

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146719.4 - phase: 0 /pseudo
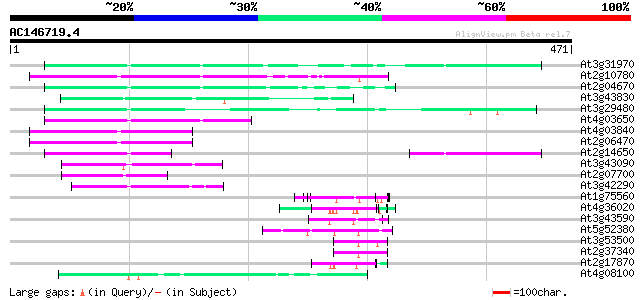
(471 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g31970 hypothetical protein 101 8e-22
At2g10780 pseudogene 99 5e-21
At2g04670 putative retroelement pol polyprotein 80 3e-15
At3g43830 putative protein 79 4e-15
At3g29480 hypothetical protein 79 4e-15
At4g03650 putative reverse transcriptase 74 2e-13
At4g03840 putative transposon protein 65 1e-10
At2g06470 putative retroelement pol polyprotein 64 2e-10
At2g14650 putative retroelement pol polyprotein 62 5e-10
At3g43090 putative protein 58 1e-08
At2g07700 putative retroelement pol polyprotein 58 1e-08
At3g42290 putative protein 56 4e-08
At1g75560 DNA-binding protein 54 1e-07
At4g36020 glycine-rich protein 54 2e-07
At3g43590 putative protein 52 5e-07
At5g52380 unknown protein 51 1e-06
At3g53500 splicing factor - like protein 51 2e-06
At2g37340 unknown protein 50 3e-06
At2g17870 putative glycine-rich, zinc-finger DNA-binding protein 50 3e-06
At4g08100 putative polyprotein 50 4e-06
>At3g31970 hypothetical protein
Length = 1329
Score = 101 bits (252), Expect = 8e-22
Identities = 103/418 (24%), Positives = 163/418 (38%), Gaps = 49/418 (11%)
Query: 30 VRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 89
+RM+E R F G P+ A W VE F +C +V H L +A W
Sbjct: 144 LRMMEQLQRIGTRYFFGGTSPEEADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLW 203
Query: 90 WVSLLPILEQDGAAVTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVE 149
W S+ Q A ++W F EF YFP++ + E FLE+ QG+ SV EY +F
Sbjct: 204 WRSVTARRRQ--AHMSWADFVAEFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNR 261
Query: 150 LAKFYPHYTTETAEFSKCIKFENGLRADIKRAIGYEKIRIFSELVSSCRIYEEDTKAHYK 209
L + + + ++ +F GLR D++ + + LV + EED +
Sbjct: 262 LLVYAGRGMQD--DQAQMRRFLRGLRPDLRVRCRVLQYATKAALVETAAEVEEDLQRQVV 319
Query: 210 VMSERRGKGQ*SRPKPYSAPTDKGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDR 269
+S + + + AP+ GK +R+ E+GH C +
Sbjct: 320 GVSP---AVKPKKTQQQVAPSKGGKPAQGQKRK----------------ERGHLRPNCPK 360
Query: 270 DEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPKKVRTGGKVFALTGTQTVN 329
++ Q + G V + HI++ P+ T
Sbjct: 361 LQRMAVAVVQPA-----VQHGAQVQQRVQQLAHIAA---APQGYTT-------------- 398
Query: 330 EDRLIRGTCFFNSTHLIAIIDTGATHCFIVVDCAYKLGLIVSDMNGEMVVETPAKGSVTT 389
R I T T + D+GA+HCFI + A + G I D ++ A G
Sbjct: 399 --REIGSTSKRAITEAHVLFDSGASHCFITPESASR-GNIRGDPGEQLGAVKVAGGQFLA 455
Query: 390 SLVCLSCL-LSMFGRDFEVDLVCLPLSGVDVIFGMNWLEYNHFHINCFSKTVHFSSAE 446
L + + + G VDL+ P+ DVI GM+WL++ H++C V F E
Sbjct: 456 VLGRAKGVDIQIAGESMPVDLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPE 513
>At2g10780 pseudogene
Length = 1611
Score = 99.0 bits (245), Expect = 5e-21
Identities = 85/315 (26%), Positives = 134/315 (41%), Gaps = 27/315 (8%)
Query: 17 VGQQPNAGAGN-DGVRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKV 75
V QPN G D + +LE R F G DP A +W ++R F+ +C E +
Sbjct: 154 VNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQR 213
Query: 76 RFGTHMLAEEADDWWVSLLPILEQDGAAV-TWVVFRREFLNMYFPEDVRGKKEIEFLEMK 134
H L +A +WW L + ++ G V ++ F EF YFP + + E +L++
