

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.5 + phase: 0 /pseudo
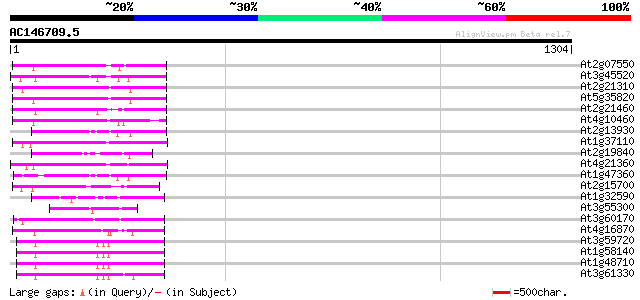
(1304 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g07550 putative retroelement pol polyprotein 229 6e-60
At3g45520 copia-like polyprotein 225 1e-58
At2g21310 putative retroelement pol polyprotein 224 3e-58
At5g35820 copia-like retrotransposable element 218 1e-56
At2g21460 putative retroelement pol polyprotein 209 1e-53
At4g10460 putative retrotransposon 201 2e-51
At2g13930 putative retroelement pol polyprotein 183 6e-46
At1g37110 179 9e-45
At2g19840 copia-like retroelement pol polyprotein 175 1e-43
At4g21360 putative transposable element 172 1e-42
At1g47360 polyprotein, putative 168 2e-41
At2g15700 copia-like retroelement pol polyprotein 126 7e-29
At1g32590 hypothetical protein, 5' partial 111 3e-24
At3g55300 putative protein 110 5e-24
At3g60170 putative protein 102 1e-21
At4g16870 retrotransposon like protein 99 1e-20
At3g59720 copia-type reverse transcriptase-like protein 98 4e-20
At1g58140 hypothetical protein 98 4e-20
At1g48710 hypothetical protein 98 4e-20
At3g61330 copia-type polyprotein 97 6e-20
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 229 bits (585), Expect = 6e-60
Identities = 137/391 (35%), Positives = 219/391 (55%), Gaps = 46/391 (11%)
Query: 7 DIEKFTGSNDFGLWKVKMRA---ILIQQKCVEALKGEAQMSAHLTPAEKT---------- 53
++EKF G D+ +WK K+ A IL ++ + + + L +++
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 54 --EMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYR 111
E KA SAI+L + D+VLR++ +E+TA +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 112 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGK-EGT 170
M E+ + + EF +II DL N++V + DED+ + LL ALP++F+ KDT+ Y +
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 171 ITLEEVQAALRTKEL---TKFKELKVDDSG---EGLNISRGRSHNKGKGKGKNSRSKSRS 224
+TL+EV AA+ +KEL + K +KV G + N ++G+ KGKGKGK +SK +
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENKGKGEQKGKGKGKKGKSKKKP 246
Query: 225 KGDGNKTQYKCFICHNPGHFKKDCPERK------------DNGGGESSVQIASKDEGYES 272
C+ C GHF+ CP + ++ GG+ ++ A+ GY
Sbjct: 247 ---------GCWTCGEEGHFRSSCPNQNKPQFKQSQVVKGESSGGKGNLAEAA---GYYV 294
Query: 273 AGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIR 332
+ AL+ T E W+LD+GCSYH+ ++E+F GG VR+GN +++G+GTIR
Sbjct: 295 SEALSSTEVHLEDEWILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGTIR 354
Query: 333 LKMFDDRDFLLKNVRYIPELKRNLISISMFD 363
+K D +L NVRYIP++ RNL+S+ F+
Sbjct: 355 VKNSDGLTIVLTNVRYIPDMDRNLLSLGTFE 385
>At3g45520 copia-like polyprotein
Length = 1363
Score = 225 bits (573), Expect = 1e-58
Identities = 142/400 (35%), Positives = 225/400 (55%), Gaps = 49/400 (12%)
Query: 3 GSKWDIEKFTGSNDFGLWKVKM---------RAILIQQKCVEALKGEAQMSAHLTPAEKT 53
G++ ++EKF G D+ +WK K+ A+L + + + +++ S E+
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEERE 62
Query: 54 EMND------KAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQL 107
+M KA S I+L + D+VLR++ +E +A +M LD LYM+K+L +R LKQ+L
Sbjct: 63 KMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKL 122
Query: 108 YFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGK 167
Y ++M E+ I + EF I+ DL N++V + DED+ + LL +LP+ F+ KDT+ Y
Sbjct: 123 YSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSS 182
Query: 168 EGTI-TLEEVQAALRTKELTKFKELK--VDDSGEGLNI-----SRGRSHNKGKGKGKNSR 219
T+ +L+EV AA+ ++EL +F +K + EGL + +RGRS K KGKGK S+
Sbjct: 183 GKTVLSLDEVAAAIYSREL-EFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGKRSK 241
Query: 220 SKSRSKGDGNKTQYKCFICHNPGHFKKDCPER-----KDNGG--GESSVQIASKDEG--- 269
SKS+ C+IC GH K CP + K+ G GESS + EG
