

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146719.10 + phase: 0 /pseudo
(112 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
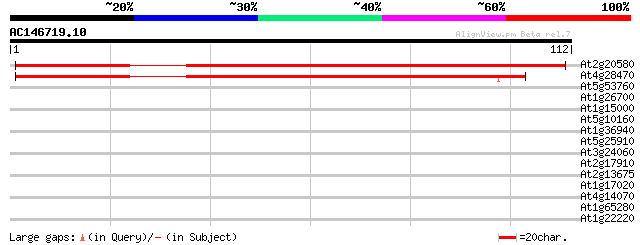
Score E
Sequences producing significant alignments: (bits) Value
At2g20580 putative 26S proteasome regulatory subunit S2 152 3e-38
At4g28470 putative protein 130 8e-32
At5g53760 unknown protein 32 0.067
At1g26700 membrane protein Mlo14 28 0.97
At1g15000 serine carboxypeptidase precursor, putative 26 2.8
At5g10160 (3R)-hydroxymyristoyl-[acyl carrier protein] dehydrata... 25 4.8
At1g36940 hypothetical protein 25 4.8
At5g25910 disease resistance protein - like 25 6.3
At3g24060 hypothetical protein 25 6.3
At2g17910 putative non-LTR retroelement reverse transcriptase 25 6.3
At2g13675 callose synthase (1,3-beta-glucan synthase) like protein 25 6.3
At1g17020 SRG1-like protein 25 6.3
At4g14070 A6 anther-specific protein (Z97335.16) 25 8.2
At1g65280 25 8.2
At1g22220 unknown protein 25 8.2
>At2g20580 putative 26S proteasome regulatory subunit S2
Length = 891
Score = 152 bits (383), Expect = 3e-38
Identities = 79/110 (71%), Positives = 89/110 (80%), Gaps = 11/110 (10%)
Query: 2 LELPSV**GKYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDV 61
L++ + GKYHYVLYFLVLAMQ P M+LTV ENLKPL VP+RVGQAVDV
Sbjct: 786 LDMKPIILGKYHYVLYFLVLAMQ-----------PRMMLTVDENLKPLSVPVRVGQAVDV 834
Query: 62 VGQAGRPKKITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVILKE 111
VGQAGRPK ITGFQTHST VLLAAG+RAELAT+KYIP+SP+LE F+ILKE
Sbjct: 835 VGQAGRPKTITGFQTHSTPVLLAAGERAELATDKYIPLSPILEGFIILKE 884
>At4g28470 putative protein
Length = 1103
Score = 130 bits (328), Expect = 8e-32
Identities = 70/103 (67%), Positives = 82/103 (78%), Gaps = 12/103 (11%)
Query: 2 LELPSV**GKYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDV 61
L++ S+ GKYHYVLYFLVLAMQ P M+LTV ++LKP+ VP+RVGQAVDV
Sbjct: 889 LDMKSIILGKYHYVLYFLVLAMQ-----------PRMMLTVDQSLKPISVPVRVGQAVDV 937
Query: 62 VGQAGRPKKITGFQTHSTSVLLAAGDRAELATEKY-IPISPVL 103
VGQAGRPK ITGFQTHST VLLAAG+RAELATEKY +P+ P+L
Sbjct: 938 VGQAGRPKTITGFQTHSTPVLLAAGERAELATEKYVVPLIPLL 980
>At5g53760 unknown protein
Length = 573
Score = 31.6 bits (70), Expect = 0.067
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 12/78 (15%)
Query: 11 KYHYVLYFLVLAMQLTLGY-------YILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVG 63
K HY++YF +L LG+ Y L ++ +G N K L+P R+ + + G
Sbjct: 402 KNHYLVYF-----RLLLGFAGQFLCSYSTLPLYALVTQMGTNYKAALIPQRIRETIRGWG 456
Query: 64 QAGRPKKITGFQTHSTSV 81
+A R K+ G ++V
Sbjct: 457 KATRRKRRHGLYGDDSTV 474
>At1g26700 membrane protein Mlo14
Length = 554
Score = 27.7 bits (60), Expect = 0.97
Identities = 18/70 (25%), Positives = 35/70 (49%), Gaps = 7/70 (10%)
Query: 19 LVLAMQLTLGY-------YILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKKI 71
L++ ++L LG+ Y L ++ +G N K L+P RV + ++ G+A R K+
Sbjct: 400 LLVYLRLILGFSGQFLCSYSTLPLYALVTQMGTNYKAALLPQRVRETINGWGKATRRKRR 459
Query: 72 TGFQTHSTSV 81
G +++
Sbjct: 460 HGLYGDDSTI 469
>At1g15000 serine carboxypeptidase precursor, putative
Length = 444
Score = 26.2 bits (56), Expect = 2.8
Identities = 25/80 (31%), Positives = 35/80 (43%), Gaps = 20/80 (25%)
Query: 27 LGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKKITGFQTHSTSV----L 82
+GYYIL +KP N K L L +G G +T QTH+ +V L
Sbjct: 179 IGYYILKEKP--------NGKVNLKGLAIGN--------GLTDPVTQVQTHAVNVYYSGL 222
Query: 83 LAAGDRAELATEKYIPISPV 102
+ A R EL + I ++ V
Sbjct: 223 VNAKQRVELQKAQEISVALV 242
>At5g10160 (3R)-hydroxymyristoyl-[acyl carrier protein] dehydratase
-like protein
Length = 219
Score = 25.4 bits (54), Expect = 4.8
