

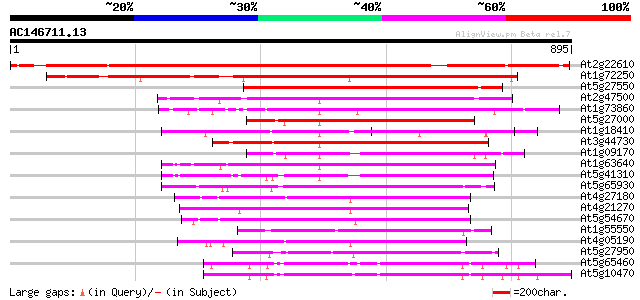
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146711.13 - phase: 0 /pseudo
(895 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g22610 putative kinesin heavy chain 999 0.0
At1g72250 kinesin, putative 655 0.0
At5g27550 kinesin-like protein 489 e-138
At2g47500 kinesin heavy chain like protein 350 3e-96
At1g73860 kinesin-related protein 339 5e-93
At5g27000 kinesin-like heavy chain (KATD) 338 6e-93
At1g18410 kinesin-related protein, putative 334 1e-91
At3g44730 kinesin-like protein heavy chain (KP1) 331 1e-90
At1g09170 putative kinesin 317 2e-86
At1g63640 kinesin-like protein 310 2e-84
At5g41310 kinesin-like protein 309 5e-84
At5g65930 kinesin-like calmodulin-binding protein 305 6e-83
At4g27180 heavy chain polypeptide of kinesin-like protein (katB) 299 4e-81
At4g21270 kinesin-related protein katA 293 4e-79
At5g54670 heavy chain polypeptide of kinesin-like protein (katC) 291 1e-78
At1g55550 290 2e-78
At4g05190 kinesin - like protein 285 1e-76
At5g27950 unknown protein 271 2e-72
At5g65460 putative protein 255 9e-68
At5g10470 TH65 protein (th65) 249 6e-66
>At2g22610 putative kinesin heavy chain
Length = 1068
Score = 999 bits (2584), Expect = 0.0
Identities = 525/898 (58%), Positives = 659/898 (72%), Gaps = 57/898 (6%)
Query: 1 MEDANDSLFDSMLVDS---SSKLI--QNGFARSQSSGLKINIMY*FVKLSVHF*RIHFEA 55
++D +D+L DSM DS S +LI ++GF + ++ I Y
Sbjct: 45 IDDCDDALGDSM--DSGFYSFRLILGEDGFNLMLTRRVRFLIQY---------------- 86
Query: 56 EECVMFVNVGGEATNEGADGVKFLSDTFFDGGDVFLTNEAIVEGGDYPSIYQSARVGSFS 115
E +MF+N GG+ + + D +F+GGDV T E+IVE GD+P IYQSARVG+F
Sbjct: 87 -ETIMFINAGGDDSKVLDSELNISRDDYFEGGDVLRTEESIVEAGDFPFIYQSARVGNFC 145
Query: 116 YRIDNLPPGQYLVDLHFVEIINVNGPKGMRVFNVYIQEEKVLSELDIYAAVGVNKPLQLI 175
Y+++NL PG+YL+D HF EIIN NGPKG+RVFNVY+Q+EK +E DI++ VG N+PL L+
Sbjct: 146 YQLNNLLPGEYLIDFHFAEIINTNGPKGIRVFNVYVQDEKA-TEFDIFSVVGANRPLLLV 204
Query: 176 DCRATVKDDGVILIRFESLNGRPIVSGICIRRASKESVPPVPSDFIECNYCAAQIEIPSS 235
D R V DDG+I +RFE +NG P+V GIC+R+A + SVP DFI+C CA +IEI +
Sbjct: 205 DLRVMVMDDGLIRVRFEGINGSPVVCGICLRKAPQVSVPRTSQDFIKCENCATEIEISPT 264
Query: 236 QIKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSF 295
+ ++M+ K+ KYE KI EL+ + E K EC+EAW SLT + ++EKV MEL+ +++
Sbjct: 265 RKRLMRAKAHDKYEKKIAELSERYEHKTNECHEAWMSLTSANEQLEKVMMELNNKIYQAR 324
Query: 296 TTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDSIP 355
+ + T QA+ L+SI+ +YE DK+ WA AI SLQEK+++MK EQS+LS +AHECV+ IP
Sbjct: 325 SLDQTVITQADCLKSITRKYENDKRHWATAIDSLQEKIEIMKREQSQLSQEAHECVEGIP 384
Query: 356 ELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSS 415
EL KMV VQ LV QCEDLK KY EE +RK+L+N +QE KGNIRVFCRCRPLN E S+
Sbjct: 385 ELYKMVGGVQALVSQCEDLKQKYSEEQAKRKELYNHIQETKGNIRVFCRCRPLNTEETST 444
Query: 416 GCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNV 475
T+VDFD AKDG LG++ +SKK F+FDRVYTPKD QVDVFADAS MV+SVLDGYNV
Sbjct: 445 KSATIVDFDGAKDGELGVITGNNSKKSFKFDRVYTPKDGQVDVFADASPMVVSVLDGYNV 504
Query: 476 CIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQI 535
CIFAYGQTGTGKTFTMEGT QNRGVNYRT+E LF V++ER ET SY+ISVSVLEVYNEQI
Sbjct: 505 CIFAYGQTGTGKTFTMEGTPQNRGVNYRTVEQLFEVARERRETISYNISVSVLEVYNEQI 564
Query: 536 RDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEH 595
RDLLAT P SK+LEIKQ+ +G HHVPG+VEA V+NI++VW VLQAGSNAR+VGSNNVNEH
Sbjct: 565 RDLLATSPGSKKLEIKQSSDGSHHVPGLVEANVENINEVWNVLQAGSNARSVGSNNVNEH 624
Query: 596 SSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSA 655
SSRSHCML IMVK KNLMNG+CTKSKLWLVDL+GSERLAKTDVQGERLKEAQNINRSLSA
Sbjct: 625 SSRSHCMLSIMVKAKNLMNGDCTKSKLWLVDLAGSERLAKTDVQGERLKEAQNINRSLSA 684
Query: 656 LGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFAT 715
LGDVI ALA KSSHIPY SPS+ DV ETLSSLNFAT
Sbjct: 685 LGDVIYALATKSSHIPY-------------------------SPSEHDVSETLSSLNFAT 719
Query: 716 RVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNI 775
RVRGVEL P +KQ+DTGE+QK KAM++KAR E R K+ES++K+EE++QN+E K KG+DN
Sbjct: 720 RVRGVELGPARKQVDTGEIQKLKAMVEKARQESRSKDESIKKMEENIQNLEGKNKGRDNS 779
Query: 776 HKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEETCCTLQHKVKELERKIKEQ 835
+++LQEK K+L+ Q+ S+ NQSEKQ +QL ERLK ++E C LQ KVKELE K++E+
Sbjct: 780 YRSLQEKNKDLQNQLD---SVHNQSEKQYAQLQERLKSRDEICSNLQQKVKELECKLRER 836
Query: 836 LQTETANFQQKVCNTFAKLLPTLFSNSLFLPKVWDLEKKLKDQLQGSESESSFLKDKV 893
Q+++A QKV + L + S+ ++ KV D E KLK+ SE S + K+
Sbjct: 837 HQSDSAANNQKVKDLENNLKESEGSSLVWQQKVKDYENKLKE----SEGNSLVWQQKI 890
>At1g72250 kinesin, putative
Length = 1195
Score = 655 bits (1689), Expect = 0.0
Identities = 367/780 (47%), Positives = 493/780 (63%), Gaps = 68/780 (8%)
Query: 59 VMFVNVGGEATNEGADGVKFLSDTFFDGGDVFLTNEAIVEGGDYPSIYQSARVGSFSYRI 118
V+ +N G +T+ + V FL D FF GG+ +T +A+V D +YQ+AR+G+F+Y+
Sbjct: 172 VISINSGSISTDVTVEDVTFLKDEFFSGGES-ITTDAVVGNEDEILLYQTARLGNFAYKF 230
Query: 119 DNLPPGQYLVDLHFVEIINVNGPKGMRVFNVYIQEEKVLSELDIYAAVGVNKPLQLIDCR 178
+L PG Y +DLHF EI GP G V+S LD+++ VG N PL + D R
Sbjct: 231 QSLDPGDYFIDLHFAEIEFTKGPPG------------VISGLDLFSQVGANTPLVIEDLR 278
Query: 179 ATVKDDGVILIRFESLNGRPIVSGICIRR-------------ASKESVPPVPSDFIECNY 225
V +G + IR E + G I+ GI IR+ A K S V S + N
Sbjct: 279 MLVGREGELSIRLEGVTGAAILCGISIRKETTATYVEETGMLAVKGSTDTVLSQQTQENL 338
Query: 226 -CAAQIEIPSSQIKV-MQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKV 283
C A+ E + Q K + ++EL ++ + K +EC EA SL+E+ E+ +
Sbjct: 339 VCRAEEEAEGMRSDCEQQRKEMEDMKRMVEELKLENQQKTRECEEALNSLSEIQNELMRK 398
Query: 284 QMELDQVTFKSFTTELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRL 343
M + S T E+ ++ + E+++ K E + + VQ
Sbjct: 399 SMHVG-----SLGTSQREEQMVLFIKRFDKKIEVEQIKLLEEATTYKHLVQ--------- 444
Query: 344 SFKAHECVDSIPELNKMVYAVQELVKQC----EDLKVKYYEEMTQRKKLFNEVQEAKGNI 399
++N+ +Q VKQ E+LKVK+ +RK+L+N++ E KGNI