Sbjct: 242 SKSKR---------GCWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVN 292
Query: 270 -YESAG-----ALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKAC 323
ESAG AL+ T E W++D+GC YH+ ++E+ E + + GG VR+GN
Sbjct: 293 FVESAGMFVSEALSSTDIHLEDEWIMDTGCIYHMTHKREWLEDFDEEAGGSVRMGNKSIS 352
Query: 324 KIQGMGTIRLKMFDDRDFLLKNVRYIPELKRNLISISMFD 363
+++G+GT+R+ + L+NVRYIP++ RNL+S+ F+
Sbjct: 353 RVKGVGTVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFE 392
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 224 bits (571), Expect = 3e-58
Identities = 137/380 (36%), Positives = 213/380 (56%), Gaps = 28/380 (7%)
Query: 7 DIEKFTGSNDFGLWKVKMRAI---------LIQQKCVEALKGEAQMSAHLTPAEKT---E 54
++EK G D+ LWK K+ A L + + +E + A+ + LT E E
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 55 MNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVE 114
KA S +IL LG+ VLR+V +E TA M LD L+M KSL +R LKQ+LY Y+M +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 115 SKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTITLE 174
S I E + +F K+I DL N+ V++ DED+ + LL +LP+ F+ KDT+ YGK T+ L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKT-TLALD 185
Query: 175 EVQAALRTKELTKFKELK-VDDSGEGLNI-SRGRSHNKGKGKGKNSRSKSRSKGDGNKTQ 232
E+ A+R+K L K + +S + L + RGRS + K +N +S+SRSK K
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSERN-KSQSRSKSREKKV- 243
Query: 233 YKCFICHNPGHFKKDC---PERKDNGGGESSVQIASKDEGYESAGALTV------TSWEP 283
C++C GHFKK C E+ G + ++ A AL V + E
Sbjct: 244 --CWVCGKEGHFKKQCYVWKEKNKKGNNSEKGESSNVIGQAADAAALAVREESNADNQEV 301
Query: 284 EKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLL 343
+ W++D+GCS+H+ PR+++F + + G V++ N +I+G+G+IR++ D+ LL
Sbjct: 302 DNEWIMDTGCSFHMTPRRDWFVEFDESQTGRVKMANQTYSEIKGIGSIRIQNDDNTTVLL 361
Query: 344 KNVRYIPELKRNLISISMFD 363
KNVRY+P + +NLIS+ +
Sbjct: 362 KNVRYVPSMSKNLISMGTLE 381
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 218 bits (556), Expect = 1e-56
Identities = 140/388 (36%), Positives = 211/388 (54%), Gaps = 37/388 (9%)
Query: 7 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEKT------------- 53
++EKF G D+ LWK K+ A + +E L E + + E +
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 54 -------EMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQ 106
E KA S IIL LG+ VLR+V ++ TA M LD L+M KSL +R LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 107 LYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYG 166
LY Y+M E+ + E + +F K+I DL N+ V + DED+ + LL +LPR F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 167 KEGTITLEEVQAALRTKELTKFKELK-VDDSGEGLNI-SRGRSHNKGKGKGKNSRSKSRS 224
K T+ LEE+ +A+R+K L K + ++ +GL + RGRS +GKG KN +S+S+S
Sbjct: 187 KT-TLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKN-KSRSKS 244
Query: 225 KGDGNKTQYKCFICHNPGHFKKDC---PERKDNGGGESSVQIASKDEGYESAGALTVT-- 279
KG G C+IC GHFKK C ER G + ++ A AL V+
Sbjct: 245 KGAGK----TCWICGKEGHFKKQCYVWKERNKQGSTSERGEASTVTARVTDAAALVVSRA 300
Query: 280 ----SWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKM 335
+ +W+LD+GCS+H+ RK++ + G VR+GN+ +++G+G +R+K
Sbjct: 301 LLGFAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKN 360
Query: 336 FDDRDFLLKNVRYIPELKRNLISISMFD 363
D LL +VRYIPE+ +NLIS+ +
Sbjct: 361 EDGSTILLTDVRYIPEMSKNLISLGTLE 388
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 209 bits (531), Expect = 1e-53
Identities = 139/380 (36%), Positives = 206/380 (53%), Gaps = 46/380 (12%)
Query: 7 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEKTEMNDK-------- 58
++EKF G D+ +WK K+ A L ALK E + + + TE +K
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 59 -------AVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYR 111
