQTSNESNDFTHIEL 174
Query: 194 QYYSGDV----IRRLLLNKRYFGNGSEDYGPGLEEIE-YVEAIFTVLQYLFAFSVSDFMP 248
+ D+ I R++ KRY+G+ + EE E + + ++ + Y A +D++P
Sbjct: 175 EPLLSDLTFNNIVRMVTGKRYYGDDVNNK----EEAELFKKLVYDIAMYSGANHSADYLP 230
Query: 249 CLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKD---- 304
L+ L G++ +K K + K D I++ + + + K E +++ LISL+
Sbjct: 231 ILK---LFGNK--FEKEVKAIGKSMDDILQRLLDECRRDK--EGNTMVNHLISLQQQQPE 283
Query: 305 -----------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEY 347
+ + +EW +A L+ PE+L+KA E+D +GK RL+ E
Sbjct: 284 YYTDVIIKGLMMSMMLAGTETSAVTLEWAMANLLRNPEVLEKARSEIDEKIGKDRLIDES 343
Query: 348 DFPKLNYVKACAKEAFRRHPICDFNLP 374
D L Y++ E FR P+ F +P
Sbjct: 344 DIAVLPYLQNVVSETFRLFPVAPFLIP 370
>At5g44620 flavonoid 3',5'-hydroxylase-like; cytochrome P450
Length = 519
Score = 125 bits (314), Expect = 4e-29
Identities = 95/365 (26%), Positives = 167/365 (45%), Gaps = 40/365 (10%)
Query: 36 KSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMMNDLNTDIACIRLGNVHVITIS 95
K ++ P LPPGP PI+GNLP + +P + + + I + LG I ++
Sbjct: 38 KCKRRSPPLPPGPWGLPIIGNLPFL---QPELHTYFQGLAKKHGPIFKLWLGAKLTIVVT 94
Query: 96 DPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLR 155
E+A+E+ D IFA+ T G +P+G +W+ ++K+ N ++
Sbjct: 95 SSEVAQEILKTNDIIFANHDVPAVGPVNTYGGTEIIWSPYGPKWRMLRKLCVNRILRNAM 154
Query: 156 HKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLN---KRYFG 212
D R E VRY+ ++ ++G VN+ G+ I ++LN + +G
Sbjct: 155 LDSSTDLRRRETRQTVRYLADQA-RVGSP--VNL--------GEQIFLMMLNVVTQMLWG 203
Query: 213 NGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKY 272
++ + E++E I + L ++SDF P L DL G + +++ + M +
Sbjct: 204 TTVKEEEREVVGAEFLEVIREMNDLLLVPNISDFFPVLSRFDLQGLAKRMRRPAQRMDQM 263
Query: 273 HDPIIEDRIQQWKNGKKIEKEDLLDVLISLKD-----------------------AVDNP 309
D II R+ ++ D LDVL+ +KD D
Sbjct: 264 FDRIINQRLGMDRDSSDGRAVDFLDVLLKVKDEEAEKTKLTMNDVKAVLMDMVLGGTDTS 323
Query: 310 SNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPIC 369
+ +E+ +AEL++ P+++K+A +E+D VVGK ++V+E KL Y+ A KE R H +
Sbjct: 324 LHVIEFAMAELLHNPDIMKRAQQEVDKVVGKEKVVEESHISKLPYILAIMKETLRLHTVA 383
Query: 370 DFNLP 374
+P
Sbjct: 384 PLLVP 388
>At4g37320 cytochrome P450-like protein
Length = 495
Score = 124 bits (311), Expect = 9e-29
Identities = 96/386 (24%), Positives = 173/386 (43%), Gaps = 39/386 (10%)
Query: 19 LMLLVYKFLIKHQRSTQKSEKPKPKLPPGPT-PWPIVGNLPEMLANRPTFRWIQKMMNDL 77
++LL + FL + KP LPP P P P++G+L L +P R + L
Sbjct: 6 ILLLSFLFLTISIKLLLTKSNRKPNLPPSPAYPLPVIGHLH--LLKQPVHRTFHSISKSL 63
Query: 78 -NTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFG 136
N I +RLGN V IS IA E K D + A+RP ++V + +G
Sbjct: 64 GNAPIFHLRLGNRLVYVISSHSIAEECFTKNDVVLANRPDIIMAKHVGYNFTNMIAASYG 123
Query: 137 EQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYY 196
+ W+ ++++ + E+ S R R +E ++ ++ + G V +