Sbjct: 445 ------------DINEFSSHIQSRVKQDAELHENLKVKFVAGEKERKELYNKILELKGNI 492
Query: 400 RVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVF 459
RVFCRCRPLN E +G + +D ++ K+G + +++ G KK F+FD V+ P Q DVF
Sbjct: 493 RVFCRCRPLNFEETEAGVSMGIDVESTKNGEVIVMSNGFPKKSFKFDSVFGPNASQADVF 552
Query: 460 ADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETF 519
D + SV+DGYNVCIFAYGQTGTGKTFTMEGT+ +RGVNYRTLE+LFR+ K R +
Sbjct: 553 EDTAPFATSVIDGYNVCIFAYGQTGTGKTFTMEGTQHDRGVNYRTLENLFRIIKAREHRY 612
Query: 520 SYDISVSVLEVYNEQIRDLLA----TGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVW 575
+Y+ISVSVLEVYNEQIRDLL + A KR EI+Q EG+HHVPG+VEA V +I +VW
Sbjct: 613 NYEISVSVLEVYNEQIRDLLVPASQSASAPKRFEIRQLSEGNHHVPGLVEAPVKSIEEVW 672
Query: 576 TVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAK 635
VL+ GSNARAVG NEHSSRSHC+ C+MVK +NL+NGECTKSKLWLVDL+GSER+AK
Sbjct: 673 DVLKTGSNARAVGKTTANEHSSRSHCIHCVMVKGENLLNGECTKSKLWLVDLAGSERVAK 732
Query: 636 TDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFV 695
T+VQGERLKE QNIN+SLSALGDVI ALA KSSHIP+RNSKLTHLLQDSLGGDSKTLMFV
Sbjct: 733 TEVQGERLKETQNINKSLSALGDVIFALANKSSHIPFRNSKLTHLLQDSLGGDSKTLMFV 792
Query: 696 QISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESL 755
QISP++ D ETL SLNFA+RVRG+EL P KKQ+D EL K K M++K + + + K+E +
Sbjct: 793 QISPNENDQSETLCSLNFASRVRGIELGPAKKQLDNTELLKYKQMVEKWKQDMKGKDEQI 852
Query: 756 RKLEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQ------SEKQVSQLCE 809
RK+EE++ +E+K K +D +K LQ+K+KELE Q+ ++ + Q +E+Q Q E
Sbjct: 853 RKMEETMYGLEAKIKERDTKNKTLQDKVKELESQLLVERKLARQHVDTKIAEQQTKQQTE 912
>At5g27550 kinesin-like protein
Length = 425
Score = 489 bits (1259), Expect = e-138
Identities = 245/415 (59%), Positives = 320/415 (77%), Gaps = 6/415 (1%)
Query: 374 LKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGI 433
L+ +Y EE ++RK+L+NEV E KGNIRVFCRCRPLN+ E+++GC +V +FD ++ L I
Sbjct: 15 LEKQYLEESSERKRLYNEVIELKGNIRVFCRCRPLNQAEIANGCASVAEFDTTQENELQI 74
Query: 434 LATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG 493
L++ SSKK F+FD V+ P D Q VFA +V SVLDGYNVCIFAYGQTGTGKTFTMEG
Sbjct: 75 LSSDSSKKHFKFDHVFKPDDGQETVFAQTKPIVTSVLDGYNVCIFAYGQTGTGKTFTMEG 134
Query: 494 TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLA--TGPASKRLEIK 551
T +NRGVNYRTLE LFR S+ +S +++SVS+LEVYNE+IRDLL + K+LE+K
Sbjct: 135 TPENRGVNYRTLEELFRCSESKSHLMKFELSVSMLEVYNEKIRDLLVDNSNQPPKKLEVK 194
Query: 552 QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKN 611
Q+ EG VPG+VEA+V N VW +L+ G R+VGS NE SSRSHC+L + VK +N
Sbjct: 195 QSAEGTQEVPGLVEAQVYNTDGVWDLLKKGYAVRSVGSTAANEQSSRSHCLLRVTVKGEN 254
Query: 612 LMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIP 671
L+NG+ T+S LWLVDL+GSER+ K +V+GERLKE+Q IN+SLSALGDVISALA+K+SHIP
Sbjct: 255 LINGQRTRSHLWLVDLAGSERVGKVEVEGERLKESQFINKSLSALGDVISALASKTSHIP 314
Query: 672 YRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDT 731
YRNSKLTH+LQ+SLGGD KTLMFVQISPS D+GETL SLNFA+RVRG+E P +KQ D
Sbjct: 315 YRNSKLTHMLQNSLGGDCKTLMFVQISPSSADLGETLCSLNFASRVRGIESGPARKQADV 374
Query: 732 GELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNIHKNLQEKIKEL 786
EL K+K M +K + E E+ +KL++++Q+++ + +++I + LQ+K+ L
Sbjct: 375 SELLKSKQMAEKLKHE----EKETKKLQDNVQSLQLRLTAREHICRGLQDKVSPL 425
>At2g47500 kinesin heavy chain like protein
Length = 983
Score = 350 bits (897), Expect = 3e-96
Identities = 219/574 (38%), Positives = 326/574 (56%), Gaps = 31/574 (5%)
Query: 237 IKVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSFT 296
I+ + +K ++EN++ T Q EL E+ +S S + E ++ +FK+
Sbjct: 250 IESLLSKVVEEFENRV---TNQYELVRAAPRESTSSQNNRSFLKPLGEREREEKSFKAI- 305
Query: 297 TELTAEKQAENLRSISNRYELDKKKWAEAIISLQEKV----QLMKSEQSRLSFKAHECVD 352
+K N + + + + + K QE + Q + + ++ + F + +
Sbjct: 306 -----KKDDHNSQILDEKMKTRQFKQLTIFNQQQEDIEGLRQTLYTTRAGMQFMQKKFQE 360
Query: 353 SIPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVE 412
L V+ + Y+ + + +KL+N+VQ+ KG+IRV+CR RP +
Sbjct: 361 EFSSLGMHVHGLAHAASG-------YHRVLEENRKLYNQVQDLKGSIRVYCRVRPFLPGQ 413
Query: 413 MSSGCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDG 472
SS +T+ + + G G S K F F++V+ P Q +VF+D ++ SVLDG
Sbjct: 414 -SSFSSTIGNMEDDTIGINTASRHGKSLKSFTFNKVFGPSATQEEVFSDMQPLIRSVLDG 472
Query: 473 YNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVL 528
YNVCIFAYGQTG+GKTFTM G TE+++GVNYR L LF ++++R +TF YDI+V ++
Sbjct: 473 YNVCIFAYGQTGSGKTFTMSGPRDLTEKSQGVNYRALGDLFLLAEQRKDTFRYDIAVQMI 532
Query: 529 EVYNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVG 588
E+YNEQ+RDLL T ++KRLEI+ + + VP V + DV +++ G RAVG
Sbjct: 533 EIYNEQVRDLLVTDGSNKRLEIRNSSQKGLSVPDASLVPVSSTFDVIDLMKTGHKNRAVG 592
Query: 589 SNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQN 648
S +N+ SSRSH L + V+ ++L +G + + LVDL+GSER+ K++V G+RLKEAQ+
Sbjct: 593 STALNDRSSRSHSCLTVHVQGRDLTSGAVLRGCMHLVDLAGSERVDKSEVTGDRLKEAQH 652
Query: 649 INRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETL 708
INRSLSALGDVI++LA K+ H+PYRNSKLT LLQDSLGG +KTLMFV ISP VGET+
Sbjct: 653 INRSLSALGDVIASLAHKNPHVPYRNSKLTQLLQDSLGGQAKTLMFVHISPEADAVGETI 712
Query: 709 SSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESK 768
S+L FA RV VEL + DT ++++ K + K RK ES QN K
Sbjct: 713 STLKFAERVATVELGAARVNNDTSDVKELKEQI------ATLKAALARKEAESQQNNILK 766
Query: 769 AKGKDNIHKNLQEKIKELEGQIKLKTSMQNQSEK 802
G HK +++ I K S + E+
Sbjct: 767 TPGGSEKHKAKTGEVEIHNNNIMTKKSESCEVEE 800
>At1g73860 kinesin-related protein
Length = 1050
Score = 339 bits (869), Expect = 5e-93
Identities = 236/682 (34%), Positives = 366/682 (53%), Gaps = 64/682 (9%)
Query: 238 KVMQTKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQ------------- 284
KV+ T A E ++KEL E KE + A +L E +++++++
Sbjct: 366 KVVNTAKNA-LEERVKEL----EQMGKEAHSAKNALEEKIKQLQQMEKETKTANTSLEGK 420