A SAI+L + D+VLR++ +E +A +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 112 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYG-KEGT 170
M E+ I + EF +II DL N +V + DED+ + LL +LP+ F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 171 ITLEEVQAALRTKELTKFKELK-VDDSGEGLNI-----SRGRSHNKG-KGKGKNSRSKSR 223
++L+EV AA+ +KEL K + EGL + +RGR+ +G K SRSKSR
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEKTETRGRTEQRGNNNNNKKSRSKSR 246
Query: 224 SKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGALTVTSWEP 283
SK C+IC ++ G S+ S+ G + AL+ T
Sbjct: 247 SKKG-------CWIC-------------GESSNGSSNY---SEANGLYVSEALSSTDIHL 283
Query: 284 EKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLL 343
E WV+D+GCSYH+ ++E+FE L GG VR+GN K++G+GTIR+K L
Sbjct: 284 EDEWVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRL 343
Query: 344 KNVRYIPELKRNLISISMFD 363
NVRYIPE+ RNL+S+ F+
Sbjct: 344 TNVRYIPEMDRNLLSLGTFE 363
>At4g10460 putative retrotransposon
Length = 1230
Score = 201 bits (511), Expect = 2e-51
Identities = 134/387 (34%), Positives = 194/387 (49%), Gaps = 54/387 (13%)
Query: 7 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEK-------------T 53
++EKF G D+ LWK K+ A + AL+ +S L E+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 54 EMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMV 113
E KA S I+L + D+VLR+ +E TA SM LD LYM+K+L +R LKQ+LY Y+M
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 114 ESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGK-EGTIT 172
E+ + + EF ++I DL N +V + DED+ + LL +LP+ F+ KDT+ YG T++
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 173 LEEVQAALRTKELTKFKELK-VDDSGEGLNIS---RGRSHNKGKGKGKNSRSKSRSKGDG 228
++EV AA+ +KEL K + EGL + R ++ K KG RS+SRSKG
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKGNKGRSRSRSKG-- 244
Query: 229 NKTQYKCFICHNPGHFKKDCPER-------KDNGGGESSVQI-----ASKDEGYESAGAL 276
C+IC GHFK CP + KD G S+ GY + AL
Sbjct: 245 ---WKGCWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSEGSGYYVSEAL 301
Query: 277 TVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMF 336
T WV+D+GC+YH+ +KE+FE L GG VR+GN K +
Sbjct: 302 HSTDVNLGNEWVMDTGCNYHMTHKKEWFEELSEDAGGTVRMGNKSTSKFR---------- 351
Query: 337 DDRDFLLKNVRYIPELKRNLISISMFD 363
V+YIP++ RNL+S+ +
Sbjct: 352 ---------VKYIPDMDRNLLSMGTLE 369
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 183 bits (464), Expect = 6e-46
Identities = 109/328 (33%), Positives = 175/328 (53%), Gaps = 24/328 (7%)
Query: 52 KTEMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYR 111
+ E DKA + I L + DKVLR++ TA W LD L+M +SL HR + Y ++
Sbjct: 42 RLERCDKAKNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFK 101
Query: 112 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLY-GKEGT 170
M E+K I E + +F KI+ DL ++ +++ DE + + LL +LP ++ +TM Y
Sbjct: 102 MQENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLVETMKYSNSREK 161
Query: 171 ITLEEVQAALRTKELTKFKELKVDDSG--EGLNISRGRSHNKGKGKGKNSRSKSRSK-GD 227
+ L++V A R KE +EL ++ EG + +RGR K +G +++SRSK D
Sbjct: 162 LRLDDVMVAARDKE----RELSQNNRPVVEG-HFARGRPDGKNNNQGNKGKNRSRSKSAD 216
Query: 228 GNKTQYKCFICHNPGHFKKDC------PERKDNGGGESSVQIASKDEGYESAGALTVT-- 279
G + C+IC GHFKK C + K G +A E + A L T
Sbjct: 217 GKRV---CWICGKEGHFKKQCYKWIERNKSKQQGSDNGESSLAKSTEAFNPAMVLLATDE 273
Query: 280 ----SWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKM 335
+ WVLD+GCS+H+ PRK++F+ + G V++GN+ ++G+G+I+++
Sbjct: 274 TLVVTDSIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGNDTYSPVKGIGSIKIRN 333
Query: 336 FDDRDFLLKNVRYIPELKRNLISISMFD 363
D +L +VRY+P + RNLIS+ +
Sbjct: 334 SDGSQVILTDVRYMPNMTRNLISLGTLE 361
>At1g37110