Sbjct: 214 DIAVHFLEGDAHNWW---LTVEKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLV 270
Query: 135 QGDMSVTEYAAKFVELAKFYPHYTTETAEFSKCIKFENGLRADIKRAIGYEKIRIFSELV 194
QG+ +V EY +F L ++ E E ++ +F GLR +I+ SELV
Sbjct: 271 QGNRTVREYDEEFNRLRRYVGRELEE--EQAQLRRFIRGLRIEIRNHCLVRTFNSVSELV 328
Query: 195 SSCRIYEEDTKAHYKVMSERRGKGQ*SRPKPYSAPTDKGKQRLNDERRPKRRDAPTEIIC 254
+ EE + + E+ R + P D K+R D+ + DA T C
Sbjct: 329 ERAAMIEEGIEEERYLNREKAP----IRNNQSTKPAD--KKRKFDKVDNTKSDAKTG-EC 381
Query: 255 FKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRGDV-----------VCYNCNEEGHI 303
CG K H S C + C RCG K H + C + + C+ C + GH+
Sbjct: 382 VTCG-KNH-SGTCWKAIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFYCGKTGHL 439
Query: 304 SSQCTQPKKVRTGGK 318
+C + + G+
Sbjct: 440 KRECPKLTAEKQAGQ 454
Score = 36.2 bits (82), Expect = 0.040
Identities = 17/45 (37%), Positives = 23/45 (50%)
Query: 402 GRDFEVDLVCLPLSGVDVIFGMNWLEYNHFHINCFSKTVHFSSAE 446
G + DL+ +PL DVI GM+WL HI+C + F E
Sbjct: 530 GVNMPADLIIVPLKKHDVILGMDWLGKYKAHIDCHRGRIQFERDE 574
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 79.7 bits (195), Expect = 3e-15
Identities = 75/295 (25%), Positives = 116/295 (38%), Gaps = 41/295 (13%)
Query: 30 VRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 89
+RM+E R F P+ A W V+R F +C +V H L +A W
Sbjct: 124 LRMMEQLQRIGTWYFSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLW 183
Query: 90 WVSLLPILEQDGAAVTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVE 149
W S+ Q A ++W F EF YFP++ + E FLE+ QG+ SV EY +F
Sbjct: 184 WRSVTARRRQ--ADMSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNR 241
Query: 150 LAKFYPHYTTETAEFSKCIKFENGLRADIKRAIGYEKIRIFSELVSSCRIYEEDTKAHYK 209
L + + + ++ +F GLR D++ + + LV + EED +
Sbjct: 242 LLAYAGRGMED--DQAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVEEDLQRQVV 299
Query: 210 VMSERRGKGQ*SRPKPYSAPTDKGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDR 269
+S Q + P+ GK +R+ D P+ + G+ G
Sbjct: 300 GVSPAV---QTKNTQQQVTPSKGGKPAQGQKRK---WDHPS-----RAGQGGRAGY---- 344
Query: 270 DEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPKKVRTGGKVFALTG 324
F CG HT+AD C E GH+ + GG+ A+ G
Sbjct: 345 -----FSCGSLDHTVAD----------CTERGHLHVKV-------AGGQFLAVLG 377
Score = 38.5 bits (88), Expect = 0.008
Identities = 16/49 (32%), Positives = 25/49 (50%)
Query: 398 LSMFGRDFEVDLVCLPLSGVDVIFGMNWLEYNHFHINCFSKTVHFSSAE 446
+ + G DL+ P+ DVI GM+WL++ H++C V F E
Sbjct: 384 IQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPE 432
>At3g43830 putative protein
Length = 329
Score = 79.3 bits (194), Expect = 4e-15
Identities = 63/249 (25%), Positives = 94/249 (37%), Gaps = 46/249 (18%)
Query: 43 TFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILEQDGA 102
TF G DP A W +E F+ M+C + L EA +WW + EQ G
Sbjct: 115 TFNGGTDPVKAYNWRNMLECKFKSMRCPVEFWRDLASCYLRGEAQEWWERVKQ-REQVGC 173
Query: 103 AVTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVELAKFYPHYTTETA 162
W F+ EF Y PE+ E++FL ++QG +V +Y +F L +F +
Sbjct: 174 VDQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVRKYEKEFHSLERF---ERRKRG 230
Query: 163 EFSKCIKFENGLRADIK---RAIGYEKIRIFSELVSSCRIYEEDTKAHYKVMSERRGKGQ 219
E KF +GLR DI+ ++ + E +S I E+ + ++ E+ G
