

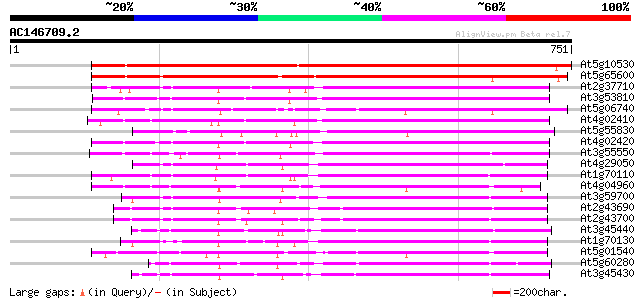
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.2 - phase: 0
(751 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g10530 lectin-like protein kinase - like 731 0.0
At5g65600 receptor protein kinase-like protein 662 0.0
At2g37710 putative receptor-like protein kinase 391 e-109
At3g53810 serine/threonine-specific kinase like protein 389 e-108
At5g06740 lectin-like protein kinase 385 e-107
At4g02410 unknown protein 382 e-106
At5g55830 serine/threonine-specific kinase like protein 373 e-103
At4g02420 373 e-103
At3g55550 probable serine/threonine-specific protein kinase 370 e-102
At4g29050 serine/threonine-specific kinase like protein 367 e-101
At1g70110 hypothetical protein 348 8e-96
At4g04960 unknown protein 341 8e-94
At3g59700 serine/threonine-specific kinase lecRK1 precursor,lect... 337 1e-92
At2g43690 putative receptor protein kinase 334 1e-91
At2g43700 putative receptor protein kinase 330 2e-90
At3g45440 receptor like protein kinase 329 4e-90
At1g70130 hypothetical protein 327 2e-89
At5g01540 receptor like protein kinase 326 3e-89
At5g60280 receptor like protein kinase 326 3e-89
At3g45430 receptor like protein kinase 324 1e-88
>At5g10530 lectin-like protein kinase - like
Length = 651
Score = 731 bits (1888), Expect = 0.0
Identities = 374/650 (57%), Positives = 480/650 (73%), Gaps = 11/650 (1%)
Query: 110 LFFLSLILLTFHAYSIHFQIPSFNPNDANIIYQGSAAPVDGEVDFNINGNYSCQVGWALY 169
+ S +L+ S+ F I F + + I YQG A +G V+ N +Y+C+ GWA Y
Sbjct: 5 ILLFSFVLVLPFVCSVQFNISRFGSDVSEIAYQGDAR-ANGAVELT-NIDYTCRAGWATY 62
Query: 170 SKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSSGGLMG 229
K+V LW+ T + +DF+T ++F I+TR YGHG AFFLA ++PPNS+GG +G
Sbjct: 63 GKQVPLWNPGTSKPSDFSTRFSFRIDTRNVGYGNYGHGFAFFLAPARIQLPPNSAGGFLG 122
Query: 230 LFNTTTMLSSSNHIVHVEFDSFANSEFS--ETTEHVGINNNSIKSSISTPWNASLHSGDT 287
LFN T SS+ +V+VEFD+F N E+ + HVGINNNS+ SS T WNA+ H+ D
Sbjct: 123 LFNGTNNQSSAFPLVYVEFDTFTNPEWDPLDVKSHVGINNNSLVSSNYTSWNATSHNQDI 182
Query: 288 AEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVLPEWVTIGFSAATGYNGEV 347
V I Y+S +NL+VSW Y TS P E ++LSY ID KVLP VTIGFSA +G E
Sbjct: 183 GRVLIFYDSARRNLSVSWTYDLTSDPLENSSLSYIIDLSKVLPSEVTIGFSATSGGVTEG 242
Query: 348 NNLLSWEFNSNLEKSDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKRKRSEKQK 407
N LLSWEF+S+LE D S++ + +I+ +S+ ++ + + +I+ KRK+ +K+
Sbjct: 243 NRLLSWEFSSSLELIDIKKSQNDKKGMIIGISVSGFVL-LTFFITSLIVFLKRKQQKKKA 301
Query: 408 EEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQV 467
EE +LTS+N+DLERGAGPR+FTYK+L A NNF+ DRKLG+GGFGAV++GY LD+ V
Sbjct: 302 EETENLTSINEDLERGAGPRKFTYKDLASAANNFADDRKLGEGGFGAVYRGYLNSLDMMV 361
Query: 468 AVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHL 527
A+KK + GS+QGK+E+VTEVK+IS LRHRNLV+L+GWCH+K EFL+IYEFMPNGSLD+HL
Sbjct: 362 AIKKFAGGSKQGKREFVTEVKIISSLRHRNLVQLIGWCHEKDEFLMIYEFMPNGSLDAHL 421
Query: 528 FGKRTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLA 587
FGK+ L+W VR KITLGLAS LLYLHEEWE+CVVHRDIK+SNVMLDS+FN KLGDFGLA
Sbjct: 422 FGKKPHLAWHVRCKITLGLASALLYLHEEWEQCVVHRDIKASNVMLDSNFNAKLGDFGLA 481
Query: 588 KLMDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEK 647
+LMDHELGPQTTGLAGTFGY+APEY+STGRASKESDVYSFG+V LEI +G+K+ + + +
Sbjct: 482 RLMDHELGPQTTGLAGTFGYMAPEYISTGRASKESDVYSFGVVTLEIVTGRKSVDRRQGR 541
Query: 648 DEE-KGMIEWVWDHYGKGELLVAMDENLR-KDFDEKQVECLMIVGLWCAHPDVSLRPSIR 705
E ++E +WD YGKGE++ A+DE LR FDEKQ ECLMIVGLWCAHPDV+ RPSI+
Sbjct: 542 VEPVTNLVEKMWDLYGKGEVITAIDEKLRIGGFDEKQAECLMIVGLWCAHPDVNTRPSIK 601
Query: 706 QAIQVLNFEVSMPNLPPKRPVATYH---APTPSVSSVEASIT-NSLQDGR 751
QAIQVLN E +P+LP K PVATYH + T SVSS A++T +S Q GR
Sbjct: 602 QAIQVLNLEAPVPHLPTKMPVATYHVSSSNTTSVSSGGATVTFSSAQHGR 651
>At5g65600 receptor protein kinase-like protein
Length = 675
Score = 662 bits (1707), Expect = 0.0
Identities = 350/658 (53%), Positives = 463/658 (70%), Gaps = 29/658 (4%)
Query: 110 LFFLSLIL-LTFHAYSIHFQIPSFNPND-ANIIYQGSAAP-VDGEVDFNINGNYSCQVGW 166
+ FLSL L L F S++F SF D +I Y G A P DG V+FN N + QVGW
Sbjct: 20 ILFLSLFLFLPFVVDSLYFNFTSFRQGDPGDIFYHGDATPDEDGTVNFN-NAEQTSQVGW 78
Query: 167 ALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSSGG 226
YSKKV +W KTG+ +DF+T ++F I+ R S GHG+ FFLA G ++P S GG
Sbjct: 79 ITYSKKVPIWSHKTGKASDFSTSFSFKIDARNLSAD--GHGICFFLAPMGAQLPAYSVGG 136
Query: 227 LMGLFNTTTMLSSSNHIVHVEFDSFANSEF--SETTEHVGINNNSIKSSISTPWNASLHS 284
+ LF SSS +VHVEFD+F N + ++ HVGINNNS+ SS T WNAS HS
Sbjct: 137 FLNLFTRKNNYSSSFPLVHVEFDTFNNPGWDPNDVGSHVGINNNSLVSSNYTSWNASSHS 196
Query: 285 GDTAEVWIRYNSTTKNLTVSWEYQ--TTSSPQEKTNLSYQIDFKKVLPEWVTIGFSAATG 342
D I Y+S TKNL+V+W Y+ TS P+E ++LSY ID KVLP V GF AA G
Sbjct: 197 QDICHAKISYDSVTKNLSVTWAYELTATSDPKESSSLSYIIDLAKVLPSDVMFGFIAAAG 256
Query: 343 YNGEVNNLLSWEFNSNLEKSDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKRKR 402
N E + LLSWE +S+L DS+ D+R+ +++ +S + ++ V++ +++R
Sbjct: 257 TNTEEHRLLSWELSSSL----DSDKADSRIGLVIGISASGFVFLTFMVITTVVVWSRKQR 312
Query: 403 SEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFAD 462
+K+++ ++ S+N DLER AGPR+F+YK+L ATN FS RKLG+GGFGAV++G +
Sbjct: 313 KKKERD-IENMISINKDLEREAGPRKFSYKDLVSATNRFSSHRKLGEGGFGAVYEGNLKE 371
Query: 463 LDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGS 522
++ VAVKK+S SRQGK E++ EVK+IS+LRHRNLV+L+GWC++K EFLLIYE +PNGS
Sbjct: 372 INTMVAVKKLSGDSRQGKNEFLNEVKIISKLRHRNLVQLIGWCNEKNEFLLIYELVPNGS 431
Query: 523 LDSHLFGKRTPL-SWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKL 581
L+SHLFGKR L SW +R+KI LGLAS LLYLHEEW++CV+HRDIK+SN+MLDS FNVKL
Sbjct: 432 LNSHLFGKRPNLLSWDIRYKIGLGLASALLYLHEEWDQCVLHRDIKASNIMLDSEFNVKL 491
Query: 582 GDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKAT 641
GDFGLA+LM+HELG TTGLAGTFGY+APEYV G ASKESD+YSFGIV+LEI +G+K+
Sbjct: 492 GDFGLARLMNHELGSHTTGLAGTFGYMAPEYVMKGSASKESDIYSFGIVLLEIVTGRKSL 551
Query: 642 EVMK------EKDEEKGMIEWVWDHYGKGELLVA-MDENLRKDFDEKQVECLMIVGLWCA 694
E + E D+EK ++E VW+ YGK EL+ + +D+ L +DFD+K+ ECL+++GLWCA
Sbjct: 552 ERTQEDNSDTESDDEKSLVEKVWELYGKQELITSCVDDKLGEDFDKKEAECLLVLGLWCA 611
Query: 695 HPDVSLRPSIRQAIQVLNFEVSMPNLPPKRPVATYHAPT------PSVSSVEASITNS 746