Length = 1356
Score = 179 bits (454), Expect = 9e-45
Identities = 113/382 (29%), Positives = 196/382 (50%), Gaps = 27/382 (7%)
Query: 7 DIEKFTGSNDFGLWKVKMRAIL------------IQQKCVEALKGEAQMSA--------- 45
+I+ F G DF LWK++++A L K V K EA+ +
Sbjct: 9 EIKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSGTK 68
Query: 46 HLTPAEKTEMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQ 105
+ K E +++A + II + D VL +V+ AT +W L+ YM SL +R +
Sbjct: 69 EVPDPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYTQL 128
Query: 106 QLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLY 165
+LY ++MV + I + + EF +I+ +L ++++ +++E + + +L +LP S K T+ Y
Sbjct: 129 KLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHTLKY 188
Query: 166 GKEGTITLEEVQAALRT--KELTKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSR 223
G + T+T+++V ++ ++ +EL + +L + RGR + KG + +SR
Sbjct: 189 GNK-TLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRPLVRNNQKGGQGKGRSR 247
Query: 224 SKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGALTVTSWEP 283
S +KT+ C+ C GH KKDC RK E + E + AL+V
Sbjct: 248 SN---SKTKVPCWYCKKEGHVKKDCYSRKKKMESEGQGEAGVITEKLVFSEALSVNEQMV 304
Query: 284 EKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLL 343
+ W+LDSGC+ H+ R+++F + + K + LG++ + + QG GTIR+ +L
Sbjct: 305 KDLWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDHSVESQGQGTIRIDTHGGTIKIL 364
Query: 344 KNVRYIPELKRNLISISMFDGL 365
+NV+Y+P L+RNLIS D L
Sbjct: 365 ENVKYVPHLRRNLISTGTLDKL 386
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 175 bits (444), Expect = 1e-43
Identities = 102/294 (34%), Positives = 168/294 (56%), Gaps = 32/294 (10%)
Query: 52 KTEMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYR 111
+ + ++KA+ I + +GDKVLR + TA W LD LY+ KSL +R L+ ++Y YR
Sbjct: 37 RIDQDEKAMDMIFINVGDKVLRNIENSKTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYR 96
Query: 112 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTI 171
M +SK + E + EF K+I DL N+ + + DE + + +L ALP S++ K+T+ YG+EG I
Sbjct: 97 MQDSKTLEENVDEFQKMISDLNNLQIQVPDEVQAILILSALPDSYDMLKETLKYGREG-I 155
Query: 172 TLEEVQAALRTKELTKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRSKG-DGNK 230
L++V +A K KEL++ DS G ++ G+G R KS+++G DG K
Sbjct: 156 KLDDVISA------AKSKELELRDSSGG---------SRPVGEGLYVRGKSQARGSDGPK 200
Query: 231 T---QYKCFICHNPGHFKKDC----PERKDNGGGESSVQIASKDEGYESAGALT-----V 278
+ + C+IC GHFK+ C + K NG GE+++ KD+ + G +
Sbjct: 201 STEGKKVCWICGKEGHFKRQCYKWLEKNKANGAGETAL---VKDDAQDLVGLVASEVNMS 257
Query: 279 TSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIR 332
+ ++ W++D+GCS+H+ PRKEY + G VR+ NN +++G+G ++
Sbjct: 258 EGKDDQEEWIMDTGCSFHMTPRKEYLMDFVEAKSGKVRMANNSFSEVKGIGKVK 311
>At4g21360 putative transposable element
Length = 1308
Score = 172 bits (435), Expect = 1e-42
Identities = 119/396 (30%), Positives = 205/396 (51%), Gaps = 43/396 (10%)
Query: 2 MGSKWDIEKFTGSNDFGLWKVKMRAILIQQKCVEAL---------------KGEAQMSAH 46
M +K +I+ F G DF LWK+++ A L AL K E++
Sbjct: 3 MSTKVEIKTFNGDRDFSLWKIRIEAQLGVLGLKPALSDFTLTKTILVVKSEKKESESEDD 62
Query: 47 LTPAEKTE---------MNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSL 97
T ++KTE +D+A + II + D VL +V TA +W L+ L+M SL
Sbjct: 63 ETDSKKTEEVPDPIKFEQSDQAKNFIINHITDTVLLKVQHCVTAAELWATLNKLFMETSL 122
Query: 98 AHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFE 157
+R + +LY ++MV++ I + EF +I+ +L ++ + + +E + + +L +LP S+
Sbjct: 123 PNRIYTQLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAILILNSLPPSYI 182
Query: 158 NFKDTMLYGKEGTITLEEVQAALRT--KELTKFKE-LKVDDSGEGLNISRGRSHNK---G 211
K T+ YG + T+++++V ++ ++ +EL++ KE ++ S RGR K G
Sbjct: 183 QLKHTLKYGNK-TLSVQDVVSSAKSLERELSEQKETIRAPASTALYTAERGRPQTKNTQG 241