Sbjct: 231 EHELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLEIGLEEEARYKRIAQEKEAMG- 289
Query: 220 *SRPKPYSAPTDKGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDRDEKKCFRCGQ 279
G+ GH C E C+RCG
Sbjct: 290 ------------------------------------SYGQTGHSKRRC--QEVTCYRCGV 311
Query: 280 KGHTLADCK 288
GH DC+
Sbjct: 312 AGHIARDCR 320
>At3g29480 hypothetical protein
Length = 718
Score = 79.3 bits (194), Expect = 4e-15
Identities = 95/428 (22%), Positives = 142/428 (32%), Gaps = 119/428 (27%)
Query: 30 VRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 89
+RM+E R F G P+ A W VER F +C +V H L +A W
Sbjct: 37 LRMMEQLQRIGIGYFSGGTSPEEADSWRSRVERNFGSSRCPVEYRVDLAVHFLEGDAHLW 96
Query: 90 WVSLLPILEQDGAAVTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVE 149
W S+ Q A ++W F EF YFP++ + E FLE+ QG+ SV EY +F
Sbjct: 97 WKSVTTRRRQ--ANMSWADFVAEFNAKYFPQEALDRMEAHFLELTQGERSVREYDREF-- 152
Query: 150 LAKFYPHYTTETAEFSKCIKFENGLRADIKRAIGYEKIRIFSELVSSCRIYEEDTKAHYK 209
++ +++E S+ + K H +
Sbjct: 153 -----------------------------------NRLLVYAECRSTRIPVVQPKKTHQQ 177
Query: 210 VMSERRGKGQ*SRPKPYSAPTDKGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDR 269
V + GK + + + P+ G+ G G
Sbjct: 178 VAPSKGGKPAQGQKRSWDHPSRAGQG----------------------GRAG-------- 207
Query: 270 DEKKCFRCGQKGHTLADC-KRGDV-VCYNCNEEGHISSQCTQPKKVRTGGKVFALTGTQT 327
CF CG H +ADC +R + Y+C + H+ C PK R
Sbjct: 208 ----CFFCGSLDHKVADCTQRAETREYYHCRKRRHLRPNC--PKLQRMA----------- 250
Query: 328 VNEDRLIRGTCFFNSTHLIAIIDTGATHCFIVVDCAYKLGLIVSDMNGEMVVETPAKG-- 385
+A++ V +L I G E
Sbjct: 251 ------------------VAVVHAAVQQGAQVQQNVQQLAHIAPAPQGYTTREIGVTNNR 292
Query: 386 SVTTSLVCLSCLLSMFGRDFEVD-----------LVCLPLSGVDVIFGMNWLEYNHFHIN 434
++T V L++ GR VD L P DVI GM+WL+Y H++
Sbjct: 293 AITAVKVVGGQFLAVLGRAKGVDIQIARESMPANLFLSPEELYDVILGMDWLDYYRVHLD 352
Query: 435 CFSKTVHF 442
C V F
Sbjct: 353 CHRGRVSF 360
>At4g03650 putative reverse transcriptase
Length = 839
Score = 73.6 bits (179), Expect = 2e-13
Identities = 51/174 (29%), Positives = 77/174 (43%), Gaps = 4/174 (2%)
Query: 30 VRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 89
+RM++ R F G P+ A W +VER F +C +V H L +A W
Sbjct: 142 LRMMKQLQRIGTEYFSGGTSPEEADSWRSQVERNFGSSRCPAEYRVDLTVHFLEGDAHLW 201
Query: 90 WVSLLPILEQDGAAVTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVE 149
W S+ Q A ++W F EF YFP + + E FLE+ QG SV EY KF
Sbjct: 202 WRSVTARRRQ--ADMSWADFMAEFNAKYFPREALDRMEARFLELTQGVRSVREYDRKFNR 259
Query: 150 LAKFYPHYTTETAEFSKCIKFENGLRADIKRAIGYEKIRIFSELVSSCRIYEED 203
L + + + ++ +F GLR +++ + + LV + EED
Sbjct: 260 LLVYAGRGMED--DQAQMRRFLRGLRPNLRVRCRVSQYATKAALVETAAEVEED 311
Score = 35.0 bits (79), Expect = 0.089
Identities = 15/49 (30%), Positives = 24/49 (48%)
Query: 398 LSMFGRDFEVDLVCLPLSGVDVIFGMNWLEYNHFHINCFSKTVHFSSAE 446
+ + G DL+ + DVI GM+WL++ H++C V F E
Sbjct: 377 IQIAGESLPADLIISHVELYDVILGMDWLDHYRVHLDCHRGRVSFERPE 425
>At4g03840 putative transposon protein
Length = 973
Score = 64.7 bits (156), Expect = 1e-10
Identities = 42/139 (30%), Positives = 67/139 (47%), Gaps = 5/139 (3%)
Query: 17 VGQQPNAGAGN-DGVRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKV 75
V QPN G D + +LE R F G DP A +W ++R F+ +C E +