Query: 285 -MELDQ--VTFKSFTTELTAEKQAENLRSISNRYELDKKKW----AEAIISLQEKVQLMK 337
EL+Q V +K+ E+ EK++E+ ++ EL K + ++A++ L+ + +K
Sbjct: 421 IQELEQNLVMWKTKVREM--EKKSESNHQRWSQKELSYKSFIDNQSQALLELRSYSRSIK 478
Query: 338 SEQSRLSFKAHECVDSIPELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKG 397
E ++ D +L K + EL E+ Y+ +T+ +KLFNE+QE KG
Sbjct: 479 QEILKVQ---ENYTDQFSQLGKKLI---ELSNAAEN----YHAVLTENRKLFNELQELKG 528
Query: 398 NIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGIL-ATGSSK---KLFRFDRVYTPKD 453
NIRVFCR RP + TVV++ +DG L + T K + F+F++VY+P
Sbjct: 529 NIRVFCRVRPF--LPAQGAANTVVEY-VGEDGELVVTNPTRPGKDGLRQFKFNKVYSPTA 585
Query: 454 DQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLF 509
Q DVF+D +V SVLDGYNVCIFAYGQTG+GKT+TM G +E++ GVNYR L LF
Sbjct: 586 SQADVFSDIRPLVRSVLDGYNVCIFAYGQTGSGKTYTMTGPDGSSEEDWGVNYRALNDLF 645
Query: 510 RVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLEIKQN-------YEGHHHVPG 562
++S+ R SY++ V ++E+YNEQ+ DLL+ + K+ N + VP
Sbjct: 646 KISQSRKGNISYEVGVQMVEIYNEQVLDLLSDDNSQKKYPFVLNPGILSTTQQNGLAVPD 705
Query: 563 VVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKL 622
V + SDV T++ G RAVGS +NE SSRSH ++ + V+ K+L G L
Sbjct: 706 ASMYPVTSTSDVITLMDIGLQNRAVGSTALNERSSRSHSIVTVHVRGKDLKTGSVLYGNL 765
Query: 623 WLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQ 682
LVDL+GSER+ +++V G+RL+EAQ+IN+SLS+LGDVI +LA+KSSH+PYRNSKLT LLQ
Sbjct: 766 HLVDLAGSERVDRSEVTGDRLREAQHINKSLSSLGDVIFSLASKSSHVPYRNSKLTQLLQ 825
Query: 683 DSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLD 742
SLGG +KTLMFVQ++P E++S+L FA RV GVEL K + +++ L
Sbjct: 826 TSLGGRAKTLMFVQLNPDATSYSESMSTLKFAERVSGVELGAAKTSKEGKDVRDLMEQLA 885
Query: 743 KARSECRCKEESLRKLEESLQNIESKAKGK------DNIHKNLQEKIKELEGQIKLKTSM 796
+ K+E + +L+ Q ++ + D+I+ + E + + S+
Sbjct: 886 SLKDTIARKDEEIERLQHQPQRLQKSMMRRKSIGHTDDINSDTGEYSSQSRYSVTDGESL 945
Query: 797 QNQSEKQVSQLCERLKGKEETCCTLQHKVKELER--KIKEQLQTETANFQQKVCNTFAKL 854
+ +E + + + + T Q + +R +I ++ ++ TA V KL
Sbjct: 946 ASSAEAEYDERLSEITSDAASMGT-QGSIDVTKRPPRISDRAKSVTAKSSTSVTRPLDKL 1004
Query: 855 LPTLFSNSLFLPKVWDLEKKLK 876
+ + KV L K
Sbjct: 1005 RKVATRTTSTVAKVTGLTSSSK 1026
Score = 42.7 bits (99), Expect = 0.001
Identities = 27/95 (28%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query: 758 LEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLKGKEET 817
LE L+ +E + K + L+E++KELE K S +N E+++ QL + K +
Sbjct: 354 LESRLKELEQEGKVVNTAKNALEERVKELEQMGKEAHSAKNALEEKIKQLQQMEKETKTA 413
Query: 818 CCTLQHKVKELER-------KIKEQLQTETANFQQ 845
+L+ K++ELE+ K++E + +N Q+
Sbjct: 414 NTSLEGKIQELEQNLVMWKTKVREMEKKSESNHQR 448
Score = 33.5 bits (75), Expect = 0.55
Identities = 26/101 (25%), Positives = 50/101 (48%), Gaps = 9/101 (8%)
Query: 738 KAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQ 797
++ L + E + + LEE ++ +E K + L+EKIK+L+ Q++ +T
Sbjct: 355 ESRLKELEQEGKVVNTAKNALEERVKELEQMGKEAHSAKNALEEKIKQLQ-QMEKETKTA 413
Query: 798 NQS-EKQVSQLCERLKGKEETCCTLQHKVKELERKIKEQLQ 837
N S E ++ +L E+ + KV+E+E+K + Q
Sbjct: 414 NTSLEGKIQEL-------EQNLVMWKTKVREMEKKSESNHQ 447
>At5g27000 kinesin-like heavy chain (KATD)
Length = 987
Score = 338 bits (868), Expect = 6e-93
Identities = 184/372 (49%), Positives = 247/372 (65%), Gaps = 11/372 (2%)
Query: 378 YYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGILATG 437
Y + + +KL+N VQ+ KGNIRV+CR RP + S G + V D D +G + I
Sbjct: 374 YKRVLEENRKLYNLVQDLKGNIRVYCRVRPFLPGQESGGLSAVEDID---EGTITIRVPS 430
Query: 438 ----SSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG 493
+ +K F F++V+ P Q +VF+D +V SVLDGYNVCIFAYGQTG+GKTFTM G
Sbjct: 431 KYGKAGQKPFMFNKVFGPSATQEEVFSDMQPLVRSVLDGYNVCIFAYGQTGSGKTFTMTG 490
Query: 494 ----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLE 549
TE++ GVNYR L LF +S +R +T SY+ISV +LE+YNEQ+RDLLA +KRLE
Sbjct: 491 PKELTEESLGVNYRALADLFLLSNQRKDTTSYEISVQMLEIYNEQVRDLLAQDGQTKRLE 550
Query: 550 IKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKT 609
I+ N +VP V + DV ++ G RAV S +N+ SSRSH + + V+
Sbjct: 551 IRNNSHNGINVPEASLVPVSSTDDVIQLMDLGHMNRAVSSTAMNDRSSRSHSCVTVHVQG 610
Query: 610 KNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSH 669
++L +G + LVDL+GSER+ K++V G+RLKEAQ+IN+SLSALGDVIS+L+ K+SH
Sbjct: 611 RDLTSGSILHGSMHLVDLAGSERVDKSEVTGDRLKEAQHINKSLSALGDVISSLSQKTSH 670
Query: 670 IPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQI 729
+PYRNSKLT LLQDSLGG +KTLMFV ISP +GET+S+L FA RV VEL +
Sbjct: 671 VPYRNSKLTQLLQDSLGGSAKTLMFVHISPEPDTLGETISTLKFAERVGSVELGAARVNK 730
Query: 730 DTGELQKTKAML 741
D E+++ K +
Sbjct: 731 DNSEVKELKEQI 742
>At1g18410 kinesin-related protein, putative
Length = 1081
Score = 334 bits (856), Expect = 1e-91
Identities = 209/584 (35%), Positives = 316/584 (53%), Gaps = 30/584 (5%)
Query: 243 KSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSFTTELTAE 302
+ T ++ + E KE TSL +RE+E+ + E V E
Sbjct: 469 QETMTVTTSLEAQNRELEQAIKETMTVNTSLEAKNRELEQSKKETMTVNTSLKAKNRELE 528
Query: 303 KQAENLRS----ISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDSIPEL- 357
+ + +S + + EL + W++ +S + + L F + I ++
Sbjct: 529 QNLVHWKSKAKEMEEKSELKNRSWSQKELSYRSFISFQCQALQELRFYSKSIKQEILKVQ 588
Query: 358 NKMVYAVQELVK---QCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMS 414
+K +L K + D Y+E +T+ +KLFNE+QE KGNIRV+CR RP + + +
Sbjct: 589 DKYTVEFSQLGKKLLELGDAAANYHEVLTENQKLFNELQELKGNIRVYCRVRPFLRGQGA 648
Query: 415 SGCTTVVDFDAAKDGCLGILATGSSK---KLFRFDRVYTPKDDQVDVFADASSMVISVLD 471
S TVV+ + + T K + FRF++VY+P Q +VF+D ++ SVLD
Sbjct: 649 S--KTVVEHIGDHGELVVLNPTKPGKDAHRKFRFNKVYSPASTQAEVFSDIKPLIRSVLD 706
Query: 472 GYNVCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSV 527
GYNVCIFAYGQTG+GKT+TM G +E+ GVNYR L LFR+S+ R +Y++ V +
Sbjct: 707 GYNVCIFAYGQTGSGKTYTMTGPDGASEEEWGVNYRALNDLFRISQSRKSNIAYEVGVQM 766
