

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.12 + phase: 0 /pseudo
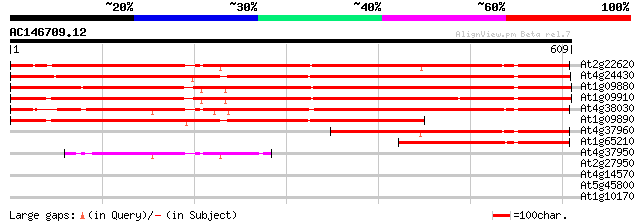
(609 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g22620 unknown protein 644 0.0
At4g24430 LG27/30-like gene 596 e-170
At1g09880 hypothetical protein 589 e-168
At1g09910 hypothetical protein 587 e-168
At4g38030 LG127/30 like gene 565 e-161
At1g09890 hypothetical protein 448 e-126
At4g37960 putative protein 283 3e-76
At1g65210 hypothetical protein 214 9e-56
At4g37950 hypothetical protein 128 8e-30
At2g27950 unknown protein 30 5.1
At4g14570 acylamino acid-releasing enzyme (aare) 29 6.7
At5g45800 receptor kinase-like protein 29 8.7
At1g10170 hypothetical protein 29 8.7
>At2g22620 unknown protein
Length = 677
Score = 644 bits (1662), Expect = 0.0
Identities = 330/623 (52%), Positives = 419/623 (66%), Gaps = 39/623 (6%)
Query: 2 ARPEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVIA 61
+ PEG I GI Y+GIDNVLD + DRGY+DVVW EP + K ++ GT F +I
Sbjct: 74 SNPEGLITGIKYHGIDNVLDDKID--DRGYWDVVWYEPEK----KQKTDKLEGTKFEIIT 127
Query: 62 ADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMPGIEVDQI 121
+E +E+SF+R WT S G VP+N+D RYI RSG SG Y Y I R +G P +++DQI
Sbjct: 128 QNEEQIEISFTRTWTISRRGSLVPLNVDKRYIIRSGVSGLYMYGILERLEGWPDVDMDQI 187
Query: 122 RFVFKLDKDRFKYMAISDTRQRNMPAMKDRNTGQVLAYPEAVLLTNPINPQFRGEVDDKY 181
R VFKL+ +F +MAISD RQR+MP+M DR + LAY EAVLLTNP NP F+GEVDDKY
Sbjct: 188 RIVFKLNPKKFDFMAISDDRQRSMPSMADRENSKSLAYKEAVLLTNPSNPMFKGEVDDKY 247
Query: 182 QYSCENKDTQFMDGLVLILIHQWVSG**HRVMSFAMVDPLSKILLL------------ML 229
YS E+KD +H W+S + F M+ P + L +
Sbjct: 248 MYSMEDKDNN---------VHGWISS--DPPVGFWMITPSDEFRLGGPIKQDLTSHAGPI 296
Query: 230 DQSLFR-THYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSENDNATLWSDAVQQLSKEV 288
S+F THYAGKE+ M ++ GE +KKVFGPV YLN+ S ++ LW DA +Q++ EV
Sbjct: 297 TLSMFTSTHYAGKEMRMDYRNGEPWKKVFGPVLAYLNSVSPKDSTLRLWRDAKRQMAAEV 356
Query: 289 QSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYANNAYVGLALPGDAGSWQRE 348
+SWPYDF S+DY +QRG + G+ L++D Y + Y A+VGLA G+AGSWQ E
Sbjct: 357 KSWPYDFITSEDYPLRHQRGTLEGQFLIKDSYVSRLKI-YGKFAFVGLAPIGEAGSWQTE 415
Query: 349 SKGYQFWTQTDAKGYFKITNVVPGHYNLFGWVSGFIGDYKYNSTITITPGGVIKLNSLVY 408
SKGYQFWT+ D +G F I NV G+Y+L+ W SGFIGDYKY ITITPG + + LVY
Sbjct: 416 SKGYQFWTKADRRGRFIIENVRAGNYSLYAWGSGFIGDYKYEQNITITPGSEMNVGPLVY 475
Query: 409 NPPRNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYT---EGRDKQYGLWERYTEMYPTD 465