HPD + RPSI+Q IQV+NFE +P+LP KRPVA Y+ T PSV+S S+T S
Sbjct: 612 HPDKNSRPSIKQGIQVMNFESPLPDLPLKRPVAMYYISTTTSSSSPSVNSNGVSVTFS 669
>At2g37710 putative receptor-like protein kinase
Length = 675
Score = 391 bits (1004), Expect = e-109
Identities = 234/647 (36%), Positives = 359/647 (55%), Gaps = 52/647 (8%)
Query: 110 LFFLSLILLTFHAYSIHFQIPSFNPNDANIIYQGSAAP-----VDGEVDFNING-----N 159
+F L + F +++ FQ S N Y P + G NG N
Sbjct: 1 MFLKLLTIFFFFFFNLIFQSSS---QSLNFAYNNGFNPPTDLSIQGITTVTPNGLLKLTN 57
Query: 160 YSCQ-VGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFE 218
+ Q G A Y+K + DS G ++ F+T + F I+++ S GHG+AF +A
Sbjct: 58 TTVQKTGHAFYTKPIRFKDSPNGTVSSFSTSFVFAIHSQIAILS--GHGIAFVVAPNA-S 114
Query: 219 IPPNSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTE-HVGINNNSIKSSISTP 277
+P + +GLFN + +NH+ VE D+ ++EF++T + HVGI+ NS+KS S+P
Sbjct: 115 LPYGNPSQYIGLFNLANNGNETNHVFAVELDTILSTEFNDTNDNHVGIDINSLKSVQSSP 174
Query: 278 ---W-------NASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKK 327
W N +L S +VW+ Y+ T + V+ P + + D
Sbjct: 175 AGYWDEKGQFKNLTLISRKPMQVWVDYDGRTNKIDVTMAPFNEDKPTRPLVTAVR-DLSS 233
Query: 328 VLPEWVTIGFSAATGYNGEVNNLLSWEFNSNLEKSDDSNSKDTRLV------VILTVSLG 381
VL + + +GFS+ATG + +L W F N + + S+ +L + +G
Sbjct: 234 VLLQDMYVGFSSATGSVLSEHYILGWSFGLNEKAPPLALSRLPKLPRFEPKRISEFYKIG 293
Query: 382 AVIIGVGALVAYV-----ILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELEL 436
+I + + +++ I++R+RK +E+ +E E+ G RF +K+L
Sbjct: 294 MPLISLFLIFSFIFLVCYIVRRRRKFAEELEE-----------WEKEFGKNRFRFKDLYY 342
Query: 437 ATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHR 496
AT F + LG GGFG+V+KG L++AVK++S SRQG KE+V E+ I ++ HR
Sbjct: 343 ATKGFKEKGLLGTGGFGSVYKGVMPGTKLEIAVKRVSHESRQGMKEFVAEIVSIGRMSHR 402
Query: 497 NLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGK-RTPLSWGVRHKITLGLASGLLYLHE 555
NLV LLG+C +GE LL+Y++MPNGSLD +L+ L+W R K+ LG+ASGL YLHE
Sbjct: 403 NLVPLLGYCRRRGELLLVYDYMPNGSLDKYLYNTPEVTLNWKQRIKVILGVASGLFYLHE 462
Query: 556 EWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVST 615
EWE+ V+HRD+K+SNV+LD N +LGDFGLA+L DH PQTT + GT GYLAPE+ T
Sbjct: 463 EWEQVVIHRDVKASNVLLDGELNGRLGDFGLARLYDHGSDPQTTHVVGTLGYLAPEHTRT 522
Query: 616 GRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLR 675
GRA+ +DV++FG +LE+ G++ E +E DE +++WV+ + KG++L A D N+
Sbjct: 523 GRATMATDVFAFGAFLLEVACGRRPIEFQQETDETFLLVDWVFGLWNKGDILAAKDPNMG 582
Query: 676 KDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPP 722
+ DEK+VE ++ +GL C+H D RPS+RQ + L + +P L P
Sbjct: 583 SECDEKEVEMVLKLGLLCSHSDPRARPSMRQVLHYLRGDAKLPELSP 629
>At3g53810 serine/threonine-specific kinase like protein
Length = 677
Score = 389 bits (998), Expect = e-108
Identities = 234/631 (37%), Positives = 357/631 (56%), Gaps = 30/631 (4%)
Query: 111 FFLSLILLTFHAYSIHFQIPSFNPNDANIIYQGSAAPVDGEVDFNINGNYSCQVGWALYS 170
FFL ++ + +++F F+P +I QG A + + + G A +
Sbjct: 11 FFLLCQIMISSSQNLNFTYNGFHPPLTDISLQGLATVTPNGL-LKLTNTSVQKTGHAFCT 69
Query: 171 KKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSSGGLMGL 230
+++ DS+ G ++ F+T + F I+++ P+ GHG+AF +A +P +GL
Sbjct: 70 ERIRFKDSQNGNVSSFSTTFVFAIHSQ--IPTLSGHGIAFVVAPT-LGLPFALPSQYIGL 126
Query: 231 FNTTTMLSSSNHIVHVEFDSFANSEFSETTE-HVGINNNSIKSS-ISTPW---------N 279
FN + + +NHI VEFD+ +SEF + + HVGI+ N ++S+ ST N
Sbjct: 127 FNISNNGNDTNHIFAVEFDTIQSSEFGDPNDNHVGIDLNGLRSANYSTAGYRDDHDKFQN 186
Query: 280 ASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVLPEWVTIGFSA 339
SL S +VWI Y++ + + V+ + P+ K +SY D +L E + +GFS+
Sbjct: 187 LSLISRKRIQVWIDYDNRSHRIDVTVAPFDSDKPR-KPLVSYVRDLSSILLEDMYVGFSS 245
Query: 340 ATGYNGEVNNLLSWEFNSNLEKSDDSNSKDTRLV------VILTVSLGAVIIGVGALVAY 393
ATG + L+ W F N E S SK +L + +G +I + + +
Sbjct: 246 ATGSVLSEHFLVGWSFRLNGEAPMLSLSKLPKLPRFEPRRISEFYKIGMPLISLSLIFSI 305
Query: 394 VILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFG 453
+ L R +K+ EE + DD E G RF +KEL AT F + LG GGFG
Sbjct: 306 IFLAFYIVRRKKKYEEEL------DDWETEFGKNRFRFKELYHATKGFKEKDLLGSGGFG 359
Query: 454 AVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLL 513
V++G L+VAVK++S S+QG KE+V E+ I ++ HRNLV LLG+C +GE LL
Sbjct: 360 RVYRGILPTTKLEVAVKRVSHDSKQGMKEFVAEIVSIGRMSHRNLVPLLGYCRRRGELLL 419
Query: 514 IYEFMPNGSLDSHLFGK-RTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVM 572
+Y++MPNGSLD +L+ T L W R I G+ASGL YLHEEWE+ V+HRD+K+SNV+
Sbjct: 420 VYDYMPNGSLDKYLYNNPETTLDWKQRSTIIKGVASGLFYLHEEWEQVVIHRDVKASNVL 479
Query: 573 LDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVL 632
LD+ FN +LGDFGLA+L DH PQTT + GT GYLAPE+ TGRA+ +DVY+FG +L
Sbjct: 480 LDADFNGRLGDFGLARLYDHGSDPQTTHVVGTLGYLAPEHSRTGRATTTTDVYAFGAFLL 539
Query: 633 EITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENL-RKDFDEKQVECLMIVGL 691
E+ SG++ E D+ ++EWV+ + +G ++ A D L +D ++VE ++ +GL
Sbjct: 540 EVVSGRRPIEFHSASDDTFLLVEWVFSLWLRGNIMEAKDPKLGSSGYDLEEVEMVLKLGL 599
Query: 692 WCAHPDVSLRPSIRQAIQVLNFEVSMPNLPP 722
C+H D RPS+RQ +Q L ++++P L P
Sbjct: 600 LCSHSDPRARPSMRQVLQYLRGDMALPELTP 630
>At5g06740 lectin-like protein kinase
Length = 652
Score = 385 bits (990), Expect = e-107
Identities = 239/654 (36%), Positives = 354/654 (53%), Gaps = 39/654 (5%)
Query: 110 LFFLSLILLTFHAYSIHFQIPSFN-PNDANIIYQGS-----AAPVDGEVDFNINGNYSCQ 163
LF + + + F P FN N+ +I S A V +V G + Q
Sbjct: 10 LFLILTCKIETQVKCLKFDFPGFNVSNELELIRDNSYIVFGAIQVTPDVTGGPGGTIANQ 69
Query: 164 VGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNS 223
G ALY K LW T + TF+IN ++ G GLAF L P NS
Sbjct: 70 AGRALYKKPFRLWSKHKSA----TFNTTFVINISNKTDPG-GEGLAFVLTPEE-TAPQNS 123
Query: 224 SGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTEHVGINNNSIKSSISTPWNA--- 280
SG +G+ N T ++ + IV VEFD+ + HV +N N+I S + +
Sbjct: 124 SGMWLGMVNERTNRNNESRIVSVEFDTRKSHSDDLDGNHVALNVNNINSVVQESLSGRGI 183
Query: 281 SLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVLPEWVTIGFSAA 340
+ SG +RY+ KNL+V Q S ID LPE V +GF+A+
Sbjct: 184 KIDSGLDLTAHVRYDG--KNLSVYVSRNLDVFEQRNLVFSRAIDLSAYLPETVYVGFTAS 241
Query: 341 TGYNGEVNNLLSWEFNSNLEKSDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKR 400
T E+N + SW F L+ D N L + +T+ + I+G+GA + + L+ +