Sbjct: 119 VNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQR 178
Query: 76 RFGTHMLAEEADDWWVSLLPILEQDGAAV-TWVVFRREFLNMYFPEDVRGKKEIEFLEMK 134
H L +A +WW L + ++ G V ++ F EF YFP + + E +L++
Sbjct: 179 DIAVHFLERDAHNWW---LTVEKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLV 235
Query: 135 QGDMSVTEYAAKFVELAKF 153
QG+ +V EY +F L ++
Sbjct: 236 QGNRTVREYDEEFNRLRRY 254
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 63.9 bits (154), Expect = 2e-10
Identities = 42/139 (30%), Positives = 67/139 (47%), Gaps = 5/139 (3%)
Query: 17 VGQQPNAGAGN-DGVRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKV 75
V QPN G D + +LE R F G DP A +W ++R F+ +C E +
Sbjct: 119 VNPQPNHGRKRGDYLSLLEHVSRLGTRHFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQR 178
Query: 76 RFGTHMLAEEADDWWVSLLPILEQDGAAV-TWVVFRREFLNMYFPEDVRGKKEIEFLEMK 134
H L +A +WW L + ++ G V ++ F EF YFP + + E +L++
Sbjct: 179 DIAVHFLEGDAHNWW---LTVEKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLV 235
Query: 135 QGDMSVTEYAAKFVELAKF 153
QG+ +V EY +F L ++
Sbjct: 236 QGNRTVREYDEEFNRLRRY 254
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 62.4 bits (150), Expect = 5e-10
Identities = 35/107 (32%), Positives = 49/107 (45%), Gaps = 2/107 (1%)
Query: 30 VRMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDW 89
+RM+E R F G P+ A W VER F +C ++ H L +A W
Sbjct: 142 LRMMEQLQRIGTGYFSGGTSPEVADSWRSRVERNFGSSRCPAEYRIDLAVHFLEGDAHLW 201
Query: 90 WVSLLPILEQDGAAVTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQG 136
W S+ Q A ++W F EF YFP++ + E FLE+ QG
Sbjct: 202 WRSVTARRRQ--ADISWADFVAEFNAKYFPQEALDRMEAHFLELTQG 246
Score = 51.2 bits (121), Expect = 1e-06
Identities = 32/112 (28%), Positives = 49/112 (43%), Gaps = 2/112 (1%)
Query: 336 GTCFFNSTHLIAIIDTGATHCFIVVDCAYKLGLIVSDMNGEMVVETPAKGSVTTSLVCLS 395
GT + D+GA+HCFI + A + G I D ++ A G L
Sbjct: 304 GTLLVGGVEAHVLFDSGASHCFITPESASR-GNIRGDPGEQLGAVKVAGGQFVAVLGRTK 362
Query: 396 CL-LSMFGRDFEVDLVCLPLSGVDVIFGMNWLEYNHFHINCFSKTVHFSSAE 446
+ + + G DL+ P+ DVI GM+WL++ H++C V F E
Sbjct: 363 GVDIQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPE 414
>At3g43090 putative protein
Length = 357
Score = 57.8 bits (138), Expect = 1e-08
Identities = 41/138 (29%), Positives = 57/138 (40%), Gaps = 9/138 (6%)
Query: 44 FKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLL---PILEQD 100
F G DP A W K +E F +C + H L EEA WW +L P+
Sbjct: 131 FTGGIDPVKADDWRKLLENNFENARCPVEFQKDIIVHFLREEASHWWDGVLGNTPVQH-- 188
Query: 101 GAAVTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVELAKFYPHYTTE 160
+ W FR EF +FP++ E +F E++Q V E + L++F
Sbjct: 189 --VIFWEDFREEFNRKFFPQEAMDSLEDDFEELRQDTKKVRENERELSHLSRF--SVRAG 244
Query: 161 TAEFSKCIKFENGLRADI 178
E S + GLR DI
Sbjct: 245 RGEQSMIRRLMRGLRPDI 262
>At2g07700 putative retroelement pol polyprotein
Length = 543
Score = 57.8 bits (138), Expect = 1e-08
Identities = 28/89 (31%), Positives = 48/89 (53%), Gaps = 2/89 (2%)
Query: 44 FKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILEQDGAA 103
FKG ++ A KWL+++E F + +C + K + + L ++A W S+ G
Sbjct: 53 FKGEHNTVVADKWLRDLEMNFEISRCPDNFKRQIAVNFLDKDARIWRESVTA--RNQGQM 110
Query: 104 VTWVVFRREFLNMYFPEDVRGKKEIEFLE 132