Query: 212 KGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDC--PERKDNGGGESSVQIASKDEG 269
+GKG+ RS S+S+ C+ C GH KKDC +RK G+ + ++
Sbjct: 242 QGKGRG-RSNSKSR-------LTCWFCKKEGHVKKDCYAGKRKLENEGQGKAGVITEKLV 293
Query: 270 YESAGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMG 329
Y A L++ E + WV+DSGC+YH+ R ++F E ++ LG++ + +G G
Sbjct: 294 YSEA--LSMYDQEAKDKWVIDSGCTYHMTSRMDWFSEFNENETTMILLGDDHTVESKGSG 351
Query: 330 TIRLKMFDDRDFLLKNVRYIPELKRNLISISMFDGL 365
T+++ +LKNVR++P L+RNLIS D L
Sbjct: 352 TVKVNTHGGSIRVLKNVRFVPNLRRNLISTGTLDKL 387
>At1g47360 polyprotein, putative
Length = 1182
Score = 168 bits (426), Expect = 2e-41
Identities = 114/372 (30%), Positives = 181/372 (48%), Gaps = 42/372 (11%)
Query: 8 IEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEKTEMNDKAVSAIILCL 67
I F S DF LWK ++ A L V LK + + +TP E D I LC
Sbjct: 6 IAVFDESGDFSLWKTRIMAHL----SVIGLK-DVVIGKTITPLTAEEEEDPE-KKIELC- 58
Query: 68 GDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNK 127
TA W LD L+M +SL HR + Y ++M E+K I E + +F K
Sbjct: 59 -----------QTAQEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLK 107
Query: 128 IIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGT-ITLEEVQAALRTKELT 186
I+ DL ++ + + DE + + LL +LP ++ +TM Y + L++V R KE
Sbjct: 108 IVADLNHLQIEVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDDVMVGARDKE-- 165
Query: 187 KFKELKVDDS--GEGLNISRGRSHNKGKGKGKNSRSKSRSKG-DGNKTQYKCFICHNPGH 243
+EL ++ EG + +RGR K +G +++SRSK DG + C+IC GH
Sbjct: 166 --RELSQNNRPVAEG-HYARGRPEGKNNNQGNKGKNRSRSKSADGKRV---CWICGKEGH 219
Query: 244 FKKDC------PERKDNGGGESSVQIASKDEGYESA------GALTVTSWEPEKSWVLDS 291
FKK C + K G +A E ++ A G + + W LD+
Sbjct: 220 FKKQCYKWLERNKSKQQGSDVGESSLAKSSEIFDPAMVIMATGETLLVTGRDADEWFLDT 279
Query: 292 GCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLLKNVRYIPE 351
GCS+H+ PR++ F+ + G V++GN+ ++G+G+I+++ D +L +VRY+P
Sbjct: 280 GCSFHMTPRRDLFKDFKELSSGFVKMGNDTYSPVKGIGSIKIRNNDGTQVILTDVRYVPN 339
Query: 352 LKRNLISISMFD 363
+ RNLIS+ +
Sbjct: 340 MARNLISLGTLE 351
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 126 bits (317), Expect = 7e-29
Identities = 103/367 (28%), Positives = 172/367 (46%), Gaps = 56/367 (15%)
Query: 7 DIEKFTGSNDFGLWKVKMRA---------------ILIQQKCVEALKGEAQMSAHLTPAE 51
++++F G+ DF LWKV+M A +L +E A H+ E
Sbjct: 8 EVDRFDGTGDFSLWKVRMLAHYGVLGLKGILNDEQLLRDPPVIEEEAAVAGRDYHVGDFE 67
Query: 52 --------KTEMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCL 103
K E ++KA I+L +G++VLR++ TA +MW+ L LYM SL +R L
Sbjct: 68 LPSNVDLIKFEKSEKAKDLIVLNVGNQVLRKIKNCETAAAMWSTLKRLYMETSLPNRIYL 127
Query: 104 KQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTM 163
+ + Y Y+M +S+ I + +F K++ DL NI V++ +E + + LL +L EN +
Sbjct: 128 QLKFYTYKMTDSRSIDGNVDDFLKLVTDLNNIGVDVTEEVQAILLLSSLYDRKENSQKVE 187
Query: 164 LYGKEGTITLEEVQAALRTKELTKFKELKVDDSGEGLNISRGRS-HNKGKGKGKNSR--S 220
EG +E + + LK + + G ++ R ++ N+G+ K +R S
Sbjct: 188 SSQSEGLYVEQEAD---------QRRGLKKEKANHGEDVLRAKADQNQGQSTIKTTRVVS 238
Query: 221 KSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGALTVTS 280
K G + K H P SS IA++ + LT +
Sbjct: 239 SVVKKDIGRGSVLKSL--HKP----------------SSSANIATEP---KQPLVLTASP 277
Query: 281 WEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRD 340
+ ++ WV+DSGCS+HI K+ L+ +GG V +GN +++G+G I++ DD
Sbjct: 278 QDTKEEWVMDSGCSFHITLDKDSLFDLQEFDGGKVLMGNMTHSEVKGIGKIKILNPDDYV 337
Query: 341 FLLKNVR 347
+L NVR
Sbjct: 338 VILTNVR 344
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 111 bits (277), Expect = 3e-24
Identities = 82/327 (25%), Positives = 153/327 (46%), Gaps = 33/327 (10%)
Query: 50 AEKTEMNDKAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYF 109