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 6/42 (14%)
Query: 65 AGRPKKITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAF 106
A RPK+ S+S++L + D ++ EK IPI EAF
Sbjct: 36 AFRPKR------RSSSLVLCSTDESKSTAEKEIPIELRYEAF 71
>At1g36940 hypothetical protein
Length = 181
Score = 25.4 bits (54), Expect = 4.8
Identities = 18/64 (28%), Positives = 29/64 (45%), Gaps = 14/64 (21%)
Query: 31 ILLQKPTML--LTVGENLKPLLVPLRVGQ------------AVDVVGQAGRPKKITGFQT 76
+ Q+PT L L ++K + +P + Q + +GQ R KITGF T
Sbjct: 8 VQFQRPTFLPMLCSRPSIKNVTLPAKSHQDDSYQQADPLSPKISCIGQVKRSNKITGFPT 67
Query: 77 HSTS 80
+T+
Sbjct: 68 TTTT 71
>At5g25910 disease resistance protein - like
Length = 811
Score = 25.0 bits (53), Expect = 6.3
Identities = 11/32 (34%), Positives = 20/32 (62%), Gaps = 3/32 (9%)
Query: 21 LAMQLTLGYYILLQKPTMLLTVGEN---LKPL 49
+++ LT+GY ++ KP L+ G N +KP+
Sbjct: 780 ISIGLTMGYILVSYKPEWLMNSGRNKRRIKPI 811
>At3g24060 hypothetical protein
Length = 147
Score = 25.0 bits (53), Expect = 6.3
Identities = 10/26 (38%), Positives = 19/26 (72%)
Query: 15 VLYFLVLAMQLTLGYYILLQKPTMLL 40
+L F+++A+ LT+ + ++LQ TM L
Sbjct: 9 LLLFMLIAISLTIIFTLMLQPQTMFL 34
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 25.0 bits (53), Expect = 6.3
Identities = 14/46 (30%), Positives = 21/46 (45%)
Query: 36 PTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKKITGFQTHSTSV 81
PT + G+ L P L L + ++ +A + KITG Q V
Sbjct: 607 PTRGIRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQDKKV 652
>At2g13675 callose synthase (1,3-beta-glucan synthase) like protein
Length = 1939
Score = 25.0 bits (53), Expect = 6.3
Identities = 17/57 (29%), Positives = 30/57 (51%), Gaps = 2/57 (3%)
Query: 14 YVLYFLVLAMQLTLGYYILLQK--PTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRP 68
+ L F +L + L +G +++ + LTVG+ ++ LL L G A+ + Q RP
Sbjct: 1814 FQLMFRLLKLFLFIGSVVIVGMLFHFLKLTVGDIMQSLLAFLPTGWALLQISQVARP 1870
>At1g17020 SRG1-like protein
Length = 358
Score = 25.0 bits (53), Expect = 6.3
Identities = 14/49 (28%), Positives = 25/49 (50%), Gaps = 7/49 (14%)
Query: 67 RPKKITGFQTHSTSV----LLAAGDRAELATEK---YIPISPVLEAFVI 108
+P ++ G HS SV L+ D L +K ++P+ P+ AF++
Sbjct: 223 QPDQVIGLTPHSDSVGLTVLMQVNDVEGLQIKKDGKWVPVKPLPNAFIV 271
>At4g14070 A6 anther-specific protein (Z97335.16)
Length = 727
Score = 24.6 bits (52), Expect = 8.2
Identities = 14/38 (36%), Positives = 23/38 (59%), Gaps = 5/38 (13%)
Query: 35 KPTMLLTVGENLKPLLVP-----LRVGQAVDVVGQAGR 67
K T++L+ GEN++PL + RV + + V+GQ R
Sbjct: 605 KDTIVLSTGENVEPLEIEEAAMRSRVIEQIVVIGQDRR 642
>At1g65280
Length = 605
Score = 24.6 bits (52), Expect = 8.2
Identities = 12/22 (54%), Positives = 15/22 (67%)
Query: 42 VGENLKPLLVPLRVGQAVDVVG 63
VG +LK LL + GQAVD+ G
Sbjct: 125 VGNDLKQLLKMIDDGQAVDIKG 146
>At1g22220 unknown protein
Length = 314
Score = 24.6 bits (52), Expect = 8.2
Identities = 13/27 (48%), Positives = 16/27 (59%)
Query: 86 GDRAELATEKYIPISPVLEAFVILKEC 112
GD AELATE ++ EA V L +C
Sbjct: 278 GDDAELATEAFVGDCMYGEAVVALLKC 304
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.149 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,158,705
Number of Sequences: 26719
Number of extensions: 72287
Number of successful extensions: 206
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 194
Number of HSP's gapped (non-prelim): 15
length of query: 112
length of database: 11,318,596
effective HSP length: 88
effective length of query: 24
effective length of database: 8,967,324
effective search space: 215215776
effective search space used: 215215776
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146719.10