Query: 528 LEVYNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAV 587
+E+YNEQ+RDLL+ + + VP V + SDV ++ G R V
Sbjct: 767 VEIYNEQVRDLLSG-------ILSTTQQNGLAVPDASMYPVTSTSDVLELMSIGLQNRVV 819
Query: 588 GSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQ 647
S +NE SSRSH ++ + V+ K+L G L LVDL+GSER+ +++V G+RLKEAQ
Sbjct: 820 SSTALNERSSRSHSIVTVHVRGKDLKTGSALYGNLHLVDLAGSERVDRSEVTGDRLKEAQ 879
Query: 648 NINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGET 707
+IN+SLSALGDVI +LA+KSSH+PYRNSKLT LLQ SLGG +KTLMFVQ++P E+
Sbjct: 880 HINKSLSALGDVIFSLASKSSHVPYRNSKLTQLLQSSLGGRAKTLMFVQLNPDITSYSES 939
Query: 708 LSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLE-----ESL 762
+S+L FA RV GVEL K D ++++ L + K++ + +L
Sbjct: 940 MSTLKFAERVSGVELGAAKSSKDGRDVRELMEQLGSLKDTIARKDDEIERLHLLKDINYP 999
Query: 763 QNIESKAKGK-DNIHKNLQEKIKELEGQIKLKTSMQNQSEKQVS 805
Q ++ K+ G+ D+ + + +E + + QS V+
Sbjct: 1000 QRLQKKSLGQSDDFNSEAGDSQLSIEDDSRFQHDYTRQSRHSVT 1043
Score = 45.8 bits (107), Expect = 1e-04
Identities = 73/282 (25%), Positives = 123/282 (42%), Gaps = 32/282 (11%)
Query: 578 LQAGSNARAVGSNNVNEHSSRSHCMLC--IMVKTKNLMNGECTKSKLWLVDLSGSERLAK 635
LQA + + SNN++ ++S + I+ +T NGE + L+ E +
Sbjct: 231 LQAAKISELMKSNNLDNAPTQSLLSIVNGILDETIERKNGELPQRVACLLRKVVQEIERR 290
Query: 636 TDVQGERLKEAQNINRS----LSALGDVISALAAKSSHI-PYRNSKLTHLLQDSLGGDSK 690
Q E L+ ++ ++ + V+ LA+ +S SKL +D K
Sbjct: 291 ISTQSEHLRTQNSVFKAREEKYQSRIKVLETLASGTSEENETEKSKLEEKKKD------K 344
Query: 691 TLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQ-KTKAMLDKARSECR 749
V I + +S+L EL+ KK + LQ ++K A E R
Sbjct: 345 EEDMVGIEKENGHYNLEISTLRR-------ELETTKKAYEQQCLQMESKTKGATAGIEDR 397
Query: 750 CKE-ESLRK--------LEESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQS 800
KE E +RK LEE ++ +E K D + NL+EK+KEL+ + K +T S
Sbjct: 398 VKELEQMRKDASVARKALEERVRELEKMGKEADAVKMNLEEKVKELQ-KYKDETITVTTS 456
Query: 801 EKQVSQLCERLKGKEETCCT-LQHKVKELERKIKEQLQTETA 841
+ ++ E+ K + T T L+ + +ELE+ IKE + T+
Sbjct: 457 IEGKNRELEQFKQETMTVTTSLEAQNRELEQAIKETMTVNTS 498
>At3g44730 kinesin-like protein heavy chain (KP1)
Length = 1087
Score = 331 bits (848), Expect = 1e-90
Identities = 193/452 (42%), Positives = 277/452 (60%), Gaps = 18/452 (3%)
Query: 324 EAIISLQEKVQLMKSE--QSRLSFKAHECVDSIPELNKMVYAVQELVKQCEDLKVKYYEE 381
EAI Q++++ +KS ++R K + E K + + VK E Y++
Sbjct: 306 EAIGLQQKELEEVKSNFVETRSQVKQMQS-----EWQKELQRIVHHVKAMEVTSSSYHKV 360
Query: 382 MTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCL---GILATGS 438
+ + + L+NEVQ+ KG IRV+CR RP + + T VD+ +
Sbjct: 361 LEENRLLYNEVQDLKGTIRVYCRVRPFFQEQKDMQST--VDYIGENGNIIINNPFKQEKD 418
Query: 439 SKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG----T 494
++K+F F++V+ Q ++ D ++ SVLDG+NVCIFAYGQTG+GKT+TM G T
Sbjct: 419 ARKIFSFNKVFGQTVSQEQIYIDTQPVIRSVLDGFNVCIFAYGQTGSGKTYTMSGPDLMT 478
Query: 495 EQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLEIKQNY 554
E GVNYR L LF++S R+ +Y+I V ++E+YNEQ+RDLL + +S+RL+I+ N
Sbjct: 479 ETTWGVNYRALRDLFQLSNARTHVVTYEIGVQMIEIYNEQVRDLLVSDGSSRRLDIRNNS 538
Query: 555 EGHH-HVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLM 613
+ + +VP V N DV +++ G RAVG+ +NE SSRSH +L + V+ K L
Sbjct: 539 QLNGLNVPDANLIPVSNTRDVLDLMRIGQKNRAVGATALNERSSRSHSVLTVHVQGKELA 598
Query: 614 NGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYR 673
+G + L LVDL+GSER+ K++ GERLKEAQ+IN+SLSALGDVI ALA KSSH+PYR
Sbjct: 599 SGSILRGCLHLVDLAGSERVEKSEAVGERLKEAQHINKSLSALGDVIYALAQKSSHVPYR 658
Query: 674 NSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGE 733
NSKLT +LQDSLGG +KTLMFV I+P VGET+S+L FA RV +EL + +TGE
Sbjct: 659 NSKLTQVLQDSLGGQAKTLMFVHINPEVNAVGETISTLKFAQRVASIELGAARSNKETGE 718
Query: 734 LQKTKAMLDKARSECRCKEESLRKLEE-SLQN 764
++ K + +S KE L +L S++N
Sbjct: 719 IRDLKDEISSLKSAMEKKEAELEQLRSGSIRN 750
>At1g09170 putative kinesin
Length = 1046
Score = 317 bits (811), Expect = 2e-86
Identities = 195/465 (41%), Positives = 274/465 (57%), Gaps = 42/465 (9%)
Query: 378 YYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGCLGILATG 437
Y + + +KL+N+VQ+ KG+IRV+CR RP + S TTV D +D L I
Sbjct: 414 YQRVLEENRKLYNQVQDLKGSIRVYCRVRPFLPGQKSV-LTTV---DHLEDSTLSIATPS 469
Query: 438 S----SKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEG 493
+K F F++V+ P Q VFAD ++ SVLDGYNVCIFAYGQTG+GKTFTM G
Sbjct: 470 KYGKEGQKTFTFNKVFGPSASQEAVFADTQPLIRSVLDGYNVCIFAYGQTGSGKTFTMMG 529
Query: 494 ----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASKRLE 549
T++ GVNYR L LF +S R ETFSY+ISV +LE+YNEQIR+ G
Sbjct: 530 PNELTDETLGVNYRALSDLFHLSSVRKETFSYNISVQMLEIYNEQIRNSTQDGI------ 583
Query: 550 IKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKT 609
+VP V SDV ++ G RAV + +N+ SSRSH L + V+
Sbjct: 584 ---------NVPEATLVPVSTTSDVIHLMNIGQKNRAVSATAMNDRSSRSHSCLTVHVQG 634
Query: 610 KNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSH 669
K+L +G + + LVDL+GSER+ K++V G+RLKEAQ+IN+SLSALGDVI++L+ K++H
Sbjct: 635 KDLTSGVTLRGSMHLVDLAGSERIDKSEVTGDRLKEAQHINKSLSALGDVIASLSQKNNH 694
Query: 670 IPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQI 729
IPYRNSKLT LLQD+LGG +KTLMF+ ISP +D+GETLS+L FA RV V+L +
Sbjct: 695 IPYRNSKLTQLLQDALGGQAKTLMFIHISPELEDLGETLSTLKFAERVATVDLGAARVNK 754
Query: 730 DTGELQKTKAM-----LDKARSECRCKEESLRK-------LEESLQNIESKAKGKDNIHK 777
DT E+++ K L AR E + L++ L + + S N +
Sbjct: 755 DTSEVKELKEQIASLKLALARKESGADQTQLQRPLTPDKLLRKKSLGVSSSFSKSANSTR 814
Query: 778 NLQEKIKELEGQIKLKTSMQNQSEKQVSQLCERLK-GKEETCCTL 821
+Q K K QI S++ + +++ +++K G+ ++ +L
Sbjct: 815 QVQTKHK--PSQIDDVNSIEVKGTRKLIHQTKQIKLGQSDSASSL 857
>At1g63640 kinesin-like protein
Length = 1071
Score = 310 bits (794), Expect = 2e-84
Identities = 193/545 (35%), Positives = 301/545 (54%), Gaps = 19/545 (3%)
Query: 243 KSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSFTTELTAE 302