AEKT + K + + + +L+ + ++ T+ +W + Y L++
Sbjct: 19 AEKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRS 78
Query: 110 YRMVESKPIMEQLTEF----NKIIDDLANIDVNLEDE-----------DKVLHLLCALPR 154
+ ++E K I E +T + +I +D+ N+ ++ D +K +++CA+
Sbjct: 79 FEVLEMK-IGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEE 137
Query: 155 SFENFKDTMLYGKEGTITLEEVQAALRTKE--LTKFKELKVDDSGEGLNISRGRSHNKGK 212
S N K+ + G + ++ + E + E + K + D G G RG S ++G+
Sbjct: 138 S-NNIKELTVDGLQSSLMVHEQNLSRHDVEERVLKAETQWRPDGGRG----RGGSPSRGR 192
Query: 213 GKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYES 272
G+G + R +G N+ +CF CH GH+K +CP E +E
Sbjct: 193 GRGGY---QGRGRGYVNRDTVECFKCHKMGHYKAECP------SWEKEANYVEMEEDLLL 243
Query: 273 AGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIR 332
+ E ++ W LDSGCS H+C +E+F L+ VRLG+++ ++G G +R
Sbjct: 244 MAHVEQIGDEEKQIWFLDSGCSNHMCGTREWFLELDSGFKQNVRLGDDRRMAVEGKGKLR 303
Query: 333 LKMFDDRDFLLKNVRYIPELKRNLISI 359
L++ D R ++ +V ++P LK NL S+
Sbjct: 304 LEV-DGRIQVISDVYFVPGLKNNLFSV 329
>At3g55300 putative protein
Length = 262
Score = 110 bits (275), Expect = 5e-24
Identities = 70/212 (33%), Positives = 111/212 (52%), Gaps = 14/212 (6%)
Query: 92 YMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCA 151
Y TK+LA+ LK + ++MVE+K I E L K++ DLA++D+N DED+ L +
Sbjct: 40 YQTKTLANMIYLKHKFTSFKMVENKSIEENLDTSLKLVADLASLDINTSDEDQALQFMAG 99
Query: 152 LPRSFENFKDTMLYGK-EGTITLEEVQAALRTKELTKFKE-----LKVDDSGEGLNISRG 205
LP +++ DT+ YG + T+TL V ++ +KE+ + KE + DS L +G
Sbjct: 100 LPPQYDSLVDTLKYGSGKDTLTLNAVISSAYSKEI-ELKEKGLLNVSKPDSEALLGDFKG 158
Query: 206 RSHNKGKGKGKNSRSKSRSKGDGN-KTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIA 264
RS+NKG + S K CFIC H+K++CP+RK + ++A
Sbjct: 159 RSNNKGNKRPCTGSSSGNGKFQSRPSNNTSCFICGKEDHWKRECPQRKKH------AELA 212
Query: 265 SKDEGYESAGALTVTSWEPEKSWVLDSGCSYH 296
+ LTV++ K WV+DSG ++H
Sbjct: 213 NIATEPPQPLVLTVSTQSTCKEWVMDSGRTFH 244
>At3g60170 putative protein
Length = 1339
Score = 102 bits (254), Expect = 1e-21
Identities = 83/370 (22%), Positives = 169/370 (45%), Gaps = 28/370 (7%)
Query: 8 IEKFTGSNDFGLWKVKMRAIL--------IQQKCVEALKGEAQMS-AHLTPAEKTEMND- 57
I +F G DF W + M L +++ + G +S A + E+ ++ D
Sbjct: 12 IPRFDGYYDF--WSMTMENFLRSRELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKLKDL 69
Query: 58 KAVSAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHR---QCLKQQLYFYRMVE 114
K + + + ++L + ++T+ ++W + Y + R Q L+++ M E
Sbjct: 70 KVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAMKE 129
Query: 115 SKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKE-GTITL 173
+ I L +++ + +E V +L +L F ++ + T+++
Sbjct: 130 GEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTLSI 189
Query: 174 EEVQAALRTKELTKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRSKGDG----N 229
+E+ +L E ++ + + L ++ ++G+G+G S+ R +G G N
Sbjct: 190 DELHGSLLVHEQRLNGHVQEE---QALKVTHEERPSQGRGRGVFRGSRGRGRGRGRSGTN 246
Query: 230 KTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGALTVTSWEPEKSWVL 289
+ +C+ CHN GHF+ +CPE + N ++ ++E + ++ W L
Sbjct: 247 RAIVECYKCHNLGHFQYECPEWEKN----ANYAELEEEEELLLMAYVEQNQANRDEVWFL 302
Query: 290 DSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLLKNVRYI 349
DSGCS H+ KE+F LE V+LGN+ + G G++++K+ + ++ V Y+
Sbjct: 303 DSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGSVKVKV-NGVTQVIPEVYYV 361
Query: 350 PELKRNLISI 359
PEL+ NL+S+
Sbjct: 362 PELRNNLLSL 371
>At4g16870 retrotransposon like protein
Length = 1474
Score = 99.4 bits (246), Expect = 1e-20
Identities = 97/395 (24%), Positives = 174/395 (43%), Gaps = 62/395 (15%)
Query: 7 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAH-LTPAEKTEMN--------- 56