++W FRREF YFP + R + E +F++
Sbjct: 111 ISWEAFRREFEKKYFPPEARDRLEQQFMK 139
>At3g42290 putative protein
Length = 462
Score = 56.2 bits (134), Expect = 4e-08
Identities = 38/127 (29%), Positives = 60/127 (46%), Gaps = 4/127 (3%)
Query: 53 AQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILEQDGAAVTWVVFRRE 112
A W V+R F +C +V H L E+A WW S+ Q A ++W F E
Sbjct: 163 ADSWRSRVKRNFSSSRCLMEYRVDLAVHFLEEDAHLWWKSVTARRRQ--ADMSWADFVVE 220
Query: 113 FLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVELAKFYPHYTTETAEFSKCIKFEN 172
F YF +D + E+ FLE+ + SV EY +F + Y + + + ++ +F
Sbjct: 221 FNAKYFLQDELDRMEVRFLELTLVERSVWEYDREFNRVL-VYAGWGMKDGQ-AELRRFMR 278
Query: 173 GLRADIK 179
GLR D++
Sbjct: 279 GLRPDLR 285
>At1g75560 DNA-binding protein
Length = 257
Score = 54.3 bits (129), Expect = 1e-07
Identities = 27/82 (32%), Positives = 40/82 (47%), Gaps = 10/82 (12%)
Query: 247 DAPTEIICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRGDV------VCYNCNEE 300
+ E C+ C E GH ++ C +E C CG+ GH DC D +C NC ++
Sbjct: 88 ECTAESRCWNCREPGHVASNCS-NEGICHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQ 146
Query: 301 GHISSQCTQP---KKVRTGGKV 319
GH+++ CT K RT G +
Sbjct: 147 GHLAADCTNDKACKNCRTSGHI 168
Score = 52.0 bits (123), Expect = 7e-07
Identities = 23/63 (36%), Positives = 31/63 (48%), Gaps = 7/63 (11%)
Query: 251 EIICFKCGEKGHKSNVCDRDEKK------CFRCGQKGHTLADCKRGDVVCYNCNEEGHIS 304
E IC CG+ GH++ C + + C C ++GH ADC D C NC GHI+
Sbjct: 111 EGICHSCGKSGHRARDCSNSDSRAGDLRLCNNCFKQGHLAADCTN-DKACKNCRTSGHIA 169
Query: 305 SQC 307
C
Sbjct: 170 RDC 172
Score = 49.3 bits (116), Expect = 5e-06
Identities = 26/79 (32%), Positives = 38/79 (47%), Gaps = 3/79 (3%)
Query: 240 ERRPKRRDAPTEIICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRGDVVCYNCNE 299
ER P+R + + C C GH + C + C CG GH A+C + C+NC E
Sbjct: 44 EREPRRAFSQGNL-CNNCKRPGHFARDCS-NVSVCNNCGLPGHIAAECT-AESRCWNCRE 100
Query: 300 EGHISSQCTQPKKVRTGGK 318
GH++S C+ + GK
Sbjct: 101 PGHVASNCSNEGICHSCGK 119
Score = 45.8 bits (107), Expect = 5e-05
Identities = 25/73 (34%), Positives = 34/73 (46%), Gaps = 10/73 (13%)
Query: 253 ICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQC----- 307
+C C ++GH + C D K C C GH DC R D VC C+ GH++ C
Sbjct: 139 LCNNCFKQGHLAADCTND-KACKNCRTSGHIARDC-RNDPVCNICSISGHVARHCPKGDS 196
Query: 308 ---TQPKKVRTGG 317
+ +VR GG
Sbjct: 197 NYSDRGSRVRDGG 209
>At4g36020 glycine-rich protein
Length = 299
Score = 53.9 bits (128), Expect = 2e-07
Identities = 36/122 (29%), Positives = 49/122 (39%), Gaps = 25/122 (20%)
Query: 227 SAPTDKGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVC------------DRDEKKC 274
+AP ++ N+ R R C+ CGE GH S C R + C
Sbjct: 75 TAPGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGGGERRSRGGEGC 134
Query: 275 FRCGQKGHTLADC------------KRGDVVCYNCNEEGHISSQCTQPKKVRTGGKVFAL 322
+ CG GH DC K G+ CY C + GH++ CTQ K V G + A+
Sbjct: 135 YNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQ-KSVGNGDQRGAV 193
Query: 323 TG 324
G
Sbjct: 194 KG 195
Score = 50.8 bits (120), Expect = 2e-06
Identities = 23/65 (35%), Positives = 34/65 (51%), Gaps = 10/65 (15%)
Query: 254 CFKCGEKGHKSNVCD---RDEKKCFRCGQKGHTLADC-KRG------DVVCYNCNEEGHI 303