K + + +I++L + +L KE +E E+ + +K + EL++ K EL
Sbjct: 319 KEKERSDAEIRQLKQELKL-VKETHE--NQCLELEAKAQKTRDELEK---KLKDAELHVV 372
Query: 303 KQAENLRSISNRYELDKKKWAEAIISLQEKVQ-----LMKSEQSRLSFKAHECVDSIPEL 357
+ ++ + + ++W + Q + L + + LS K HE V + +
Sbjct: 373 DSSRKVKELEKLCQSKSQRWEKKECIYQNFIDNHSGALQELSATSLSIK-HEVVRTQRKY 431
Query: 358 NKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGC 417
+ + +K D Y+ + + ++L+NEVQE KGNIRV+CR RP + S
Sbjct: 432 FEDLNYYGLKLKGVADAAKNYHVVLEENRRLYNEVQELKGNIRVYCRIRPFLPGQNSRQT 491
Query: 418 TTVVDFDAAKDGCLGILATGS-SKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVC 476
T + + G + +LF+F++V+ Q +VF D ++ S+LDGYNVC
Sbjct: 492 TIEYIGETGELVVANPFKQGKDTHRLFKFNKVFDQAATQEEVFLDTRPLIRSILDGYNVC 551
Query: 477 IFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYN 532
IFAYGQTG+GKT+TM G ++++ GVNYR L LF +++ R T Y++ V ++E+YN
Sbjct: 552 IFAYGQTGSGKTYTMSGPSITSKEDWGVNYRALNDLFLLTQSRQNTVMYEVGVQMVEIYN 611
Query: 533 EQIRDLLATGPASKRLEIKQN-YEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNN 591
EQ+RD+L+ G +S+R I VP V + DV ++ G R VG+
Sbjct: 612 EQVRDILSDGGSSRRYRIWNTALPNGLAVPDASMHCVRSTEDVLELMNIGLMNRTVGATA 671
Query: 592 VNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINR 651
+NE SSRSHC+L + V+ ++ + L LVDL+GSER+ +++ GERLKEAQ+IN+
Sbjct: 672 LNERSSRSHCVLSVHVRGVDVETDSILRGSLHLVDLAGSERVDRSEATGERLKEAQHINK 731
Query: 652 SLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSL 711
SLSALGDVI ALA K+ H+PYRNSKLT +LQ SLGG +KTLMFVQ++P ET+S+L
Sbjct: 732 SLSALGDVIFALAHKNPHVPYRNSKLTQVLQSSLGGQAKTLMFVQVNPDGDSYAETVSTL 791
Query: 712 NFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEE-SLQNIESKAK 770
FA RV GVEL K + ++++ + + K+E L+ ++ N S +
Sbjct: 792 KFAERVSGVELGAAKSSKEGRDVRQLMEQVSNLKDVIAKKDEELQNFQKVKGNNATSLKR 851
Query: 771 GKDNI 775
G N+
Sbjct: 852 GLSNL 856
>At5g41310 kinesin-like protein
Length = 967
Score = 309 bits (791), Expect = 5e-84
Identities = 192/542 (35%), Positives = 303/542 (55%), Gaps = 43/542 (7%)
Query: 243 KSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSFTTELTAE 302
K + ++ +L + E+ KE +E E+ +K ++EL++ S + A+
Sbjct: 279 KGKERSNAELSKLKQELEI-VKETHEK--QFLELKLNAQKAKVELERQVKNSELRVVEAK 335
Query: 303 KQAENLRSISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVDSIPELNKMVY 362
+ + + + R+E ++ + I E +Q +K+ L + ++ L+ Y
Sbjct: 336 ELEKLCETKTKRWEKKEQTYKRFINHQTEALQELKATSMSLKHDVLKIGENY-FLDLTYY 394
Query: 363 AVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPL----NKVEMSSGCT 418
++ ++ Y + + ++L+NEVQE KGNIRV+CR RP NK + S T
Sbjct: 395 GIK--LRGVAHAAKNYQIIIEENRRLYNEVQELKGNIRVYCRIRPFLQGQNKKQTSIEYT 452
Query: 419 ----TVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYN 474
+V + K G + +LF+F++V+ P+ Q +VF D M+ S+LDGYN
Sbjct: 453 GENGELVVANPLKQG-------KDTYRLFKFNKVFGPESTQEEVFLDTRPMIRSILDGYN 505
Query: 475 VCIFAYGQTGTGKTFTMEG----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEV 530
VCIFAYGQTG+GKT+TM G +E++RGVNYR L LF +++ R + Y++ V ++E+
Sbjct: 506 VCIFAYGQTGSGKTYTMSGPSITSEEDRGVNYRALNDLFHLTQSRQNSVMYEVGVQMVEI 565
Query: 531 YNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSN 590
YNEQ+RDLL+ VP V + DV ++ G R VG+
Sbjct: 566 YNEQVRDLLS-----------------QDVPDASMHSVRSTEDVLELMNIGLMNRTVGAT 608
Query: 591 NVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNIN 650
+NE SSRSH +L + V+ ++ + L LVDL+GSER+ +++V GERLKEAQ+IN
Sbjct: 609 TLNEKSSRSHSVLSVHVRGVDVKTESVLRGSLHLVDLAGSERVGRSEVTGERLKEAQHIN 668
Query: 651 RSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSS 710
+SLSALGDVI ALA K+ H+PYRNSKLT +LQ+SLGG +KTLMFVQI+P + ET+S+
Sbjct: 669 KSLSALGDVIFALAHKNPHVPYRNSKLTQVLQNSLGGQAKTLMFVQINPDEDSYAETVST 728
Query: 711 LNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAK 770
L FA RV GVEL + + ++++ + + K+E L+K +++ I+ +
Sbjct: 729 LKFAERVSGVELGAARSYKEGRDVRQLMEQVSNLKDMIAKKDEELQKF-QNINGIQKRGL 787
Query: 771 GK 772
K
Sbjct: 788 SK 789
>At5g65930 kinesin-like calmodulin-binding protein
Length = 1260
Score = 305 bits (782), Expect = 6e-83
Identities = 193/558 (34%), Positives = 308/558 (54%), Gaps = 41/558 (7%)
Query: 242 TKSTAKYENKIKELTMQCELKAKECYEAWTSLTEMSREVEKVQMELDQVTFKSFTTELTA 301
TKS+ + ++++ E+ Q K ++ E ++ + K + +++ ++ E
Sbjct: 714 TKSSKETKSELAEMNNQILYKIQKELEVRNKELHVAVDNSKRLLSENKILEQNLNIE--- 770
Query: 302 EKQAENLRSISNRYELDKKKWAEAIISLQEKVQLMK----SEQSRLSFK----------- 346
+K+ E + RYE +KK + L+ K++++ S +S + K
Sbjct: 771 KKKKEEVEIHQKRYEQEKKVLKLRVSELENKLEVLAQDLDSAESTIESKNSDMLLLQNNL 830
Query: 347 -----AHECVDSIPELNKMVYAVQELV-KQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIR 400
E + I N+ A+ ++ Q +L++ Y EE RK+ +N +++ KG IR
Sbjct: 831 KELEELREMKEDIDRKNEQTAAILKMQGAQLAELEILYKEEQVLRKRYYNTIEDMKGKIR 890
Query: 401 VFCRCRPLNKVEMSSG----CTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQV 456
V+CR RPLN+ E S TTV +F +K +DRV+ + Q
Sbjct: 891 VYCRIRPLNEKESSEREKQMLTTVDEFTVEHPW------KDDKRKQHIYDRVFDMRASQD 944
Query: 457 DVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERS 516
D+F D +V S +DGYNVCIFAYGQTG+GKTFT+ G E N G+ R + LF + K S
Sbjct: 945 DIFEDTKYLVQSAVDGYNVCIFAYGQTGSGKTFTIYGHESNPGLTPRATKELFNILKRDS 1004
Query: 517 ETFSYDISVSVLEVYNEQIRDLLATGPASK-RLEIKQNYEGHHHVPGVVEAKVDNISDVW 575
+ FS+ + ++E+Y + + DLL A + +LEIK++ +G V V + + ++
Sbjct: 1005 KRFSFSLKAYMVELYQDTLVDLLLPKSARRLKLEIKKDSKGMVFVENVTTIPISTLEELR 1064
Query: 576 TVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAK 635
+L+ GS R V N+NE SSRSH +L +++++ +L + KL VDL+GSER+ K
Sbjct: 1065 MILERGSERRHVSGTNMNEESSRSHLILSVVIESIDLQTQSAARGKLSFVDLAGSERVKK 1124
Query: 636 TDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFV 695
+ G +LKEAQ+IN+SLSALGDVI AL++ + HIPYRN KLT L+ DSLGG++KTLMFV
Sbjct: 1125 SGSAGCQLKEAQSINKSLSALGDVIGALSSGNQHIPYRNHKLTMLMSDSLGGNAKTLMFV 1184