Sbjct: 242 TSNFTELNCVRSWSFEG-LKIDGDGNM----LWLWITIPI-VFIVGIGAFLGALYLRSRS 295
Query: 401 KRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYF 460
K E + L + A P++F +EL+ AT NF + KLGQGGFG VFKG +
Sbjct: 296 KAGETNPDIEAELDNC------AANPQKFKLRELKRATGNFGAENKLGQGGFGMVFKGKW 349
Query: 461 ADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPN 520
D +AVK++S S QGK+E++ E+ I L HRNLVKLLGWC+++ E+LL+YE+MPN
Sbjct: 350 QGRD--IAVKRVSEKSHQGKQEFIAEITTIGNLNHRNLVKLLGWCYERKEYLLVYEYMPN 407
Query: 521 GSLDSHLF---GKRTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSF 577
GSLD +LF R+ L+W R I GL+ L YLH E+ ++HRDIK+SNVMLDS F
Sbjct: 408 GSLDKYLFLEDKSRSNLTWETRKNIITGLSQALEYLHNGCEKRILHRDIKASNVMLDSDF 467
Query: 578 NVKLGDFGLAKLMDHE--LGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEIT 635
N KLGDFGLA+++ T +AGT GY+APE GRA+ E+DVY+FG+++LE+
Sbjct: 468 NAKLGDFGLARMIQQSEMTHHSTKEIAGTPGYMAPETFLNGRATVETDVYAFGVLMLEVV 527
Query: 636 SGKKATEVM---KEKDEEKGMIEWVWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLW 692
SGKK + V+ + + ++ W+W+ Y G + A D + FD+++++ ++++GL
Sbjct: 528 SGKKPSYVLVKDNQNNYNNSIVNWLWELYRNGTITDAADPGMGNLFDKEEMKSVLLLGLA 587
Query: 693 CAHPDVSLRPSIRQAIQVLNFEVSMPNLPPKRPVATYHAPTPSVSSVEASITNS 746
C HP+ + RPS++ ++VL E S P++P +RP + A PS S ++ S+T S
Sbjct: 588 CCHPNPNQRPSMKTVLKVLTGETSPPDVPTERPAFVWPAMPPSFSDIDYSLTGS 641
>At4g02410 unknown protein
Length = 674
Score = 382 bits (982), Expect = e-106
Identities = 232/646 (35%), Positives = 358/646 (54%), Gaps = 39/646 (6%)
Query: 105 FYTLPLFFLSLILLT----FHAYSIHFQIPSFNPNDANIIYQGSAAPVDGEVDFNINGNY 160
F +FF +ILL+ + S++F SF+ NI QG A + +
Sbjct: 3 FKLFTIFFFFIILLSKPLNSSSQSLNFTYNSFHRPPTNISIQGIATVTSNGI-LKLTDKT 61
Query: 161 SCQVGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIP 220
G A Y++ + DS ++ F+T TF+I P+ GHG+AFF+A +
Sbjct: 62 VISTGHAFYTEPIRFKDSPNDTVSSFST--TFVIGIYSGIPTISGHGMAFFIAPNPV-LS 118
Query: 221 PNSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTE-HVGINNN---SIKSSIST 276
+ +GLF++T + +NHI+ VEFD+ N EF +T + HVGIN N S+KSS+
Sbjct: 119 SAMASQYLGLFSSTNNGNDTNHILAVEFDTIMNPEFDDTNDNHVGININSLTSVKSSLVG 178
Query: 277 PW-------NASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVL 329
W N +L S +VW+ Y+ T + V+ P+ K +S D V
Sbjct: 179 YWDEINQFNNLTLISRKRMQVWVDYDDRTNQIDVTMAPFGEVKPR-KALVSVVRDLSSVF 237
Query: 330 PEWVTIGFSAATGYNGEVNNLLSWEFNSNLEKSDD-SNSKDTRLVVILTVSL-------- 380
+ + +GFSAATGY + + W F + + + SK + + SL
Sbjct: 238 LQDMYLGFSAATGYVLSEHFVFGWSFMVKGKTAPPLTLSKVPKFPRVGPTSLQRFYKNRM 297
Query: 381 ---GAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELA 437
++I V +V+ + L R R ++ E +D E G R +K+L A
Sbjct: 298 PLFSLLLIPVLFVVSLIFLVRFIVRRRRKFAEEF------EDWETEFGKNRLRFKDLYYA 351
Query: 438 TNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRN 497
T F LG GGFG V++G ++AVK++S SRQG KE+V E+ I ++ HRN
Sbjct: 352 TKGFKDKDLLGSGGFGRVYRGVMPTTKKEIAVKRVSNESRQGLKEFVAEIVSIGRMSHRN 411
Query: 498 LVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFG-KRTPLSWGVRHKITLGLASGLLYLHEE 556
LV LLG+C + E LL+Y++MPNGSLD +L+ L W R + +G+ASGL YLHEE
Sbjct: 412 LVPLLGYCRRRDELLLVYDYMPNGSLDKYLYDCPEVTLDWKQRFNVIIGVASGLFYLHEE 471
Query: 557 WERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTG 616
WE+ V+HRDIK+SNV+LD+ +N +LGDFGLA+L DH PQTT + GT+GYLAP++V TG
Sbjct: 472 WEQVVIHRDIKASNVLLDAEYNGRLGDFGLARLCDHGSDPQTTRVVGTWGYLAPDHVRTG 531
Query: 617 RASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLRK 676
RA+ +DV++FG+++LE+ G++ E+ E DE +++ V+ + +G +L A D NL
Sbjct: 532 RATTATDVFAFGVLLLEVACGRRPIEIEIESDESVLLVDSVFGFWIEGNILDATDPNLGS 591
Query: 677 DFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPP 722
+D+++VE ++ +GL C+H D +RP++RQ +Q L + ++P+L P
Sbjct: 592 VYDQREVETVLKLGLLCSHSDPQVRPTMRQVLQYLRGDATLPDLSP 637
>At5g55830 serine/threonine-specific kinase like protein
Length = 681
Score = 373 bits (958), Expect = e-103
Identities = 219/601 (36%), Positives = 337/601 (55%), Gaps = 49/601 (8%)
Query: 165 GWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSS 224
G +Y+ + +D + F+TH++F + P+ G GLAFFL+ + S
Sbjct: 70 GTVIYNNPIRFYDPDSNTTASFSTHFSFTVQNLNPDPTSAGDGLAFFLSHDNDTL--GSP 127
Query: 225 GGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSETT-EHVGINNNSIKS-SISTPWNAS- 281
GG +GL N++ + N V +EFD+ + F++ H+G++ +S+ S S S P +S
Sbjct: 128 GGYLGLVNSSQPMK--NRFVAIEFDTKLDPHFNDPNGNHIGLDVDSLNSISTSDPLLSSQ 185
Query: 282 --LHSGDTAEVWIRYNSTTKNLTVSWEYQ---TTSSPQEKTNLSYQIDFKKVLPEWVTIG 336
L SG + WI Y + + L V Y TT+ EK LS ID L + +G
Sbjct: 186 IDLKSGKSITSWIDYKNDLRLLNVFLSYTDPVTTTKKPEKPLLSVNIDLSPFLNGEMYVG 245
Query: 337 FSAATGYNGEVNNLLSWEF-----------NSNLEKSDDSNSKDTRLVVI---------- 375
FS +T + E++ + +W F +++L DS+ + VVI
Sbjct: 246 FSGSTEGSTEIHLIENWSFKTSGFLPVRSKSNHLHNVSDSSVVNDDPVVIPSKKRRHRHN 305
Query: 376 LTVSLGA---VIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYK 432
L + LG V+I + V +K K + +KE L + G R F+YK
Sbjct: 306 LAIGLGISCPVLICLALFVFGYFTLKKWKSVKAEKELKTELIT---------GLREFSYK 356
Query: 433 ELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQ 492
EL AT F R +G+G FG V++ F AVK+ S +GK E++ E+ +I+
Sbjct: 357 ELYTATKGFHSSRVIGRGAFGNVYRAMFVSSGTISAVKRSRHNSTEGKTEFLAELSIIAC 416
Query: 493 LRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKR----TPLSWGVRHKITLGLAS 548
LRH+NLV+L GWC++KGE LL+YEFMPNGSLD L+ + L W R I +GLAS
Sbjct: 417 LRHKNLVQLQGWCNEKGELLLVYEFMPNGSLDKILYQESQTGAVALDWSHRLNIAIGLAS 476
Query: 549 GLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYL 608
L YLH E E+ VVHRDIK+SN+MLD +FN +LGDFGLA+L +H+ P +T AGT GYL
Sbjct: 477 ALSYLHHECEQQVVHRDIKTSNIMLDINFNARLGDFGLARLTEHDKSPVSTLTAGTMGYL 536
Query: 609 APEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLV 668
APEY+ G A++++D +S+G+V+LE+ G++ + E + +++WVW + +G +L
Sbjct: 537 APEYLQYGTATEKTDAFSYGVVILEVACGRRPIDKEPESQKTVNLVDWVWRLHSEGRVLE 596
Query: 669 AMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPPKRPVAT 728
A+DE L+ +FDE+ ++ L++VGL CAHPD + RPS+R+ +Q+LN E+ +P +P +
Sbjct: 597 AVDERLKGEFDEEMMKKLLLVGLKCAHPDSNERPSMRRVLQILNNEIEPSPVPKMKPTLS 656
Query: 729 Y 729
+
Sbjct: 657 F 657
>At4g02420
Length = 669
Score = 373 bits (957), Expect = e-103