C+ CG GH + C + + C++CG GH DC +RG D CY C +EGH
Sbjct: 232 CYSCGGVGHIARDCATKRQPSRGCYQCGGSGHLARDCDQRGSGGGGNDNACYKCGKEGHF 291
Query: 304 SSQCT 308
+ +C+
Sbjct: 292 ARECS 296
Score = 47.8 bits (112), Expect = 1e-05
Identities = 29/93 (31%), Positives = 37/93 (39%), Gaps = 29/93 (31%)
Query: 254 CFKCGEKGHKSNVC-------DRDEKK-----CFRCGQKGHTLADC-------------- 287
C+ CG+ GH + C R K C+ CG GH DC
Sbjct: 134 CYNCGDTGHFARDCTSAGNGDQRGATKGGNDGCYTCGDVGHVARDCTQKSVGNGDQRGAV 193
Query: 288 KRGDVVCYNCNEEGHISSQCTQ---PKKVRTGG 317
K G+ CY C + GH + CTQ VR+GG
Sbjct: 194 KGGNDGCYTCGDVGHFARDCTQKVAAGNVRSGG 226
Score = 42.4 bits (98), Expect = 6e-04
Identities = 23/89 (25%), Positives = 35/89 (38%), Gaps = 26/89 (29%)
Query: 254 CFKCGEKGHKSNVCDR------DEKK--------CFRCGQKGHTLADCKR---------- 289
C+ CG+ GH + C + D++ C+ CG GH DC +
Sbjct: 166 CYTCGDVGHVARDCTQKSVGNGDQRGAVKGGNDGCYTCGDVGHFARDCTQKVAAGNVRSG 225
Query: 290 --GDVVCYNCNEEGHISSQCTQPKKVRTG 316
G CY+C GHI+ C ++ G
Sbjct: 226 GGGSGTCYSCGGVGHIARDCATKRQPSRG 254
Score = 42.4 bits (98), Expect = 6e-04
Identities = 20/71 (28%), Positives = 29/71 (40%), Gaps = 15/71 (21%)
Query: 254 CFKCGEKGHKSNVCDRD------------EKKCFRCGQKGHTLADC---KRGDVVCYNCN 298
C+ CG+ GH + C + C+ CG GH DC ++ CY C
Sbjct: 200 CYTCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCGGVGHIARDCATKRQPSRGCYQCG 259
Query: 299 EEGHISSQCTQ 309
GH++ C Q
Sbjct: 260 GSGHLARDCDQ 270
>At3g43590 putative protein
Length = 551
Score = 52.4 bits (124), Expect = 5e-07
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Query: 252 IICFKCGEKGHKSNVC---DRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCT 308
+ C+ CGE+GH S C + K CF CG H C +G CY C + GH + C
Sbjct: 166 VSCYSCGEQGHTSFNCPTPTKRRKPCFICGSLEHGAKQCSKGH-DCYICKKTGHRAKDC- 223
Query: 309 QPKKVRTGGK 318
P K + G K
Sbjct: 224 -PDKYKNGSK 232
Score = 50.8 bits (120), Expect = 2e-06
Identities = 22/93 (23%), Positives = 37/93 (39%), Gaps = 31/93 (33%)
Query: 252 IICFKCGEKGHKSNVC--------------------DRDEKKCFRCGQKGHTLADC---- 287
+ C++CG+ GH C R+ +C+RCG++GH +C
Sbjct: 285 VSCYRCGQLGHSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSS 344
Query: 288 -------KRGDVVCYNCNEEGHISSQCTQPKKV 313
+ +CY CN GH + +C +V
Sbjct: 345 SISTSHGRESQTLCYRCNGSGHFARECPNSSQV 377
Score = 41.6 bits (96), Expect = 0.001
Identities = 23/70 (32%), Positives = 30/70 (42%), Gaps = 15/70 (21%)
Query: 254 CFKCGEKGHKSNVCDRDEKK------CFRCGQKGHTLADCK-------RGDVVCYNCNEE 300
C+ C + GH++ C K C RCG GH + CK DV CY C
Sbjct: 210 CYICKKTGHRAKDCPDKYKNGSKGAVCLRCGDFGHDMILCKYEYSKEDLKDVQCYICKSF 269
Query: 301 GHISSQCTQP 310
GH+ C +P
Sbjct: 270 GHLC--CVEP 277
Score = 39.7 bits (91), Expect = 0.004
Identities = 26/123 (21%), Positives = 38/123 (30%), Gaps = 51/123 (41%)
Query: 244 KRRDAPTEIICFKCGEKGHKSNVCDRDEKK------------------------------ 273
K ++ +C +CG+ GH +C + K
Sbjct: 226 KYKNGSKGAVCLRCGDFGHDMILCKYEYSKEDLKDVQCYICKSFGHLCCVEPGNSLSWAV 285
Query: 274 -CFRCGQKGHTLADC--------------------KRGDVVCYNCNEEGHISSQCTQPKK 312
C+RCGQ GH+ C R CY C EEGH + +C
Sbjct: 286 SCYRCGQLGHSGLACGRHYEESNENDSATPERLFNSREASECYRCGEEGHFARECPNSSS 345
Query: 313 VRT 315
+ T
Sbjct: 346 IST 348
Score = 36.2 bits (82), Expect = 0.040