Query: 696 QISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESL 755
+SP++ ++ ET +SL +A+RVR + DP K I + E+ + K ++ + + K E
Sbjct: 1185 NVSPAESNLDETYNSLLYASRVRTIVNDP-SKHISSKEMVRLKKLVAYWKEQAGKKGE-- 1241
Query: 756 RKLEESLQNIESKAKGKD 773
EE L +IE KD
Sbjct: 1242 ---EEDLVDIEEDRTRKD 1256
>At4g27180 heavy chain polypeptide of kinesin-like protein (katB)
Length = 745
Score = 299 bits (766), Expect = 4e-81
Identities = 189/487 (38%), Positives = 273/487 (55%), Gaps = 21/487 (4%)
Query: 264 KECYEAWTSLTEMSREVEKVQMELDQVTFKSFTTELTAEKQAENLRSISNRYELDKKKWA 323
K+ E + + E+++V+ + D+ + T + A KQ + + N E
Sbjct: 257 KQKDELVNEIVSLKVEIQQVKDDRDRHITEIETLQAEATKQ-NDFKDTINELESKCSVQN 315
Query: 324 EAIISLQEKVQLMKSEQSRLSFKAHECVDSIPELNKMVYAVQELVKQCEDLKVKYYEEMT 383
+ I LQ+ QL+ SE+ +L + + E + ++ EL + E+ ++K E
Sbjct: 316 KEIEELQD--QLVASER-KLQVADLSTFEKMNEFEEQKESIMELKGRLEEAELKLIEGEK 372
Query: 384 QRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGC---LGILATGSSK 440
RKKL N +QE KGNIRVFCR RPL E SS + + + + + +L G S
Sbjct: 373 LRKKLHNTIQELKGNIRVFCRVRPLLSGENSSEEAKTISYPTSLEALGRGIDLLQNGQSH 432
Query: 441 KLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEGTEQN--- 497
F FD+V+ P Q DVF + S +V S LDGY VCIFAYGQTG+GKT+TM G N
Sbjct: 433 -CFTFDKVFVPSASQEDVFVEISQLVQSALDGYKVCIFAYGQTGSGKTYTMMGRPGNPDE 491
Query: 498 RGVNYRTLEHLFRVSKE-RSETFSYDISVSVLEVYNEQIRDLLAT---------GPASKR 547
+G+ R LE +F+ + RS+ + Y++ VS+LE+YNE IRDLL+T G + ++
Sbjct: 492 KGLIPRCLEQIFQTRQSLRSQGWKYELQVSMLEIYNETIRDLLSTNKEAVRADNGVSPQK 551
Query: 548 LEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMV 607
IK + G+ HV + V + V +L + R+VG +NE SSRSH + + +
Sbjct: 552 YAIKHDASGNTHVVELTVVDVRSSKQVSFLLDHAARNRSVGKTAMNEQSSRSHFVFTLKI 611
Query: 608 KTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKS 667
N + + L L+DL+GSERL+K+ G+RLKE Q IN+SLS+LGDVI ALA K
Sbjct: 612 SGFNESTEQQVQGVLNLIDLAGSERLSKSGSTGDRLKETQAINKSLSSLGDVIFALAKKE 671
Query: 668 SHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKK 727
H+P+RNSKLT+LLQ LGGDSKTLMFV I+P GE+L SL FA RV E+ +
Sbjct: 672 DHVPFRNSKLTYLLQPCLGGDSKTLMFVNITPEPSSTGESLCSLRFAARVNACEIGTAHR 731
Query: 728 QIDTGEL 734
++ L
Sbjct: 732 HVNARPL 738
Score = 35.0 bits (79), Expect = 0.19
Identities = 23/92 (25%), Positives = 46/92 (50%), Gaps = 2/92 (2%)
Query: 703 DVGETLSSLNFATRVRGVELDPVKKQIDTGE--LQKTKAMLDKARSECRCKEESLRKLEE 760
D +T++ L V+ E++ ++ Q+ E LQ + +E ++ES+ +L+
Sbjct: 299 DFKDTINELESKCSVQNKEIEELQDQLVASERKLQVADLSTFEKMNEFEEQKESIMELKG 358
Query: 761 SLQNIESKAKGKDNIHKNLQEKIKELEGQIKL 792
L+ E K + + K L I+EL+G I++
Sbjct: 359 RLEEAELKLIEGEKLRKKLHNTIQELKGNIRV 390
>At4g21270 kinesin-related protein katA
Length = 793
Score = 293 bits (749), Expect = 4e-79
Identities = 184/487 (37%), Positives = 272/487 (55%), Gaps = 26/487 (5%)
Query: 271 TSLTEMSREVEKVQMELDQVTFKSFTTELTAEKQAENLRSISNRYEL--DKKKWAEAIIS 328
+ +T + E+++V+ + D+ +S K EN+ S ++ K E S
Sbjct: 297 SEVTNLRNELQQVRDDRDRQVVQSQKLSEEIRKYQENVGKSSQELDILTAKSGSLEETCS 356
Query: 329 LQEKVQLMKSEQSRLSFKAHECVDSIPELNKMVYAVQ-----ELVKQCEDLKVKYYEEMT 383
LQ++ M +Q ++ + + D+ L + + Q EL + D++ + E
Sbjct: 357 LQKERLNMLEQQLAIANERQKMADASVSLTRTEFEEQKHLLCELQDRLADMEHQLCEGEL 416
Query: 384 QRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGC-LGI-LATGSSKK 441
RKKL N + E KGNIRVFCR RPL + TV+ + + + G+ L +K
Sbjct: 417 LRKKLHNTILELKGNIRVFCRVRPLLPDDGGRHEATVIAYPTSTEAQGRGVDLVQSGNKH 476
Query: 442 LFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEGTEQ---NR 498
F FD+V+ + Q +VF + S +V S LDGY VCIFAYGQTG+GKT+TM G + +
Sbjct: 477 PFTFDKVFNHEASQEEVFFEISQLVQSALDGYKVCIFAYGQTGSGKTYTMMGRPEAPDQK 536
Query: 499 GVNYRTLEHLFRVSKER-SETFSYDISVSVLEVYNEQIRDLLAT-------------GPA 544
G+ R+LE +F+ S+ ++ + Y + VS+LE+YNE IRDLL+T G +
Sbjct: 537 GLIPRSLEQIFQASQSLGAQGWKYKMQVSMLEIYNETIRDLLSTNRTTSMDLVRADSGTS 596
Query: 545 SKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLC 604
K+ I + GH HV + V ++ + ++LQ + +R+VG +NE SSRSH +
Sbjct: 597 GKQYTITHDVNGHTHVSDLTIFDVCSVGKISSLLQQAAQSRSVGKTQMNEQSSRSHFVFT 656
Query: 605 IMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALA 664
+ + N + + L L+DL+GSERL+K+ G+RLKE Q IN+SLSAL DVI ALA
Sbjct: 657 MRISGVNESTEQQVQGVLNLIDLAGSERLSKSGATGDRLKETQAINKSLSALSDVIFALA 716
Query: 665 AKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDP 724
K H+P+RNSKLT+LLQ LGGDSKTLMFV ISP GE+L SL FA RV E+
Sbjct: 717 KKEDHVPFRNSKLTYLLQPCLGGDSKTLMFVNISPDPTSAGESLCSLRFAARVNACEIGI 776
Query: 725 VKKQIDT 731
++Q T
Sbjct: 777 PRRQTST 783
>At5g54670 heavy chain polypeptide of kinesin-like protein (katC)
Length = 754
Score = 291 bits (744), Expect = 1e-78
Identities = 186/485 (38%), Positives = 267/485 (54%), Gaps = 30/485 (6%)
Query: 275 EMSREVEKVQMELDQVT---------FKSFTTELTAEKQAENLRSISNRYELDKKKWAEA 325
E+ E+ +++EL QV K+ TE T + + + E +
Sbjct: 270 ELVNEIASLKVELQQVKDDRDRHLVEVKTLQTEAT---KYNDFKDAITELETTCSSQSTQ 326
Query: 326 IISLQEKVQLMKSEQSRLSFKAHECVDSIPELNKMVYAVQELVKQCEDLKVKYYEEMTQR 385
I LQ++ L+ SE+ RL + + E ++ +L + E+ ++K E R
Sbjct: 327 IRQLQDR--LVNSER-RLQVSDLSTFEKMNEYEDQKQSIIDLKSRVEEAELKLVEGEKLR 383
Query: 386 KKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGC-LGI-LATGSSKKLF 443
KKL N + E KGNIRVFCR RPL E + + + + + GI L + K F
Sbjct: 384 KKLHNTILELKGNIRVFCRVRPLLPGENNGDEGKTISYPTSLEALGRGIDLMQNAQKHAF 443
Query: 444 RFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEGTEQN---RGV 500
FD+V+ P Q DVF + S +V S LDGY VCIFAYGQTG+GKT+TM G N +G+
Sbjct: 444 TFDKVFAPTASQEDVFTEISQLVQSALDGYKVCIFAYGQTGSGKTYTMMGRPGNVEEKGL 503
Query: 501 NYRTLEHLFRVSKE-RSETFSYDISVSVLEVYNEQIRDLLATGPASKRLE---------I 550
R LE +F + RS+ + Y++ VS+LE+YNE IRDLL+T + R + I
Sbjct: 504 IPRCLEQIFETRQSLRSQGWKYELQVSMLEIYNETIRDLLSTNKEAVRTDSGVSPQKHAI 563
Query: 551 KQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTK 610