Identities = 229/642 (35%), Positives = 346/642 (53%), Gaps = 47/642 (7%)
Query: 110 LFFLSLILLTFHAYS--IHFQIPSFNPNDANIIYQGSAAPVDGEVDFNINGNYSCQVGWA 167
+FFLS + + S I F F P +I G A + + G A
Sbjct: 9 IFFLSFFWQSLKSSSQIIDFTYNGFRPPPTDISILGIATITPNGL-LKLTNTTMQSTGHA 67
Query: 168 LYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSSGGL 227
Y+K + DS G ++ F+T + F I+ S HG+AF +A +P S
Sbjct: 68 FYTKPIRFKDSPNGTVSSFSTTFVFAIH----SQIPIAHGMAFVIAPNP-RLPFGSPLQY 122
Query: 228 MGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTE-HVGINNNSIKSSISTP---W----- 278
+GLFN T + NH+ VE D+ N EF++T HVGI+ NS+ S S+P W
Sbjct: 123 LGLFNVTNNGNVRNHVFAVELDTIMNIEFNDTNNNHVGIDINSLNSVKSSPAGYWDENDQ 182
Query: 279 --NASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVLPEWVTIG 336
N +L S +VW+ ++ T + V+ P+ K +S D VL + + +G
Sbjct: 183 FHNLTLISSKRMQVWVDFDGPTHLIDVTMAPFGEVKPR-KPLVSIVRDLSSVLLQDMFVG 241
Query: 337 FSAATGYNGEVNNLLSWEFNSNLEKSDDSNSKDTRLVV---------------ILTVSLG 381
FS+ATG +L W F N E + SK RL V + +SL
Sbjct: 242 FSSATGNIVSEIFVLGWSFGVNGEAQPLALSKLPRLPVWDLKPTRVYRFYKNWVPLISLL 301
Query: 382 AVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNF 441
+ + + I+KR+RK +E+ +D E G R +K+L AT F
Sbjct: 302 LIPFLLIIFLVRFIMKRRRKFAEEV-----------EDWETEFGKNRLRFKDLYYATKGF 350
Query: 442 SKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKL 501
LG GGFG+V+KG ++AVK++S SRQG KE+V E+ I Q+ HRNLV L
Sbjct: 351 KDKNILGSGGFGSVYKGIMPKTKKEIAVKRVSNESRQGLKEFVAEIVSIGQMSHRNLVPL 410
Query: 502 LGWCHDKGEFLLIYEFMPNGSLDSHLFGK-RTPLSWGVRHKITLGLASGLLYLHEEWERC 560
+G+C + E LL+Y++MPNGSLD +L+ L W R K+ G+AS L YLHEEWE+
Sbjct: 411 VGYCRRRDELLLVYDYMPNGSLDKYLYNSPEVTLDWKQRFKVINGVASALFYLHEEWEQV 470
Query: 561 VVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTGRASK 620
V+HRD+K+SNV+LD+ N +LGDFGLA+L DH PQTT + GT+GYLAP+++ TGRA+
Sbjct: 471 VIHRDVKASNVLLDAELNGRLGDFGLAQLCDHGSDPQTTRVVGTWGYLAPDHIRTGRATT 530
Query: 621 ESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLRKDFDE 680
+DV++FG+++LE+ G++ E+ + E +++WV+ + + +L A D NL ++D+
Sbjct: 531 TTDVFAFGVLLLEVACGRRPIEINNQSGERVVLVDWVFRFWMEANILDAKDPNLGSEYDQ 590
Query: 681 KQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPP 722
K+VE ++ +GL C+H D RP++RQ +Q L + +P+L P
Sbjct: 591 KEVEMVLKLGLLCSHSDPLARPTMRQVLQYLRGDAMLPDLSP 632
>At3g55550 probable serine/threonine-specific protein kinase
Length = 684
Score = 370 bits (949), Expect = e-102
Identities = 239/649 (36%), Positives = 341/649 (51%), Gaps = 57/649 (8%)
Query: 107 TLPLFFLSLILLTFHAYSI--HFQIPSFNPNDANIIYQGSA--APVDGEVDFNINGNYSC 162
T + L LI LT S+ F F N+ G A AP +
Sbjct: 4 TFAVILLLLIFLTHLVSSLIQDFSFIGFKKASPNLTLNGVAEIAPTGA---IRLTTETQR 60
Query: 163 QVGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPN 222
+G A YS + + F+T +F I + GHGLAF + P+
Sbjct: 61 VIGHAFYSLPIRFKPIGVNRALSFST--SFAIAMVPEFVTLGGHGLAFAITP-----TPD 113
Query: 223 SSGGL----MGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTE-HVGINNNSIKSSISTP 277
G L +GL N++ + + S+H VEFD+ + EF + + HVGI+ NS++SSISTP
Sbjct: 114 LRGSLPSQYLGLLNSSRV-NFSSHFFAVEFDTVRDLEFEDINDNHVGIDINSMESSISTP 172
Query: 278 WN----------ASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKK 327
L G + WI Y+S K L V + S + + LSY +D
Sbjct: 173 AGYFLANSTKKELFLDGGRVIQAWIDYDSNKKRLDV--KLSPFSEKPKLSLLSYDVDLSS 230
Query: 328 VLPEWVTIGFSAATGYNGEVNNLLSWEFNSNLEK------------SDDSNSKDTRLVVI 375
VL + + +GFSA+TG + +L W FN + E S K R +I
Sbjct: 231 VLGDEMYVGFSASTGLLASSHYILGWNFNMSGEAFSLSLPSLPRIPSSIKKRKKKRQSLI 290
Query: 376 LTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELE 435
L VSL ++ LVA + ++ + E + EE E GP RF+Y+EL+
Sbjct: 291 LGVSLLCSLLIFAVLVAASLFVVRKVKDEDRVEE----------WELDFGPHRFSYRELK 340
Query: 436 LATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRH 495
ATN F LG GGFG V+KG D VAVK+IS SRQG +E+++EV I LRH
Sbjct: 341 KATNGFGDKELLGSGGFGKVYKGKLPGSDEFVAVKRISHESRQGVREFMSEVSSIGHLRH 400
Query: 496 RNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTP--LSWGVRHKITLGLASGLLYL 553
RNLV+LLGWC + + LL+Y+FMPNGSLD +LF + L+W R KI G+ASGLLYL
Sbjct: 401 RNLVQLLGWCRRRDDLLLVYDFMPNGSLDMYLFDENPEVILTWKQRFKIIKGVASGLLYL 460
Query: 554 HEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYV 613
HE WE+ V+HRDIK++NV+LDS N ++GDFGLAKL +H P T + GTFGYLAPE
Sbjct: 461 HEGWEQTVIHRDIKAANVLLDSEMNGRVGDFGLAKLYEHGSDPGATRVVGTFGYLAPELT 520
Query: 614 STGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDEN 673
+G+ + +DVY+FG V+LE+ G++ E EE M++WVW + G++ +D
Sbjct: 521 KSGKLTTSTDVYAFGAVLLEVACGRRPIET-SALPEELVMVDWVWSRWQSGDIRDVVDRR 579
Query: 674 LRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPP 722
L +FDE++V ++ +GL C++ +RP++RQ + L + P + P
Sbjct: 580 LNGEFDEEEVVMVIKLGLLCSNNSPEVRPTMRQVVMYLEKQFPSPEVVP 628
>At4g29050 serine/threonine-specific kinase like protein
Length = 669
Score = 367 bits (943), Expect = e-101
Identities = 218/581 (37%), Positives = 328/581 (55%), Gaps = 42/581 (7%)
Query: 165 GWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSS 224
G Y+ V +S G ++ F+T + F I + + GHGLAF ++ +P +SS
Sbjct: 60 GHVFYNSPVRFKNSPNGTVSSFSTTFVFAIVSNVNALD--GHGLAFVISPTK-GLPYSSS 116
Query: 225 GGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSET-TEHVGINNNSIKSSIS-------- 275
+GLFN T SNHIV VEFD+F N EF + HVGI+ NS+ S +
Sbjct: 117 SQYLGLFNLTNNGDPSNHIVAVEFDTFQNQEFDDMDNNHVGIDINSLSSEKASTAGYYED 176
Query: 276 ---TPWNASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVLPEW 332
T N L + + WI Y+S+ + L V+ P+ LS D L +
Sbjct: 177 DDGTFKNIRLINQKPIQAWIEYDSSRRQLNVTIHPIHLPKPKIPL-LSLTKDLSPYLFDS 235
Query: 333 VTIGFSAATGYNGEVNNLLSWEFNSNLEKSD-----------DSNSKDTRLVVILTVSLG 381
+ +GF++ATG + +L W F N S+ DS S + ++ +++SL
Sbjct: 236 MYVGFTSATGRLRSSHYILGWTFKLNGTASNIDISRLPKLPRDSRSTSVKKILAISLSLT 295
Query: 382 AVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNF 441
++ I V ++Y++ +++K L + +D E GP RF YK+L +AT F
Sbjct: 296 SLAILVFLTISYMLFLKRKK-----------LMEVLEDWEVQFGPHRFAYKDLYIATKGF 344
Query: 442 SKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKL 501
LG+GGFG V+KG + ++ +AVKK+S SRQG +E+V E+ I +LRH NLV+L
Sbjct: 345 RNSELLGKGGFGKVYKGTLSTSNMDIAVKKVSHDSRQGMREFVAEIATIGRLRHPNLVRL 404