Identities = 22/87 (25%), Positives = 32/87 (36%), Gaps = 12/87 (13%)
Query: 238 NDERRPKRRDAPTEII-CFKCGEKGHKSNVCDRD-----------EKKCFRCGQKGHTLA 285
ND P+R E C++CGE+GH + C + C+RC GH
Sbjct: 310 NDSATPERLFNSREASECYRCGEEGHFARECPNSSSISTSHGRESQTLCYRCNGSGHFAR 369
Query: 286 DCKRGDVVCYNCNEEGHISSQCTQPKK 312
+C V E S + + K
Sbjct: 370 ECPNSSQVSKRDRETSTTSHKSRKKNK 396
Score = 32.3 bits (72), Expect = 0.58
Identities = 13/25 (52%), Positives = 15/25 (60%)
Query: 290 GDVVCYNCNEEGHISSQCTQPKKVR 314
G V CY+C E+GH S C P K R
Sbjct: 164 GWVSCYSCGEQGHTSFNCPTPTKRR 188
>At5g52380 unknown protein
Length = 268
Score = 51.2 bits (121), Expect = 1e-06
Identities = 37/122 (30%), Positives = 57/122 (46%), Gaps = 17/122 (13%)
Query: 213 ERRGKGQ*SRPKPYSAPTDKGKQRLNDERRPKRRDA--PTEIICFKCGEKGHKSNVCDRD 270
+++ K + KP S+ TD+ ++ + R P R P E CF C K H + +C
Sbjct: 35 KKKKKSLFKKKKPGSS-TDRPQRTGSSTRHPLRVPGMKPGEG-CFICHSKTHIAKLCPEK 92
Query: 271 E-----KKCFRCGQKGHTLADCKRGD------VVCYNCNEEGHISSQCTQPKKVRTGGKV 319
K C +C ++GH+L +C + +CYNC + GH S C P + GG
Sbjct: 93 SEWERNKICLQCRRRGHSLKNCPEKNNESSEKKLCYNCGDTGHSLSHCPYP--MEDGGTK 150
Query: 320 FA 321
FA
Sbjct: 151 FA 152
>At3g53500 splicing factor - like protein
Length = 243
Score = 50.8 bits (120), Expect = 2e-06
Identities = 24/50 (48%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Query: 273 KCFRCGQKGHTLADCKRGD--VVCYNCNEEGHISSQC---TQPKKVRTGG 317
+CF CG GH DC GD CY C E GHI C PKK R GG
Sbjct: 59 RCFNCGVDGHWARDCTAGDWKNKCYRCGERGHIERNCKNSPSPKKARQGG 108
>At2g37340 unknown protein
Length = 249
Score = 50.1 bits (118), Expect = 3e-06
Identities = 23/48 (47%), Positives = 25/48 (51%), Gaps = 3/48 (6%)
Query: 273 KCFRCGQKGHTLADCKRGD--VVCYNCNEEGHISSQC-TQPKKVRTGG 317
+CF CG GH DC GD CY C E GHI C PKK+R G
Sbjct: 59 RCFNCGVDGHWARDCTAGDWKNKCYRCGERGHIERNCKNSPKKLRRSG 106
Score = 41.6 bits (96), Expect = 0.001
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query: 254 CFKCGEKGHKSNVCDRDE--KKCFRCGQKGHTLADCK 288
CF CG GH + C + KC+RCG++GH +CK
Sbjct: 60 CFNCGVDGHWARDCTAGDWKNKCYRCGERGHIERNCK 96
Score = 30.4 bits (67), Expect = 2.2
Identities = 11/25 (44%), Positives = 14/25 (56%)
Query: 254 CFKCGEKGHKSNVCDRDEKKCFRCG 278
C++CGE+GH C KK R G
Sbjct: 82 CYRCGERGHIERNCKNSPKKLRRSG 106
>At2g17870 putative glycine-rich, zinc-finger DNA-binding protein
Length = 301
Score = 50.1 bits (118), Expect = 3e-06
Identities = 25/81 (30%), Positives = 33/81 (39%), Gaps = 27/81 (33%)
Query: 254 CFKCGEKGHKSNVCD----------------RDEKKCFRCGQKGHTLADCKR-------- 289
CF CGE GH + CD E +C+ CG GH DC++
Sbjct: 96 CFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGEGECYMCGDVGHFARDCRQSGGGNSGG 155
Query: 290 ---GDVVCYNCNEEGHISSQC 307
G CY+C E GH++ C
Sbjct: 156 GGGGGRPCYSCGEVGHLAKDC 176
Score = 47.8 bits (112), Expect = 1e-05
Identities = 23/73 (31%), Positives = 31/73 (41%), Gaps = 18/73 (24%)
Query: 254 CFKCGEKGHKSNVCDRD--------EKKCFRCGQKGHTLADCKR----------GDVVCY 295
C+ CG GH + VC + C+ CG GH DC R G C+
Sbjct: 226 CYTCGGVGHIAKVCTSKIPSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNKCF 285
Query: 296 NCNEEGHISSQCT 308
C +EGH + +CT
Sbjct: 286 ICGKEGHFARECT 298
Score = 45.8 bits (107), Expect = 5e-05