K + G+ HV + V + +V +L + R+VG +NE SSRSH + + +
Sbjct: 564 KHDASGNTHVAELTILDVKSSREVSFLLDHAARNRSVGKTQMNEQSSRSHFVFTLRISGV 623
Query: 611 NLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHI 670
N + + L L+DL+GSERL+K+ G+RLKE Q IN+SLS+LGDVI ALA K H+
Sbjct: 624 NESTEQQVQGVLNLIDLAGSERLSKSGSTGDRLKETQAINKSLSSLGDVIFALAKKEDHV 683
Query: 671 PYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQID 730
P+RNSKLT+LLQ LGGD+KTLMFV I+P GE+L SL FA RV E+ ++Q +
Sbjct: 684 PFRNSKLTYLLQPCLGGDAKTLMFVNIAPESSSTGESLCSLRFAARVNACEIGTPRRQTN 743
Query: 731 TGELQ 735
L+
Sbjct: 744 IKPLE 748
>At1g55550
Length = 887
Score = 290 bits (743), Expect = 2e-78
Identities = 174/419 (41%), Positives = 242/419 (57%), Gaps = 26/419 (6%)
Query: 364 VQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDF 423
+Q + Q L V+ ++ R+++ NE + KGNIRVFCR +PL G T +
Sbjct: 57 LQSIRDQLSALTVQVNDQNKLRRQILNEFLDLKGNIRVFCRVKPL-------GATEKLRP 109
Query: 424 DAAKDGCLGILATGSSK-KLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQ 482
A D I+ +K K + FDRV+ P Q DVF + ++ SV+DGYN CIFAYGQ
Sbjct: 110 PVASDTRNVIIKLSETKRKTYNFDRVFQPDSSQDDVFLEIEPVIKSVIDGYNACIFAYGQ 169
Query: 483 TGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLL--- 539
TGTGKT+TMEG + G+ R ++ LF+ +E + F+ I S+LE+Y ++DLL
Sbjct: 170 TGTGKTYTMEGLPNSPGIVPRAIKGLFKQVEESNHMFT--IHFSMLEIYMGNLKDLLLSE 227
Query: 540 ATGPASK---RLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHS 596
AT P S L I + G + +V+ KVD+ +++ + + G +RA S N N S
Sbjct: 228 ATKPISPIPPSLSIHTDPNGEIDIENLVKLKVDDFNEILRLYKVGCRSRATASTNSNSVS 287
Query: 597 SRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSAL 656
SRSHCM+ + V + +K+WLVDL GSER+ KT G R E + IN SLSAL
Sbjct: 288 SRSHCMIRVSVTSLGAPERRRETNKIWLVDLGGSERVLKTRATGRRFDEGKAINLSLSAL 347
Query: 657 GDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATR 716
GDVI++L K+SHIPYRNSKLT +L+DSLG DSKTLM V ISP + D+ ET+ SLNFATR
Sbjct: 348 GDVINSLQRKNSHIPYRNSKLTQVLKDSLGQDSKTLMLVHISPKEDDLCETICSLNFATR 407
Query: 717 VRGVEL-------DPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESK 768
+ + L + KK+ LQK M++K E +R L E+L+ + K
Sbjct: 408 AKNIHLGQDESTEEQAKKEAVMMNLQK---MMEKIEQEREMSLRKMRNLNETLEKLTGK 463
>At4g05190 kinesin - like protein
Length = 777
Score = 285 bits (728), Expect = 1e-76
Identities = 184/495 (37%), Positives = 274/495 (55%), Gaps = 35/495 (7%)
Query: 268 EAWTSLTEMSREVEKVQMELDQVTFKSFTTELTAEKQAENLRSIS-----NRYELD---- 318
EA + EV +Q EL QV + ++K A + + +ELD
Sbjct: 271 EAVKQKDSLLMEVNNLQSELQQVRDDRDRHVVQSQKLAGEILMYKESVGKSSHELDILIA 330
Query: 319 KKKWAEAIISLQ-EKVQLMKSE----QSRLSFKAHECVDSIPELNKMVYAVQELVKQCED 373
K E SLQ E++++++ E + +L ++ E + + EL + D
Sbjct: 331 KSGSLEETCSLQKERIKMLEQELAFAKEKLKMVDLSMSHTMTEFEEQKQCMHELQDRLAD 390
Query: 374 LKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDFDAAKDGC--- 430
+ + +E RKKL N + E KGNIRVFCR RPL + +V+ + + +
Sbjct: 391 TERQLFEGELLRKKLHNTILELKGNIRVFCRVRPLLPDDGGRQEASVIAYPTSTESLGRG 450
Query: 431 LGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFT 490
+ ++ +G+ K F FD+V+ Q +VF + S +V S LDGY VCIFAYGQTG+GKT+T
Sbjct: 451 IDVVQSGN-KHPFTFDKVFDHGASQEEVFFEISQLVQSALDGYKVCIFAYGQTGSGKTYT 509
Query: 491 MEG---TEQNRGVNYRTLEHLFRVSKERS-ETFSYDISVSVLEVYNEQIRDLLATG---- 542
M G T + +G+ R+LE +F+ S+ S + + Y + VS+LE+YNE IRDLL+T
Sbjct: 510 MMGRPETPEQKGLIPRSLEQIFKTSQSLSTQGWKYKMQVSMLEIYNESIRDLLSTSRTIA 569
Query: 543 ---------PASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVN 593
+ ++ I + G+ HV + V +I + ++LQ + +R+VG ++N
Sbjct: 570 IESVRADSSTSGRQYTITHDVNGNTHVSDLTIVDVCSIGQISSLLQQAAQSRSVGKTHMN 629
Query: 594 EHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSL 653
E SSRSH + + + N + + L L+DL+GSERL+++ G+RLKE Q IN+SL
Sbjct: 630 EQSSRSHFVFTLRISGVNESTEQQVQGVLNLIDLAGSERLSRSGATGDRLKETQAINKSL 689
Query: 654 SALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNF 713
SAL DVI ALA K H+P+RNSKLT+LLQ LGGDSKTLMFV ISP GE+L SL F
Sbjct: 690 SALSDVIFALAKKEDHVPFRNSKLTYLLQPCLGGDSKTLMFVNISPDPSSTGESLCSLRF 749
Query: 714 ATRVRGVELDPVKKQ 728
A RV E+ ++Q
Sbjct: 750 AARVNACEIGIPRRQ 764
Score = 31.6 bits (70), Expect = 2.1
Identities = 32/163 (19%), Positives = 71/163 (42%), Gaps = 19/163 (11%)
Query: 737 TKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNIHKNLQEKI---KELEGQIKLK 793
+K++ D+ S ++E++++ + L + + + + + ++L G+I +
Sbjct: 255 SKSLQDQLASSRVSQDEAVKQKDSLLMEVNNLQSELQQVRDDRDRHVVQSQKLAGEILMY 314
Query: 794 TSMQNQSEKQVSQLCERLKGKEETCCTLQHKVKELERKI---KEQLQ--------TETAN 842
+S ++ L + EETC + ++K LE+++ KE+L+ T T
Sbjct: 315 KESVGKSSHELDILIAKSGSLEETCSLQKERIKMLEQELAFAKEKLKMVDLSMSHTMTEF 374
Query: 843 FQQKVC-----NTFAKLLPTLFSNSLFLPKVWDLEKKLKDQLQ 880
+QK C + A LF L K+ + +LK ++
Sbjct: 375 EEQKQCMHELQDRLADTERQLFEGELLRKKLHNTILELKGNIR 417
>At5g27950 unknown protein
Length = 625
Score = 271 bits (692), Expect = 2e-72
Identities = 172/441 (39%), Positives = 246/441 (55%), Gaps = 38/441 (8%)
Query: 356 ELNKMVYAVQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVE--- 412
E N++ ++ L ++ +LK+K +RK++ N++ + KG+IRVFCR RP E
Sbjct: 37 ESNQLEKSISNLEEEVFELKLKLKSLDEKRKQVLNKIIDTKGSIRVFCRVRPFLLTERRP 96
Query: 413 ----MSSGCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVIS 468
+S G VV I + GSSK+ F FD+V+ Q +VF + ++ S
Sbjct: 97 IREPVSFGPDNVV-----------IRSAGSSKE-FEFDKVFHQSATQEEVFGEVKPILRS 144
Query: 469 VLDGYNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVL 528
LDG+NVC+ AYGQTGTGKTFTM+GT + G+ R ++ LF + +T S +S+L
Sbjct: 145 ALDGHNVCVLAYGQTGTGKTFTMDGTSEQPGLAPRAIKELFNEAS-MDQTHSVTFRMSML 203
Query: 529 EVYNEQIRDLLATGPASK--------RLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQA 580
E+Y ++DLL+ + K L I+ + +G + G+ E +V + +
Sbjct: 204 EIYMGNLKDLLSARQSLKSYEASAKCNLNIQVDSKGSVEIEGLTEVEVMDFTKARWWYNK 263