Query: 502 LGWCHDKGEFLLIYEFMPNGSLDSHLFGK-RTPLSWGVRHKITLGLASGLLYLHEEWERC 560
LG+C KGE L+Y+ MP GSLD L+ + L W R KI +ASGL YLH +W +
Sbjct: 405 LGYCRRKGELYLVYDCMPKGSLDKFLYHQPEQSLDWSQRFKIIKDVASGLCYLHHQWVQV 464
Query: 561 VVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTGRASK 620
++HRDIK +NV+LD S N KLGDFGLAKL +H PQT+ +AGTFGY++PE TG+AS
Sbjct: 465 IIHRDIKPANVLLDDSMNGKLGDFGLAKLCEHGFDPQTSNVAGTFGYISPELSRTGKAST 524
Query: 621 ESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLRKD--F 678
SDV++FGI++LEIT G++ E + +WV D + + ++L +DE +++D +
Sbjct: 525 SSDVFAFGILMLEITCGRRPVLPRASSPSEMVLTDWVLDCW-EDDILQVVDERVKQDDKY 583
Query: 679 DEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPN 719
E+QV ++ +GL+C+HP ++RPS+ IQ L+ +PN
Sbjct: 584 LEEQVALVLKLGLFCSHPVAAVRPSMSSVIQFLDGVAQLPN 624
>At1g70110 hypothetical protein
Length = 666
Score = 348 bits (892), Expect = 8e-96
Identities = 210/639 (32%), Positives = 345/639 (53%), Gaps = 46/639 (7%)
Query: 110 LFFLSLILLTFHAYSIHFQIPSFNP---NDANIIYQGSAAPVDGEVDFNINGNYSCQVGW 166
+ L ++L F S+ Q P+ N N + +Y +A ++ + + G
Sbjct: 1 MVLLLFLVLFFVPESVVCQRPNPNGVEFNTSGNMYTSGSAYINNNGLIRLTNSTPQTTGQ 60
Query: 167 ALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSSGG 226
Y+ ++ +S G ++ F+T TF+ + + + G+G+AF + ++ P
Sbjct: 61 VFYNDQLRFKNSVNGTVSSFST--TFVFSIEFHNGIYGGYGIAFVICPTR-DLSPTFPTT 117
Query: 227 LMGLFNTTTMLSSSNHIVHVEFDSFANSEFSET-TEHVGINNNSIKSSI----------S 275
+GLFN + M NHIV VE D+ + +F + HVGI+ N++ S
Sbjct: 118 YLGLFNRSNMGDPKNHIVAVELDTKVDQQFEDKDANHVGIDINTLVSDTVALAGYYMDNG 177
Query: 276 TPWNASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVLPEWVTI 335
T + L+SG ++WI Y+S K + V+ P+ LS + D L E + +
Sbjct: 178 TFRSLLLNSGQPMQIWIEYDSKQKQINVTLHPLYVPKPKIPL-LSLEKDLSPYLLELMYV 236
Query: 336 GFSAATGYNGEVNNLLSWEFNSNLEKSDDSNSKDTRLVV-----------ILTVSL---G 381
GF++ TG + +L W F N D S+ ++ ILT+SL G
Sbjct: 237 GFTSTTGDLTASHYILGWTFKMNGTTPDIDPSRLPKIPRYNQPWIQSPNGILTISLTVSG 296
Query: 382 AVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNF 441
+I+ + +L ++ LKRK+ L + +D E GP RF +K+L +AT F
Sbjct: 297 VIILIILSLSLWLFLKRKK------------LLEVLEDWEVQFGPHRFAFKDLHIATKGF 344
Query: 442 SKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKL 501
LG+GGFG V+KG ++++AVK +S SRQG +E++ E+ I +LRH NLV+L
Sbjct: 345 KDTEVLGKGGFGKVYKGTLPVSNVEIAVKMVSHDSRQGMREFIAEIATIGRLRHPNLVRL 404
Query: 502 LGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTP-LSWGVRHKITLGLASGLLYLHEEWERC 560
G+C KGE L+Y+ M GSLD L+ ++T L W R KI +ASGL YLH++W +
Sbjct: 405 QGYCRHKGELYLVYDCMAKGSLDKFLYHQQTGNLDWSQRFKIIKDVASGLYYLHQQWVQV 464
Query: 561 VVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTGRASK 620
++HRDIK +N++LD++ N KLGDFGLAKL DH PQT+ +AGT GY++PE TG+AS
Sbjct: 465 IIHRDIKPANILLDANMNAKLGDFGLAKLCDHGTDPQTSHVAGTLGYISPELSRTGKAST 524
Query: 621 ESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLRKDFDE 680
SDV++FGIV+LEI G+K + + E + +WV + + +++ +D + +++ E
Sbjct: 525 RSDVFAFGIVMLEIACGRKPI-LPRASQREMVLTDWVLECWENEDIMQVLDHKIGQEYVE 583
Query: 681 KQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPN 719
+Q ++ +GL+C+HP ++RP++ IQ+L+ +P+
Sbjct: 584 EQAALVLKLGLFCSHPVAAIRPNMSSVIQLLDSVAQLPH 622
>At4g04960 unknown protein
Length = 686
Score = 341 bits (875), Expect = 8e-94
Identities = 231/638 (36%), Positives = 346/638 (54%), Gaps = 60/638 (9%)
Query: 110 LFFLSLILLTFHAYS-IHFQIPSFNPNDANIIYQGSAAPVDGEVDFNINGNYSCQVGWAL 168
LF L+L L+ + S I F FN + +N+ G A ++ ++ + S G AL
Sbjct: 5 LFLLTLFLILPNPISAIDFIFNGFNDSSSNVSLFGIAT-IESKI-LTLTNQTSFATGRAL 62
Query: 169 YSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFEIPPNSSGGLM 228
Y++ + D T + F+T +FI + GHG+ F A I +SS +
Sbjct: 63 YNRTIRTKDPITSSVLPFST--SFIFTMAPYKNTLPGHGIVFLFAP-STGINGSSSAQHL 119
Query: 229 GLFNTTTMLSSSNHIVHVEFDSFANSEFSET-TEHVGINNNSIKSSISTP---WN----- 279
GLFN T + SNHI VEFD FAN EFS+ HVGI+ NS+ S S W+
Sbjct: 120 GLFNLTNNGNPSNHIFGVEFDVFANQEFSDIDANHVGIDVNSLHSVYSNTSGYWSDDGVV 179
Query: 280 ---ASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNL-SYQIDFKKVLPEWVTI 335
L+ G +VWI Y N+T+ Q + K L S ++ V+ + + +
Sbjct: 180 FKPLKLNDGRNYQVWIDYRDFVVNVTM----QVAGKIRPKIPLLSTSLNLSDVVEDEMFV 235
Query: 336 GFSAATGYNGEVNNLLSWEF-NSNLEKSD--------------DSNSKDTRLVVILTVSL 380
GF+AATG + + +L+W F NSN S+ DS K V +L +
Sbjct: 236 GFTAATGRLVQSHKILAWSFSNSNFSLSNSLITTGLPSFVLPKDSIVKAKWFVFVLVLIC 295
Query: 381 GAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNN 440
V+ VG LV + +++++ +R+ K+ ++ +D E P R Y+E+E T
Sbjct: 296 FLVVALVG-LVLFAVVRKRLERARKR--------ALMEDWEMEYWPHRIPYEEIESGTKG 346
Query: 441 FSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVK 500
F + +G GG G V+KG ++VAVK+IS+ S G +E+V E+ + +L+HRNLV
Sbjct: 347 FDEKNVIGIGGNGKVYKGLLQGGVVEVAVKRISQESSDGMREFVAEISSLGRLKHRNLVS 406
Query: 501 LLGWCHDK-GEFLLIYEFMPNGSLDSHLFG---KRTPLSWGVRHKITLGLASGLLYLHEE 556
L GWC + G F+L+Y++M NGSLD +F K T LS R +I G+ASG+LYLHE
Sbjct: 407 LRGWCKKEVGSFMLVYDYMENGSLDRWIFENDEKITTLSCEERIRILKGVASGILYLHEG 466
Query: 557 WERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTG 616
WE V+HRDIK+SNV+LD +L DFGLA++ HE +TT + GT GYLAPE V TG
Sbjct: 467 WESKVLHRDIKASNVLLDRDMIPRLSDFGLARVHGHEQPVRTTRVVGTAGYLAPEVVKTG 526
Query: 617 RASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLRK 676
RAS ++DV+++GI+VLE+ G++ E + +K +++WVW +GE+L +D +
Sbjct: 527 RASTQTDVFAYGILVLEVMCGRRPIE-----EGKKPLMDWVWGLMERGEILNGLDPQMMM 581
Query: 677 DFDEKQV----ECLMIVGLWCAHPDVSLRPSIRQAIQV 710
+V E ++ +GL CAHPD + RPS+RQ +QV
Sbjct: 582 TQGVTEVIDEAERVLQLGLLCAHPDPAKRPSMRQVVQV 619
>At3g59700 serine/threonine-specific kinase lecRK1 precursor,lectin
receptor-like
Length = 661
Score = 337 bits (865), Expect = 1e-92
Identities = 203/604 (33%), Positives = 325/604 (53%), Gaps = 51/604 (8%)
Query: 150 GEVDFNINGNYSCQ------VGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSF 203
G V NG ++ G A ++ V + +S TG ++ F+ ++ F I
Sbjct: 31 GSVGIGFNGYFTLTNTTKHTFGQAFENEHVEIKNSSTGVISSFSVNFFFAIVPEHNQQG- 89
Query: 204 YGHGLAFFLAAYGFEIPPNSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTE-H 262
HG+ F ++ +P SS +G+FN T +SN+++ +E D + EF + + H