Identities = 26/87 (29%), Positives = 34/87 (38%), Gaps = 25/87 (28%)
Query: 254 CFKCGEKGHKSNVCDRDE---------------KKCFRCGQKGHTLADCKR--------G 290
C+ CGE GH + C C+ CG GH DC++ G
Sbjct: 163 CYSCGEVGHLAKDCRGGSGGNRYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGGG 222
Query: 291 DVVCYNCNEEGHISSQCTQPKKVRTGG 317
CY C GHI+ CT K+ +GG
Sbjct: 223 GSTCYTCGGVGHIAKVCT--SKIPSGG 247
Score = 41.2 bits (95), Expect = 0.001
Identities = 23/90 (25%), Positives = 32/90 (35%), Gaps = 26/90 (28%)
Query: 254 CFKCGEKGHKSNVCDRDE-----------KKCFRCGQKGHTLADCK-------------- 288
C+ CG+ GH + C + + C+ CG+ GH DC+
Sbjct: 132 CYMCGDVGHFARDCRQSGGGNSGGGGGGGRPCYSCGEVGHLAKDCRGGSGGNRYGGGGGR 191
Query: 289 -RGDVVCYNCNEEGHISSQCTQPKKVRTGG 317
G CY C GH + C Q GG
Sbjct: 192 GSGGDGCYMCGGVGHFARDCRQNGGGNVGG 221
Score = 39.3 bits (90), Expect = 0.005
Identities = 21/80 (26%), Positives = 30/80 (37%), Gaps = 16/80 (20%)
Query: 254 CFKCGEKGHKSNVCDRDE--------KKCFRCGQKGHTLADCKR--------GDVVCYNC 297
C+ CG GH + C ++ C+ CG GH C G CY C
Sbjct: 198 CYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTSKIPSGGGGGGRACYEC 257
Query: 298 NEEGHISSQCTQPKKVRTGG 317
GH++ C + +GG
Sbjct: 258 GGTGHLARDCDRRGSGSSGG 277
Score = 38.5 bits (88), Expect = 0.008
Identities = 21/76 (27%), Positives = 26/76 (33%), Gaps = 16/76 (21%)
Query: 258 GEKGHKSNVCDRDEKKCFRCGQKGHTLADC----------------KRGDVVCYNCNEEG 301
G K N CF CG+ GH DC G+ CY C + G
Sbjct: 80 GSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGEGECYMCGDVG 139
Query: 302 HISSQCTQPKKVRTGG 317
H + C Q +GG
Sbjct: 140 HFARDCRQSGGGNSGG 155
>At4g08100 putative polyprotein
Length = 1054
Score = 49.7 bits (117), Expect = 4e-06
Identities = 53/266 (19%), Positives = 101/266 (37%), Gaps = 23/266 (8%)
Query: 42 PTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWWVSLLPILE--Q 99
P F G +PD +W +++E +F +C + KV+ A WW L+ +
Sbjct: 97 PAFHGTDNPDTYLEWEQKIELVFLCQECLQSNKVKIAATKFYNYALSWWDQLVTSRRRTR 156
Query: 100 DGAAVTW----VVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVELAKFYP 155
D TW V R+ F+ Y+ ++ + + QG +V EY F+E+
Sbjct: 157 DYPIKTWNQLKFVMRKRFVPSYYHRELHQR----LRNLVQGSKTVEEY---FLEMETLML 209
Query: 156 HYTTETAEFSKCIKFENGLRADIKRAIGYEKIRIFSELVSSCRIYEEDTKAHYKVMSERR 215
+ + +F GL +I+ + + E++ ++E+ K S +
Sbjct: 210 RADLQEDGEAVMSRFMGGLNREIQDRLETQHYVELEEMLHKAVMFEQQIKRKNARSSHTK 269
Query: 216 GKGQ*SRPKPYSAPTDKGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDR-DEKKC 274
+P Y G Q ++ +P + P ++ KG V R + +C
Sbjct: 270 TNYSSGKPS-YQKEEKFGYQ---NDSKPFVKPKPVDL-----DPKGKGKEVITRARDIRC 320
Query: 275 FRCGQKGHTLADCKRGDVVCYNCNEE 300
F+ H + C ++ + N E
Sbjct: 321 FKSQGLRHYASKCSNKRIMVHRDNGE 346
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,132,854
Number of Sequences: 26719
Number of extensions: 499695
Number of successful extensions: 1606
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 1276
Number of HSP's gapped (non-prelim): 231
length of query: 471
length of database: 11,318,596
effective HSP length: 103
effective length of query: 368
effective length of database: 8,566,539
effective search space: 3152486352
effective search space used: 3152486352
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146719.4