++ K T SN++ +W +++ A+L + L G + A LT N
Sbjct: 30 NVTKLT-SNNYLMWSLQIHALLDGYELAGHLDGSIETPAPTLTTNNVVSANPQYTLWKRQ 88
Query: 57 DKAV-SAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVES 115
D+ + SA+I + V VSR A +W L + Y S H + L+ Q+ + +
Sbjct: 89 DRLIFSALIGAISPPVQPLVSRATKASQIWKTLTNTYAKSSYDHIKQLRTQIKQLKK-GT 147
Query: 116 KPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTITLEE 175
K I E + ++D LA + +E E++V +L LP ++ D + GK+ T ++ E
Sbjct: 148 KTIDEYVLSHTTLLDQLAILGKPMEHEEQVERILEGLPEDYKTVVD-QIEGKDNTPSITE 206
Query: 176 VQAALRTKELTKFKELKVDDSGEGL--NISRGRSHNKGKGKGKNSRSKSRSKGD------ 227
+ L E + S + N+++ R HN + N+++K+R++G+
Sbjct: 207 IHERLINHEAKLLSTAALSSSSLPMSANVAQQRHHNNNRN---NNQNKNRTQGNTYTNNW 263
Query: 228 ----GNKTQY--------KCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGA 275
NK+ KC IC+ GH + CP+ + ++Q +S S+ A
Sbjct: 264 QPSANNKSGQRPFKPYLGKCQICNVQGHSARRCPQLQ-------AMQPSS------SSSA 310
Query: 276 LTVTSWEPE-----------KSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACK 324
T T W+P +W+LDSG ++HI + G V + + + K
Sbjct: 311 STFTPWQPRANLAMGAPYTANNWLLDSGATHHITSDLNALALHQPYNGDDVMIADGTSLK 370
Query: 325 IQGMGTIRLKMFDDRDFLLKNVRYIPELKRNLISI 359
I G+ L + RD L V Y+P++++NL+S+
Sbjct: 371 ITKTGSTFLPS-NARDLTLNKVLYVPDIQKNLVSV 404
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 97.8 bits (242), Expect = 4e-20
Identities = 90/392 (22%), Positives = 166/392 (41%), Gaps = 47/392 (11%)
Query: 15 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMSAHLTPAEKTEMND------KAVSAIIL 65
+++ W ++M+AIL E + KG E + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 66 CLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 122
L + +V +A W KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 123 TEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKE-GTITLEEVQAALR 181
+ + ++L L+D + +L +L FE+ + K+ +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 182 TKELTKFKELKVDDSGEGLNIS----------RGRSHNKGKGKG--------------KN 217
E K K+ + + + I+ RG +G+G+G N
Sbjct: 196 AYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 218 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDC--PERKDNGGGESSVQIASKD 267
R ++ S+G G +K+ KC+ C GH+ +C P K + V+ ++
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFKEKANYVEEKIQE 315
Query: 268 EGYESAGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQG 327
E + E W LDSG S H+C RK F L+ G V LG+ +++G
Sbjct: 316 EDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKG 375
Query: 328 MGTIRLKMFDDRDFLLKNVRYIPELKRNLISI 359
G I +++ + + NV YIP +K N++S+
Sbjct: 376 KGNILIRLKNGDHQFISNVYYIPSMKTNILSL 407
>At1g58140 hypothetical protein
Length = 1320
Score = 97.8 bits (242), Expect = 4e-20
Identities = 90/392 (22%), Positives = 166/392 (41%), Gaps = 47/392 (11%)
Query: 15 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMSAHLTPAEKTEMND------KAVSAIIL 65
+++ W ++M+AIL E + KG E + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 66 CLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 122
L + +V +A W KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 123 TEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKE-GTITLEEVQAALR 181
+ + ++L L+D + +L +L FE+ + K+ +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 182 TKELTKFKELKVDDSGEGLNIS----------RGRSHNKGKGKG--------------KN 217
E K K+ + + + I+ RG +G+G+G N
Sbjct: 196 AYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 218 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDC--PERKDNGGGESSVQIASKD 267
R ++ S+G G +K+ KC+ C GH+ +C P K + V+ ++
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKANYVEEKIQE 315