Sbjct: 90 -SHGMTFVISPTR-GLPGASSDQYLGIFNKTNNGKASNNVIAIELDIHKDEEFGDIDDNH 147
Query: 263 VGINNNSIKSSISTPWN-----------ASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTS 311
VGIN N ++S S SL S + + I Y+ + L V+
Sbjct: 148 VGININGLRSVASASAGYYDDKDGSFKKLSLISREVMRLSIVYSQPDQQLNVTLFPAEIP 207
Query: 312 SPQEKTNLSYQIDFKKVLPEWVTIGFSAATGYNGEVNNLLSWEFNSNLE----------- 360
P K LS D L E + +GF+A+TG G ++ L+ W N +E
Sbjct: 208 VPPLKPLLSLNRDLSPYLLEKMYLGFTASTGSVGAIHYLMGWLVNGVIEYPRLELSIPVL 267
Query: 361 ---KSDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMN 417
SN T L V LTVS+ A V + + +V R +K E +E +
Sbjct: 268 PPYPKKTSNRTKTVLAVCLTVSVFAAF--VASWIGFVFYLRHKKVKEVLEEWEIQY---- 321
Query: 418 DDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSR 477
GP RF YKEL AT F + + LG+GGFG V+KG D ++AVK+ S SR
Sbjct: 322 -------GPHRFAYKELFNATKGFKEKQLLGKGGFGQVYKGTLPGSDAEIAVKRTSHDSR 374
Query: 478 QGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTP--LS 535
QG E++ E+ I +LRH NLV+LLG+C K L+Y++MPNGSLD +L L+
Sbjct: 375 QGMSEFLAEISTIGRLRHPNLVRLLGYCRHKENLYLVYDYMPNGSLDKYLNRSENQERLT 434
Query: 536 WGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELG 595
W R +I +A+ LL+LH+EW + ++HRDIK +NV++D+ N +LGDFGLAKL D
Sbjct: 435 WEQRFRIIKDVATALLHLHQEWVQVIIHRDIKPANVLIDNEMNARLGDFGLAKLYDQGFD 494
Query: 596 PQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIE 655
P+T+ +AGTFGY+APE++ TGRA+ +DVY+FG+V+LE+ G++ E + + E+ +++
Sbjct: 495 PETSKVAGTFGYIAPEFLRTGRATTSTDVYAFGLVMLEVVCGRRIIE-RRAAENEEYLVD 553
Query: 656 WVWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEV 715
W+ + + G++ A +E++R++ + QVE ++ +G+ C+H S+RP++ +++LN
Sbjct: 554 WILELWENGKIFDAAEESIRQEQNRGQVELVLKLGVLCSHQAASIRPAMSVVMRILNGVS 613
Query: 716 SMPN 719
+P+
Sbjct: 614 QLPD 617
>At2g43690 putative receptor protein kinase
Length = 664
Score = 334 bits (856), Expect = 1e-91
Identities = 211/611 (34%), Positives = 324/611 (52%), Gaps = 54/611 (8%)
Query: 139 IIYQGSAAPVDGEVDFNINGNYSCQVGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRG 198
++++GSA ++G ++ G A + +S G +T F+ + F I
Sbjct: 28 LVFEGSAGLMNGFTTLTNTKKHA--YGQAFNDEPFPFKNSVNGNMTSFSFTFFFAIVPEH 85
Query: 199 RSPSFYGHGLAFFLAAYGFEIPPNSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSE 258
HG+AF ++ IP S+ +G+FN T +SSNHI+ VE D + EF +
Sbjct: 86 IDKG--SHGIAFVISPTR-GIPGASADQYLGIFNDTNDGNSSNHIIAVELDIHKDDEFGD 142
Query: 259 TTE-HVGINNNSIKSSISTPW----------NASLHSGDTAEVWIRYNSTTKNLTVSWEY 307
+ HVGIN N ++S +S P N SL SG+ V I Y+ K L V
Sbjct: 143 IDDNHVGININGMRSIVSAPAGYYDQNGQFKNLSLISGNLLRVTILYSQEEKQLNV---- 198
Query: 308 QTTSSPQEKTN------LSYQIDFKKVLPEWVTIGFSAATGYNGEVNNLLSW-------- 353
T SP E+ N LS D L + + IGF+A+TG G ++ + W
Sbjct: 199 --TLSPAEEANVPKWPLLSLNKDLSPYLSKNMYIGFTASTGSVGAIHYMWMWYVFTFIIV 256
Query: 354 ---EFNSNLEKSDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEA 410
+F+ +L+V++T A+ + + A V ++ K+
Sbjct: 257 PKLDFDIPTFPPYPKAESQVKLIVLVTFLTLALFVALAASALIVFFYKRHKK-------- 308
Query: 411 MHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVK 470
L + ++ E GP RF+YKEL ATN F + LG+GGFG VFKG + + ++AVK
Sbjct: 309 --LLEVLEEWEVECGPHRFSYKELFNATNGFKQ--LLGEGGFGPVFKGTLSGSNAKIAVK 364
Query: 471 KISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGK 530
++S S QG +E + E+ I +LRH NLV+LLG+C K E L+Y+F+PNGSLD +L+G
Sbjct: 365 RVSHDSSQGMRELLAEISTIGRLRHPNLVRLLGYCRYKEELYLVYDFLPNGSLDKYLYGT 424
Query: 531 --RTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAK 588
+ LSW R KI +AS L YLH W V+HRDIK +NV++D N LGDFGLAK
Sbjct: 425 SDQKQLSWSQRFKIIKDVASALSYLHHGWIHVVIHRDIKPANVLIDDKMNASLGDFGLAK 484
Query: 589 LMDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKD 648
+ D PQT+ +AGTFGY+APE + TGR + +DVY+FG+ +LE++ +K E E
Sbjct: 485 VYDQGYDPQTSRVAGTFGYMAPEIMRTGRPTMGTDVYAFGMFMLEVSCDRKLFEPRAE-S 543
Query: 649 EEKGMIEWVWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAI 708
EE + W + + G+++ A E +R+D D+ Q+E ++ +G+ C+H +RP + +
Sbjct: 544 EEAILTNWAINCWENGDIVEAATERIRQDNDKGQLELVLKLGVLCSHEAEEVRPDMATVV 603
Query: 709 QVLNFEVSMPN 719
++LN +P+
Sbjct: 604 KILNGVSELPD 614
>At2g43700 putative receptor protein kinase
Length = 658
Score = 330 bits (845), Expect = 2e-90
Identities = 213/609 (34%), Positives = 331/609 (53%), Gaps = 50/609 (8%)
Query: 139 IIYQGSAAPVDGEVDFNINGNYSCQVGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRG 198
++ QGSA G ++ G A + V +S +T F+ + F I
Sbjct: 27 LVMQGSAGFFKGYRTLTSTKKHA--YGQAFEDEIVPFKNSANDTVTSFSVTFFFAIAPED 84
Query: 199 RSPSFYGHGLAFFLAAYGFEIPPNSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSE 258
+ HG+AF ++ I S+ +G+FN SSNH++ VE D + EF +
Sbjct: 85 KHKG--AHGMAFVISPTR-GITGASADQYLGIFNKANNGDSSNHVIAVELDINKDEEFGD 141
Query: 259 TTE-HVGINNNSIKSSISTPW----------NASLHSGDTAEVWIRYNSTTKNLTVSWEY 307
+ HVGIN N ++S P + SL SG V I Y+ K L V
Sbjct: 142 INDNHVGININGMRSIKFAPAGYYDQEGQFKDLSLISGSLLRVTILYSQMEKQLNV---- 197
Query: 308 QTTSSPQE-----KTNLSYQIDFKKVLPEWVTIGFSAATGYNGEVNNLLSWEFNSNLEKS 362
T SSP+E K LS D + E + +GFSA+TG ++ +LSW + ++
Sbjct: 198 -TLSSPEEAYYPNKPLLSLNQDLSPYILENMYVGFSASTGSVRAMHYMLSWFVHGGVDVP 256
Query: 363 D--------DSNSKDTRLV--VILTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEAMH 412
+ K+ LV ++L SL V+ ALVA + +R +K KE
Sbjct: 257 NLDLGIPTFPPYPKEKSLVYRIVLVTSLALVLFV--ALVASALSIFFYRRHKKVKE---- 310
Query: 413 LTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKI 472
+ ++ E GP RF YKEL AT F + LG+GGFG VFKG D ++AVK+I
Sbjct: 311 ---VLEEWEIQCGPHRFAYKELFKATKGFKQ--LLGKGGFGQVFKGTLPGSDAEIAVKRI 365
Query: 473 SRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGK-- 530
S S+QG +E++ E+ I +LRH+NLV+L G+C K E L+Y+FMPNGSLD +L+ +
Sbjct: 366 SHDSKQGMQEFLAEISTIGRLRHQNLVRLQGYCRYKEELYLVYDFMPNGSLDKYLYHRAN 425
Query: 531 RTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLM 590
+ L+W R KI +AS L YLH EW + V+HRDIK +NV++D N +LGDFGLAKL
Sbjct: 426 QEQLTWNQRFKIIKDIASALCYLHHEWVQVVIHRDIKPANVLIDHQMNARLGDFGLAKLY 485
Query: 591 DHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEE 650
D PQT+ +AGTF Y+APE + +GRA+ +DVY+FG+ +LE++ G++ E + +E
Sbjct: 486 DQGYDPQTSRVAGTFWYIAPELIRSGRATTGTDVYAFGLFMLEVSCGRRLIE-RRTASDE 544