Query: 581 GSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTK-SKLWLVDLSGSERLAKTDVQ 639
G R+ NVNE SSRSHC+ I + + G T+ SKLW++DL GSERL KT
Sbjct: 264 GRRVRSTSWTNVNETSSRSHCLTRITIFRRGDAVGSKTEVSKLWMIDLGGSERLLKTGAI 323
Query: 640 GERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISP 699
G+ + E + IN SLSALGDVI+AL K H+PYRNSKLT +L+DSLG SK LM V ISP
Sbjct: 324 GQTMDEGRAINLSLSALGDVIAALRRKKGHVPYRNSKLTQILKDSLGTRSKVLMLVHISP 383
Query: 700 SDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTK-AMLDKARSECRCKEESLRKL 758
D+DVGET+ SL+F R R VE + T ELQK + + + E +E +K+
Sbjct: 384 RDEDVGETICSLSFTKRARAVE----SNRGLTAELQKLREKKISELEEEMEETQEGCKKI 439
Query: 759 EESLQNIESKAKGKDNIHKNL 779
+ LQ +E N HK L
Sbjct: 440 KARLQEVECLV----NEHKKL 456
>At5g65460 putative protein
Length = 1264
Score = 255 bits (651), Expect = 9e-68
Identities = 192/574 (33%), Positives = 291/574 (50%), Gaps = 60/574 (10%)
Query: 310 SISNRYELDKK-------KWAEAIISLQEKVQLMKSEQSRLSFKAHECVD-SIPELNKMV 361
S+ RY + K + A + SL++KVQL K + L +A + + S +L ++
Sbjct: 35 SMLRRYSIPKNSLPPHSSELASKVQSLKDKVQLAKDDYVGLRQEATDLQEYSNAKLERVT 94
Query: 362 YAVQELVKQCEDLKVKYYEE-------MTQRKKLFNEVQEAKGNIRVFCRCRPLNK---- 410
+ L + L E + ++K+LFN++ KGN++VFCR RPL +
Sbjct: 95 RYLGVLADKSRKLDQYALETEARISPLINEKKRLFNDLLTTKGNVKVFCRARPLFEDEGP 154
Query: 411 --VEMSSGCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVIS 468
+E CT V+ D L + KK F FDRVY P+ Q +F+D V S
Sbjct: 155 SIIEFPDNCTIRVN---TSDDTLS-----NPKKEFEFDRVYGPQVGQASLFSDVQPFVQS 206
Query: 469 VLDGYNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFS-YDISVSV 527
LDG NV IFAYGQT GKT+TMEG+ Q+RG+ R E L ++ S + S + SVSV
Sbjct: 207 ALDGSNVSIFAYGQTHAGKTYTMEGSNQDRGLYARCFEELMDLANSDSTSASQFSFSVSV 266
Query: 528 LEVYNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAV 587
E+YNEQ+RDLL+ G S +I V + + KVDN S+ VL + R
Sbjct: 267 FELYNEQVRDLLS-GCQSNLPKINMGLR--ESVIELSQEKVDNPSEFMRVLNSAFQNRG- 322
Query: 588 GSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQ 647
N S+ +H ++ I + N + E SKL LVDL+GSE L D G+ + +
Sbjct: 323 ---NDKSKSTVTHLIVSIHICYSNTITRENVISKLSLVDLAGSEGLTVEDDNGDHVTDLL 379
Query: 648 NINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGET 707
++ S+SALGDV+S+L +K IPY NS LT +L DSLGG SKTLM V I PS +++ E
Sbjct: 380 HVTNSISALGDVLSSLTSKRDTIPYENSFLTRILADSLGGSSKTLMIVNICPSARNLSEI 439
Query: 708 LSSLNFATRVRGV-----ELDPVKKQIDTGELQKTKAMLDKARSECRCKEE------SLR 756
+S LN+A R R D +KK D K +L+K R R K+E +L+
Sbjct: 440 MSCLNYAARARNTVPSLGNRDTIKKWRDVAN-DARKEVLEKERENQRLKQEVTGLKQALK 498
Query: 757 KLEESLQNIESKAKGKDNIHKNLQEKIKELEGQI----KLKTSMQNQSEKQVSQLCE--- 809
+ + + ++ + + LQ +K + K++ Q Q++QL +
Sbjct: 499 EANDQCVLLYNEVQRAWRVSFTLQSDLKSENAMVVDKHKIEKEQNFQLRNQIAQLLQLEQ 558
Query: 810 ----RLKGKEETCCTLQHKVKELERKIKEQLQTE 839
+ + ++ T LQ KVK+LE ++ + L+++
Sbjct: 559 EQKLQAQQQDSTIQNLQSKVKDLESQLSKALKSD 592
>At5g10470 TH65 protein (th65)
Length = 1273
Score = 249 bits (635), Expect = 6e-66
Identities = 197/628 (31%), Positives = 310/628 (48%), Gaps = 58/628 (9%)
Query: 310 SISNRYELDKKKWAEAIISLQEKVQLMKSEQSRLSFKAHECVD-SIPELNKMVYAVQELV 368
SIS K+ A + L+EKV+L K + L +A + + S +L+++ + L
Sbjct: 46 SISTPSLPPKQAIASKVNGLKEKVKLAKEDYLELRQEATDLQEYSNAKLDRVTRYLGVLA 105
Query: 369 KQCEDLKVKYYEE-------MTQRKKLFNEVQEAKGNIRVFCRCRPL------NKVEMSS 415
++ L E + ++K+LFN++ AKGNI+VFCR RPL + +E
Sbjct: 106 EKSRKLDQFVLETEARISPLINEKKRLFNDLLTAKGNIKVFCRARPLFEDEGPSVIEFPG 165
Query: 416 GCTTVVDFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNV 475
CT V+ D L + KK F FDRVY P Q +F+D V S LDG NV
Sbjct: 166 DCTICVN---TSDDTLS-----NPKKDFEFDRVYGPHVGQAALFSDVQPFVQSALDGSNV 217
Query: 476 CIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFS-YDISVSVLEVYNEQ 534
I +YGQT GKT+TMEG+ +RG+ R E LF ++ S + S + S+SV E+YNEQ
Sbjct: 218 SILSYGQTNAGKTYTMEGSNHDRGLYARCFEELFDLANSDSTSTSRFSFSLSVFEIYNEQ 277
Query: 535 IRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNE 594
IRDLL+ ++ N + H V + + KVDN + VL++ R N
Sbjct: 278 IRDLLSETQSNLP---NINMDLHESVIELGQEKVDNPLEFLGVLKSAFLNRG----NYKS 330
Query: 595 HSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLS 654
+ +H ++ I + N + GE SKL LVDL+GSE L + G+ + + ++ S+S
Sbjct: 331 KFNVTHLIVSIHIYYSNTITGENIYSKLSLVDLAGSEGLIMENDSGDHVTDLLHVMNSIS 390
Query: 655 ALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFA 714
ALGDV+S+L + IPY NS LT +L DSLGG SKTLM V I PS Q + ET+S LN+A
Sbjct: 391 ALGDVLSSLTSGKDSIPYDNSILTRVLADSLGGSSKTLMIVNICPSVQTLSETISCLNYA 450
Query: 715 TRVRGV-----ELDPVKKQIDTGELQKTKAMLDKARSECRCKEE------SLRKLEESLQ 763
R R D +KK D K +L+K R K+E +L+ +
Sbjct: 451 ARARNTVPSLGNRDTIKKWRDVAS-DARKELLEKERENQNLKQEVVGLKKALKDANDQCV 509
Query: 764 NIESKAKGKDNIHKNLQEKIKE----LEGQIKLKTSMQNQSEKQVSQLCE-------RLK 812
+ S+ + + LQ +K L + +L+ +Q Q++Q + +++
Sbjct: 510 LLYSEVQRAWKVSFTLQSDLKSENIMLVDKHRLEKEQNSQLRNQIAQFLQLDQEQKLQMQ 569
Query: 813 GKEETCCTLQHKVKELERKIKEQLQTET-----ANFQQKVCNTFAKLLPTLFSNSLFLPK 867
++ LQ K+ +LE ++ E ++++T A Q + + K + +S K
Sbjct: 570 QQDSAIQNLQAKITDLESQVSEAVRSDTTRTGDALQSQDIFSPIPKAVEGTTDSSSVTKK 629
Query: 868 VWDLEKKLKDQLQGSESESSFLKDKVVK 895
+ + KK ++ E+ L D++ +
Sbjct: 630 LEEELKKRDALIERLHEENEKLFDRLTE 657
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,608,766
Number of Sequences: 26719
Number of extensions: 789371
Number of successful extensions: 4372
Number of sequences better than 10.0: 304
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 196
Number of HSP's that attempted gapping in prelim test: 3344
Number of HSP's gapped (non-prelim): 845
length of query: 895
length of database: 11,318,596
effective HSP length: 108
effective length of query: 787
effective length of database: 8,432,944
effective search space: 6636726928
effective search space used: 6636726928
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146711.13