Sbjct: 124 DHWRNLRRIAAVEIFSSHRISTFSSIRKDEIRRLITHLSRD--SLHGFVEVELKSLLTNL 181
Query: 197 SGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYV-EAIFTVLQYLFAFSVSDFMPCLRGLDL 255
+ + I ++ KRY+G G+ED +E + V E I ++ + +++D++P + +
Sbjct: 182 AFNNIIMMVAGKRYYGTGTEDN----DEAKLVRELIAEIMAGAGSGNLADYLPSINWVTN 237
Query: 256 DGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKKIEKEDLLDVLISLKDA---------- 305
+ KI+ D +++ + + K +K + + L+D L+S ++
Sbjct: 238 ------FENQTKILGNRLDRVLQKLVDE-KRAEKEKGQTLIDHLLSFQETEPEYYTDVII 290
Query: 306 -----------VDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNY 354
D S +EW ++ L+N PE+L+KA E+D +G RLV+E D L+Y
Sbjct: 291 KGIILALVLAGTDTSSVTLEWAMSNLLNHPEILEKARAEIDDKIGSDRLVEESDIVNLHY 350
Query: 355 VKACAKEAFRRHPICDFNLPHVRCNE 380
++ E R +P LPH +E
Sbjct: 351 LQNIVSETLRLYPAVPLLLPHFSSDE 376
>At5g06900 cytochrome P450
Length = 507
Score = 123 bits (309), Expect = 1e-28
Identities = 97/378 (25%), Positives = 173/378 (45%), Gaps = 41/378 (10%)
Query: 15 WYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPTPWPIVGNLPEMLANRPTFRWIQKMM 74
++ +++L+ + Q T + P LPP PT PI+G++ + P +
Sbjct: 6 YFSVIILVCLGITVLIQAITNRLRDRLP-LPPSPTALPIIGHIHLL---GPIAHQALHKL 61
Query: 75 NDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTP 134
+ + + +G++ + +S E+A E+ + F +RP+ + +Y+T G P
Sbjct: 62 SIRYGPLMYLFIGSIPNLIVSSAEMANEILKSNELNFLNRPTMQNVDYLTYGSADFFSAP 121
Query: 135 FGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQ 194
+G WK +K++ EL S R EE ++ V K + VN+ +
Sbjct: 122 YGLHWKFMKRICMVELFSSRALDSFVSVRSEELKKLLIRVLKKAE---AEESVNLGEQLK 178
Query: 195 YYSGDVIRRLLLNKRYFGNGSEDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLD 254
+ ++I R++ K D G + E ++ + + + F+VS+ L+ LD
Sbjct: 179 ELTSNIITRMMFRKM-----QSDSDGGEKSEEVIKMVVELNELAGFFNVSETFWFLKRLD 233
Query: 255 LDGHERIIKKACKIMKKYHDPIIEDRIQQWKNGKK--IEKEDLLDVLISLKD-------- 304
L G +KK K + +D IIE +++ ++ KK + ++LDVL+ + +
Sbjct: 234 LQG----LKKRLKNARDKYDVIIERIMEEHESSKKNATGERNMLDVLLDIYEDKNAEMKL 289
Query: 305 ---------------AVDNPSNAVEWGLAELINQPELLKKATEELDSVVGKGRLVQEYDF 349
D + VEW LAELIN PE++KKA +E++ VVG R+V+E D
Sbjct: 290 TRENIKAFIMNIYGGGTDTSAITVEWALAELINHPEIMKKAQQEIEQVVGNKRVVEESDL 349
Query: 350 PKLNYVKACAKEAFRRHP 367
L+Y +A KE R HP
Sbjct: 350 CNLSYTQAVVKETMRLHP 367
>At4g31950 cytochrome P450-like protein
Length = 512
Score = 123 bits (309), Expect = 1e-28
Identities = 106/390 (27%), Positives = 186/390 (47%), Gaps = 50/390 (12%)
Query: 10 LSTTLWYLILMLLVYKFLIKHQRSTQKSEKPKPKLPPGPT-PWPIVGNLPEMLANRPT-- 66
+ T+L+ L + +LV+ F+ + KS+KPK P P+ WPI+G+L +L +
Sbjct: 1 MDTSLFSLFVSILVFVFIALFK----KSKKPKYVKAPAPSGAWPIIGHL-HLLGGKEQLL 55
Query: 67 FRWIQKMMNDLNTDIACIRLGNVHVITISDPEIARELCIKQDAIFASRPSSWSNEYVTNG 126
+R + KM + ++ +RLG+ S E+A++ D AS ++ + G
Sbjct: 56 YRTLGKMADHYGPAMS-LRLGSSETFVGSSFEVAKDCFTVNDKALASLMTAAAKHM---G 111
Query: 127 YLTTALTPFGEQWKKVKKVISNELVSPLRHKWLHDKRVEEADNIVRYVYNKCTKIGGDGI 186