Query: 651 KGMIEWVWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQV 710
+ EW + G++L A+++ +R + + +Q+E ++ +G+ C+H V++RP + + +Q+
Sbjct: 545 VVLAEWTLKCWENGDILEAVNDGIRHEDNREQLELVLKLGVLCSHQAVAIRPDMSKVVQI 604
Query: 711 LNFEVSMPN 719
L ++ +P+
Sbjct: 605 LGGDLQLPD 613
>At3g45440 receptor like protein kinase
Length = 669
Score = 329 bits (843), Expect = 4e-90
Identities = 211/592 (35%), Positives = 326/592 (54%), Gaps = 56/592 (9%)
Query: 163 QVGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYG-HGLAFFLAAYGFEIPP 221
++G A + K ++ K+ + F+TH+ + + P F G HG+AF L+A ++
Sbjct: 60 KMGHAFFKKP---FEFKSPRSFSFSTHFVCALVPK---PGFIGGHGIAFVLSA-SMDLTQ 112
Query: 222 NSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSET-TEHVGINNNSIKSSISTPW-- 278
+ +GLFN +T S S+H+V VE D+ ++EF + HVGI+ NS+ S STP
Sbjct: 113 ADATQFLGLFNISTQGSPSSHLVAVELDTALSAEFDDIDANHVGIDVNSLMSIASTPAAY 172
Query: 279 ---------NASLHSGDTAEVWIRYNSTTKNLTVS-WEYQTTSSPQEKTNLSYQIDFKKV 328
+ L SGD +VW+ Y N+T++ + Q S P LS I+ +
Sbjct: 173 FSEIDGENKSIKLLSGDPIQVWVDYGGNVLNVTLAPLKIQKPSRPL----LSRSINLSET 228
Query: 329 LPEW-VTIGFSAATGYNGEVNNLLSWEFNSN---LEKSD----------DSNSKDTRLVV 374
P+ +GFS ATG +L W + N L+ D + +K +V+
Sbjct: 229 FPDRKFFLGFSGATGTLISYQYILGWSLSRNKVSLQTLDVTKLPRVPRHRAKNKGPSVVL 288
Query: 375 ILTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKEL 434
I+ + L A+I+ + AYV R+RK +E + ++ E+ GP RF+YK+L
Sbjct: 289 IVLLILLAIIVFLALGAAYVY--RRRKYAE-----------IREEWEKEYGPHRFSYKDL 335
Query: 435 ELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLR 494
+ATN F+KD LG+GGFG V+KG Q+AVK++S + +G K++V E+ + L+
Sbjct: 336 YIATNGFNKDGLLGKGGFGKVYKGTLPSKG-QIAVKRVSHDAEEGMKQFVAEIVSMGNLK 394
Query: 495 HRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFG-KRTPLSWGVRHKITLGLASGLLYL 553
H+N+V LLG+C KGE LL+ E+MPNGSLD +LF ++ P SW R I +A+ L Y+
Sbjct: 395 HKNMVPLLGYCRRKGELLLVSEYMPNGSLDQYLFNDEKPPFSWRRRLLIIKDIATALNYM 454
Query: 554 HEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYV 613
H + V+HRDIK+SNVMLD+ FN +LGDFG+A+ DH P TT GT GY+APE
Sbjct: 455 HTGAPQVVLHRDIKASNVMLDTEFNGRLGDFGMARFHDHGKDPATTAAVGTIGYMAPELA 514
Query: 614 STGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDEN 673
+ G A +DVY FG +LE+T G++ E E +++WV + + LL A D
Sbjct: 515 TVG-ACTATDVYGFGAFLLEVTCGRRPVEPGLSA-ERWYIVKWVCECWKMASLLGARDPR 572
Query: 674 LRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPPKRP 725
+R + ++VE ++ +GL C + LRPS+ +Q LN + +P++ P P
Sbjct: 573 MRGEISAEEVEMVLKLGLLCTNGVPDLRPSMEDIVQYLNGSLELPDISPNSP 624
>At1g70130 hypothetical protein
Length = 656
Score = 327 bits (838), Expect = 2e-89
Identities = 212/603 (35%), Positives = 314/603 (51%), Gaps = 62/603 (10%)
Query: 149 DGEVDFNINGNYSC-------QVGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSP 201
DG D N +G + G LY + +S G ++ F+T + F I ++
Sbjct: 40 DGVADLNPDGLFKLITSKTQGGAGQVLYQFPLQFKNSPNGTVSSFSTTFVFAIVAVRKTI 99
Query: 202 SFYGHGLAFFLAAYGFEIPPNSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSE-FSETT 260
+ G GL+F I P N+ + SNH V V F + + + E
Sbjct: 100 A--GCGLSF-------NISPTKG------LNSVPNIDHSNHSVSVGFHTAKSDKPDGEDV 144
Query: 261 EHVGINNNSIKSSISTPW----------NASLHSGDTAEVWIRYNSTTKNLTVSWEYQTT 310
VGIN +S K + N + SG +VWI YN++TK L V+
Sbjct: 145 NLVGINIDSSKMDRNCSAGYYKDDGRLVNLDIASGKPIQVWIEYNNSTKQLDVTMHSIKI 204
Query: 311 SSPQEKTNLSYQIDFKKVLPEWVTIGFSAATGYNGEVNNLLSWEFN----------SNLE 360
S P+ LS + D L E++ IGF++ G + +L W FN S L
Sbjct: 205 SKPKIPL-LSMRKDLSPYLHEYMYIGFTSV-GSPTSSHYILGWSFNNKGAVSDINLSRLP 262
Query: 361 KSDDSNSKDTRLVVILTVSL---GAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMN 417
K D + + + IL +SL G ++ V L + LKRK+ +
Sbjct: 263 KVPDEDQERSLSSKILAISLSISGVTLVIVLILGVMLFLKRKK------------FLEVI 310
Query: 418 DDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSR 477
+D E GP +FTYK+L +AT F LG+GGFG VFKG + +AVKKIS SR
Sbjct: 311 EDWEVQFGPHKFTYKDLFIATKGFKNSEVLGKGGFGKVFKGILPLSSIPIAVKKISHDSR 370
Query: 478 QGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTP-LSW 536
QG +E++ E+ I +LRH +LV+LLG+C KGE L+Y+FMP GSLD L+ + L W
Sbjct: 371 QGMREFLAEIATIGRLRHPDLVRLLGYCRRKGELYLVYDFMPKGSLDKFLYNQPNQILDW 430
Query: 537 GVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGP 596
R I +ASGL YLH++W + ++HRDIK +N++LD + N KLGDFGLAKL DH +
Sbjct: 431 SQRFNIIKDVASGLCYLHQQWVQVIIHRDIKPANILLDENMNAKLGDFGLAKLCDHGIDS 490
Query: 597 QTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEW 656
QT+ +AGTFGY++PE TG++S SDV++FG+ +LEIT G++ + E + +W
Sbjct: 491 QTSNVAGTFGYISPELSRTGKSSTSSDVFAFGVFMLEITCGRRPIG-PRGSPSEMVLTDW 549
Query: 657 VWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVS 716
V D + G++L +DE L + +QV ++ +GL C+HP + RPS+ IQ L+ +
Sbjct: 550 VLDCWDSGDILQVVDEKLGHRYLAEQVTLVLKLGLLCSHPVAATRPSMSSVIQFLDGVAT 609
Query: 717 MPN 719
+P+
Sbjct: 610 LPH 612
>At5g01540 receptor like protein kinase
Length = 682
Score = 326 bits (836), Expect = 3e-89
Identities = 218/663 (32%), Positives = 337/663 (49%), Gaps = 77/663 (11%)
Query: 110 LFFLSLILLTFHAYSIH---------FQIPSFNPNDANIIYQGSAAPVDGEVDFNINGNY 160
+F +S + F S+H F FN N + I +G AA + + +
Sbjct: 7 MFIVSFLFKLFLFLSVHVRAQRTTTNFAFRGFNGNQSKIRIEG-AAMIKPDGLLRLTDRK 65
Query: 161 SCQVGWALYSKKVLLWDSKTGQLT--DFTTHYTFIINTRGRSPSFYGHGLAFFLAAYGFE 218
S G A Y K V L + + +T F+T + F+I S S G G F L+ +
Sbjct: 66 SNVTGTAFYHKPVRLLNRNSTNVTIRSFSTSFVFVIIPS--SSSNKGFGFTFTLSPTPYR 123
Query: 219 IPPNSSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTE---HVGINNNSIKSSIS 275
+ S+ L G+FN NH+ VEFD+ S T +G+N NS S +
Sbjct: 124 LNAGSAQYL-GVFNKENNGDPRNHVFAVEFDTVQGSRDDNTDRIGNDIGLNYNSRTSDLQ 182
Query: 276 TPW------------NASLHSGDTAEVWIRYNSTTKNLTVSWEYQTTSSPQEKTNLSYQI 323
P + L SG+ + + Y+ T+ L V+ K +S +
Sbjct: 183 EPVVYYNNDDHNKKEDFQLESGNPIQALLEYDGATQMLNVTVYPARLGFKPTKPLISQHV 242
Query: 324 D-FKKVLPEWVTIGFSAATGYN-GEVNNLLSWEFNS--------------------NLEK 361
+++ E + +GF+A+TG + ++ W F+S N K
Sbjct: 243 PKLLEIVQEEMYVGFTASTGKGQSSAHYVMGWSFSSGGERPIADVLILSELPPPPPNKAK 302
Query: 362 SDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLE 421
+ NS+ V+++ V+L AV++ + L+ + ++ +KR E+ E+ +++
Sbjct: 303 KEGLNSQ----VIVMIVALSAVMLVMLVLLFFFVMYKKRLGQEETLEDW--------EID 350