Query: 268 EGYESAGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQG 327
E + E W LDSG S H+C RK F L+ G V LG+ +++G
Sbjct: 316 EDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKG 375
Query: 328 MGTIRLKMFDDRDFLLKNVRYIPELKRNLISI 359
G I +++ + + NV YIP +K N++S+
Sbjct: 376 KGNILIRLKNGDHQFISNVYYIPSMKTNILSL 407
>At1g48710 hypothetical protein
Length = 1352
Score = 97.8 bits (242), Expect = 4e-20
Identities = 90/392 (22%), Positives = 166/392 (41%), Gaps = 47/392 (11%)
Query: 15 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMSAHLTPAEKTEMND------KAVSAIIL 65
+++ W ++M+AIL E + KG E + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 66 CLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 122
L + +V +A W KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 123 TEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKE-GTITLEEVQAALR 181
+ + ++L L+D + +L +L FE+ + K+ +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 182 TKELTKFKELKVDDSGEGLNIS----------RGRSHNKGKGKG--------------KN 217
E K K+ + + + I+ RG +G+G+G N
Sbjct: 196 AYEEKKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 218 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDC--PERKDNGGGESSVQIASKD 267
R ++ S+G G +K+ KC+ C GH+ +C P K + V+ ++
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKANYVEEKIQE 315
Query: 268 EGYESAGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQG 327
E + E W LDSG S H+C RK F L+ G V LG+ +++G
Sbjct: 316 EDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKG 375
Query: 328 MGTIRLKMFDDRDFLLKNVRYIPELKRNLISI 359
G I +++ + + NV YIP +K N++S+
Sbjct: 376 KGNILIRLKNGDHQFISNVYYIPSMKTNILSL 407
>At3g61330 copia-type polyprotein
Length = 1352
Score = 97.1 bits (240), Expect = 6e-20
Identities = 89/395 (22%), Positives = 170/395 (42%), Gaps = 53/395 (13%)
Query: 15 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMSAHLTPAEKTEMND------KAVSAIIL 65
+++ W ++M+AIL E + KG E + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 66 CLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 122
L + +V +A W KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 123 TEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKE-GTITLEEVQAALR 181
+ + ++L L+D + +L +L FE+ + K+ +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 182 TKELTKFKELKVDDSGEGLNIS----------RGRSHNKGKGKG--------------KN 217
E K K+ + + + I+ RG +G+G+G N
Sbjct: 196 AYEEKKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 218 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEG 269
R ++ S+G G +K+ KC+ C GH+ +C + E + + +E
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKAHYV---EEK 312
Query: 270 YESAGALTVTSWEPEKS-----WVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACK 324
+ L + S++ ++ W LDSG S H+C RK F L+ G V LG+ +
Sbjct: 313 IQEEDMLLMASYKKDEQKENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKME 372
Query: 325 IQGMGTIRLKMFDDRDFLLKNVRYIPELKRNLISI 359
++G G I +++ + + NV YIP +K N++S+
Sbjct: 373 VKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSL 407
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.357 0.158 0.567
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,451,174
Number of Sequences: 26719
Number of extensions: 909725
Number of successful extensions: 5584
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 76
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 5200
Number of HSP's gapped (non-prelim): 231
length of query: 1304
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1193
effective length of database: 8,352,787
effective search space: 9964874891
effective search space used: 9964874891
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146709.5