Y+ W +++K+ EL+S R + L++ RV E V+ +Y+ K GG
Sbjct: 112 YVF---------WLEMRKIAMIELLSNRRLQMLNNVRVSEISMGVKDLYSLWVKKGGSEP 162
Query: 187 VNVSVAA--QYYSGDVIRRLLLNKRYFGNGSEDYGPGLEEI-EYVEAIFTVLQYLFAFSV 243
V V + + + ++I R++ KRYFG G + EE ++ + I + F+V
Sbjct: 163 VMVDLKSWLEDMIANMIMRMVAGKRYFGGGGAESSEHTEEARQWRKGIAKFFHLVGIFTV 222
Query: 244 SDFMPCLRGLDLDGHERIIKKACKIMKKYHDPIIEDRIQQWK-NGKKIEKEDLLDVLISL 302
SD P L LDL GHE+ +K+ + + + IE+ QQ K +G K D +DV++SL
Sbjct: 223 SDAFPKLGWLDLQGHEKEMKQTRRELDVILERWIENHRQQRKVSGTKHNDSDFVDVMLSL 282
Query: 303 KD-------------------------AVDNPSNAVEWGLAELINQPELLKKATEELDSV 337
+ + + + W ++ L+N ++LKK +E+D
Sbjct: 283 AEQGKLSHLQYDANTCIKTTCLALILGGSETSPSTLTWAISLLLNNKDMLKKVQDEIDIH 342
Query: 338 VGKGRLVQEYDFPKLNYVKACAKEAFRRHP 367
VG+ R V++ D L Y++A KE R +P
Sbjct: 343 VGRDRNVEDSDIKNLVYLQAIIKETLRLYP 372
>At4g37340 cytochrome P450-like protein
Length = 500
Score = 122 bits (306), Expect = 3e-28
Identities = 92/362 (25%), Positives = 176/362 (48%), Gaps = 38/362 (10%)
Query: 39 KPKPKLPPGPTPW--PIVGNLPEMLANRPTFRWIQKMMNDL-NTDIACIRLGNVHVITIS 95
K +P LPP P+ W P++G+L L P R + L + I +RLGN V +S
Sbjct: 25 KRRPNLPPSPS-WALPVIGHL--RLLKPPLHRVFLSVSESLGDAPIISLRLGNRLVFVVS 81
Query: 96 DPEIARELCIKQDAIFASRPSSWSNEYVTNGYLTTALTPFGEQWKKVKKVISNELVSPLR 155
+A E K D + A+R +S ++++++ G T +G+ W+ ++++ + E+ S R
Sbjct: 82 SHSLAEECFTKNDVVLANRFNSLASKHISYGCTTVVTASYGDHWRNLRRIGAVEIFSAHR 141
Query: 156 HKWLHDKRVEEADNIVRYVYNKCTKIGGDGIVNVSVAAQYYSGDVIRRLLLNKRYFGNGS 215
R +E ++ + ++ + + + S+ + ++IR +L K Y+G+G+
Sbjct: 142 LNSFSSIRRDEIHRLIACL-SRNSSLEFTKVEMKSMFSNLTFNNIIR-MLAGKCYYGDGA 199
Query: 216 EDYGPGLEEIEYVEAIFTVLQYLFAFSVSDFMPCLRGLDLDGHERIIKKACKIMKKYHDP 275
ED P + + E I + A + +D++P L + G E+ IKK + ++
Sbjct: 200 ED-DPEAKRVR--ELIAEGMGCFGAGNTADYLPILTWIT--GSEKRIKKIASRLDEFLQG 254
Query: 276 IIEDRIQQWKNGKKIEKEDLLDVLISLKDA---------------------VDNPSNAVE 314
++++R + GK+ + ++D L+ L++ D + +E
Sbjct: 255 LVDER----REGKEKRQNTMVDHLLCLQETQPEYYTDNIIKGIMLSLILAGTDTSAVTLE 310
Query: 315 WGLAELINQPELLKKATEELDSVVGKGRLVQEYDFPKLNYVKACAKEAFRRHPICDFNLP 374
W L+ L+N P++L KA +E+D+ VG RLV+E D L Y++ E+ R +P +P
Sbjct: 311 WTLSALLNHPQILSKARDEIDNKVGLNRLVEESDLSHLPYLQNIVSESLRLYPASPLLVP 370
Query: 375 HV 376
HV
Sbjct: 371 HV 372
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,495,917
Number of Sequences: 26719
Number of extensions: 436429
Number of successful extensions: 1931
Number of sequences better than 10.0: 195
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 1391
Number of HSP's gapped (non-prelim): 249
length of query: 384
length of database: 11,318,596
effective HSP length: 101
effective length of query: 283
effective length of database: 8,619,977
effective search space: 2439453491
effective search space used: 2439453491
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146720.6