Query: 422 RGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKK 481
PRR Y++L +AT+ F K +G GGFG VFKG + D +AVKKI SRQG +
Sbjct: 351 H---PRRLRYRDLYVATDGFKKTGIIGTGGFGTVFKGKLPNSD-PIAVKKIIPSSRQGVR 406
Query: 482 EYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFG----KRTPLSWG 537
E+V E++ + +LRH+NLV L GWC K + LLIY+++PNGSLDS L+ LSW
Sbjct: 407 EFVAEIESLGKLRHKNLVNLQGWCKHKNDLLLIYDYIPNGSLDSLLYTVPRRSGAVLSWN 466
Query: 538 VRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQ 597
R +I G+ASGLLYLHEEWE+ V+HRD+K SNV++DS N +LGDFGLA+L + +
Sbjct: 467 ARFQIAKGIASGLLYLHEEWEKIVIHRDVKPSNVLIDSKMNPRLGDFGLARLYERGTLSE 526
Query: 598 TTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWV 657
TT L GT GY+APE G S SDV++FG+++LEI G+K T+ +++WV
Sbjct: 527 TTALVGTIGYMAPELSRNGNPSSASDVFAFGVLLLEIVCGRKPTD-----SGTFFLVDWV 581
Query: 658 WDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSM 717
+ + GE+L A+D L +D + + VGL C H + RPS+R ++ LN E ++
Sbjct: 582 MELHANGEILSAIDPRLGSGYDGGEARLALAVGLLCCHQKPASRPSMRIVLRYLNGEENV 641
Query: 718 PNL 720
P +
Sbjct: 642 PEI 644
>At5g60280 receptor like protein kinase
Length = 657
Score = 326 bits (835), Expect = 3e-89
Identities = 205/565 (36%), Positives = 315/565 (55%), Gaps = 51/565 (9%)
Query: 186 FTTHYTFIINTRGRSPSFYG-HGLAFFLAAYGFEIPPNSSGGLMGLFNTTTMLSSSNHIV 244
F+TH+ + R P G +G+AFFL+ ++ + +GLFNTTT S S+HI
Sbjct: 81 FSTHF---VCAMVRKPGVTGGNGIAFFLSP-SMDLTNADATQYLGLFNTTTNRSPSSHIF 136
Query: 245 HVEFDSFANSEFSET-TEHVGINNNSIKSSISTPWN-----------ASLHSGDTAEVWI 292
+E D+ ++EF + HVGI+ NS+ S S P + SL SGD+ +VW+
Sbjct: 137 AIELDTVQSAEFDDIDNNHVGIDVNSLTSVESAPASYFSDKKGLNKSISLLSGDSIQVWV 196
Query: 293 RYNSTTKNLTVSWEYQTTSSPQEKTNLSYQIDFKKVLPEWVTIGFSAATGYNGEVNNLLS 352
++ T N++++ ++ +S ++ +V+ + + +GFSAATG + +L
Sbjct: 197 DFDGTVLNVSLA---PLGIRKPSQSLISRSMNLSEVIQDRMFVGFSAATGQLANNHYILG 253
Query: 353 WEFN-----------SNLEKSDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKRK 401
W F+ S L + K + L+++L + LG +++ L+ L R+ K
Sbjct: 254 WSFSRSKASLQSLDISKLPQVPHPKMKTSLLLILLLIVLGIILL---VLLVGAYLYRRNK 310
Query: 402 RSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFA 461
+E ++E E+ GP R++YK L AT F KD LG+GGFG V+KG
Sbjct: 311 YAEVREE-----------WEKEYGPHRYSYKSLYKATKGFHKDGFLGKGGFGEVYKGTLP 359
Query: 462 DLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNG 521
D +AVK+ S +G K++V E+ + L HRNLV L G+C KGEFLL+ ++MPNG
Sbjct: 360 QED--IAVKRFSHHGERGMKQFVAEIASMGCLDHRNLVPLFGYCRRKGEFLLVSKYMPNG 417
Query: 522 SLDSHLFGKRTP-LSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVK 580
SLD LF R P L+W R I G+AS L YLH E + V+HRDIK+SNVMLD+ F K
Sbjct: 418 SLDQFLFHNREPSLTWSKRLGILKGIASALKYLHTEATQVVLHRDIKASNVMLDTDFTGK 477
Query: 581 LGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKA 640
LGDFG+A+ DH P TTG GT GY+ PE S G AS ++DVY+FG ++LE+T G++
Sbjct: 478 LGDFGMARFHDHGANPTTTGAVGTVGYMGPELTSMG-ASTKTDVYAFGALILEVTCGRRP 536
Query: 641 TEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSL 700
E E++ +++WV D + + +L+ A D L + Q+E ++ +GL C +
Sbjct: 537 VE-PNLPIEKQLLVKWVCDCWKRKDLISARDPKLSGEL-IPQIEMVLKLGLLCTNLVPES 594
Query: 701 RPSIRQAIQVLNFEVSMPNLPPKRP 725
RP + + +Q L+ +VS+P+ P P
Sbjct: 595 RPDMVKVVQYLDRQVSLPDFSPDSP 619
>At3g45430 receptor like protein kinase
Length = 613
Score = 324 bits (830), Expect = 1e-88
Identities = 213/589 (36%), Positives = 320/589 (54%), Gaps = 57/589 (9%)
Query: 164 VGWALYSKKVLLWDSKTGQLTDFTTHYTFIINTRGRSPSFYG-HGLAFFLAAYGFEIPPN 222
+G A K + D + + F+TH+ + + P F G HG+ F ++ +
Sbjct: 1 MGHAFIKKPI---DFSSSKPLSFSTHFVCALVPK---PGFEGGHGITFVISPT-VDFTRA 53
Query: 223 SSGGLMGLFNTTTMLSSSNHIVHVEFDSFANSEFSETTE-HVGINNNSIKSSISTPWN-- 279
MG+FN +T S S+H+ VE D+ N +F ET H+GI+ N+ S S P +
Sbjct: 54 QPTRYMGIFNASTNGSPSSHLFAVELDTVRNPDFRETNNNHIGIDVNNPISVESAPASYF 113
Query: 280 ---------ASLHSGDTAEVWIRYNSTTKNLTVS-WEYQTTSSPQEKTNLSYQIDFKKVL 329
+L SG +VW+ Y+ N++V+ E + S P +++ F +
Sbjct: 114 SKTAQKNVSINLSSGKPIQVWVDYHGNVLNVSVAPLEAEKPSLPLLSRSMNLSEIFSR-- 171
Query: 330 PEWVTIGFSAATGYNGEVNNLLSWEFNSNLEKSD---------------DSNSKDTRLVV 374
+ +GF+AATG + + LL W F++N E S + L++
Sbjct: 172 -RRLFVGFAAATGTSISYHYLLGWSFSTNRELSQLLDFSKLPQVPRPRAEHKKVQFALII 230
Query: 375 ILTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKEL 434
L V L V++ V A V Y RK+K +E +++ E+ G RF+YK L
Sbjct: 231 ALPVILAIVVMAVLAGVYY---HRKKKYAE-----------VSEPWEKKYGTHRFSYKSL 276
Query: 435 ELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLR 494
+AT F KDR LG+GGFG V++G L+ VAVK++S QG K++V EV + L+
Sbjct: 277 YIATKGFHKDRFLGRGGFGEVYRGDLP-LNKTVAVKRVSHDGEQGMKQFVAEVVSMKSLK 335
Query: 495 HRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTP-LSWGVRHKITLGLASGLLYL 553
HRNLV LLG+C KGE LL+ E+MPNGSLD HLF ++P LSW R I G+AS L YL
Sbjct: 336 HRNLVPLLGYCRRKGELLLVSEYMPNGSLDQHLFDDQSPVLSWSQRFVILKGIASALFYL 395
Query: 554 HEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYV 613
H E E+ V+HRDIK+SNVMLD+ N +LGDFG+A+ DH TT GT GY+APE +
Sbjct: 396 HTEAEQVVLHRDIKASNVMLDAELNGRLGDFGMARFHDHGGNAATTAAVGTVGYMAPELI 455
Query: 614 STGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDEN 673
+ G AS +DVY+FG+ +LE+ G+K E + E++ +I+WV + + K LL A D
Sbjct: 456 TMG-ASTITDVYAFGVFLLEVACGRKPVE-FGVQVEKRFLIKWVCECWKKDSLLDAKDPR 513
Query: 674 LRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPP 722
L ++F ++VE +M +GL C + RP++ Q + L+ + +P+ P
Sbjct: 514 LGEEFVPEEVELVMKLGLLCTNIVPESRPAMGQVVLYLSGNLPLPDFSP 562
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,769,191
Number of Sequences: 26719
Number of extensions: 816349
Number of successful extensions: 5890
Number of sequences better than 10.0: 1014
Number of HSP's better than 10.0 without gapping: 870
Number of HSP's successfully gapped in prelim test: 144
Number of HSP's that attempted gapping in prelim test: 2252
Number of HSP's gapped (non-prelim): 1194
length of query: 751
length of database: 11,318,596
effective HSP length: 107
effective length of query: 644
effective length of database: 8,459,663
effective search space: 5448022972
effective search space used: 5448022972
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146709.2