PPRNGPT+WEIG+PDR EF++P+PYPTLMNKLY + R +QYGLW+RY ++YP +
Sbjct: 476 EPPRNGPTLWEIGVPDRTAGEFYIPDPYPTLMNKLYVNPLQDRFRQYGLWDRYADLYPQN 535
Query: 466 DLIYTLGVNK-NKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGNYTLQLALASAAD 524
DL+YT+GV+ DWF+AHV RN N+TY+PTTWQIIF ++ + G YTL++ALASAAD
Sbjct: 536 DLVYTIGVSDYRSDWFFAHVARNVGNDTYQPTTWQIIFNLKNVNRIGRYTLRIALASAAD 595
Query: 525 AYLQVRFNDRSVYPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKGKNIIY 584
+ LQ+R ND F TG IG+ DN I RHGIHGLYRL+SIDV NLL G N I+
Sbjct: 596 SELQIRINDPK-SDAIFTTGFIGK---DNAIARHGIHGLYRLYSIDVAGNLLSVGDNTIF 651
Query: 585 LTQTNADTPFQGVMYDYIRLEQP 607
LTQT + TPFQG+MYDYIRLE P
Sbjct: 652 LTQTRSRTPFQGIMYDYIRLESP 674
>At4g24430 LG27/30-like gene
Length = 646
Score = 596 bits (1536), Expect = e-170
Identities = 306/626 (48%), Positives = 419/626 (66%), Gaps = 30/626 (4%)
Query: 1 MARPEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVI 60
+++P+G++ GISY G+DN+L++ N++ +RGY+D+VW++ G + + +RI GT+F V+
Sbjct: 27 ISKPDGFVTGISYQGVDNLLETHNEDFNRGYWDLVWSDEGTPGTTGKS-ERIKGTSFEVV 85
Query: 61 AADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMPGIEVDQ 120
+E +VE+SFSR W S+ PIN+D R+I R +GFYSYAIF P + Q
Sbjct: 86 VENEELVEISFSRKWDSSLQDSIAPINVDKRFIMRKDVTGFYSYAIFEHLAEWPAFNLPQ 145
Query: 121 IRFVFKLDKDRFKYMAISDTRQRNMPAMKDR--NTGQVLAYPEAVLLTNPINPQFRGEVD 178
R V+KL KD+FKYMAI+D RQR MP +DR G+ LAYPEAVLL +P+ +F+GEVD
Sbjct: 146 TRIVYKLRKDKFKYMAIADNRQRKMPLPEDRLGKRGRPLAYPEAVLLVHPVEDEFKGEVD 205
Query: 179 DKYQYSCENKDTQFMDGLV---------LILIHQWVSG**HRVMSFAMVDPLSKILLLML 229
DKY+YS ENKD + + +I +++ SG + + V P+S + L
Sbjct: 206 DKYEYSSENKDLKVHGWISHNLDLGCWQIIPSNEFRSGGLSKQNLTSHVGPISLAMFLS- 264
Query: 230 DQSLFRTHYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSE-NDNATLWSDAVQQLSKEV 288
HYAG+++ M + G+++KKVFGPVF YLN + +D +LW DA Q+ EV
Sbjct: 265 ------AHYAGEDMVMKVKAGDSWKKVFGPVFTYLNCLPDKTSDPLSLWQDAKNQMLTEV 318
Query: 289 QSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYANNAYVGLALPGDAGSWQRE 348
QSWPYDFP S+D+ ++RG + GRLLV D + + AN A+VGLA PG+ GSWQ E
Sbjct: 319 QSWPYDFPASEDFPVSDKRGCISGRLLVCDKFLSDD-FLPANGAFVGLAPPGEVGSWQLE 377
Query: 349 SKGYQFWTQTDAKGYFKITNVVPGHYNLFGWVSGFIGDYKYNSTITITPGGVIKLNSLVY 408
SKGYQFWT+ D+ GYF I ++ G YNL G+V+G+IGDY+Y I IT G I + ++VY
Sbjct: 378 SKGYQFWTEADSDGYFAINDIREGEYNLNGYVTGWIGDYQYEQLINITAGCDIDVGNIVY 437
Query: 409 NPPRNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYT--EGRDKQYGLWERYTEMYPTDD 466
PPR+GPT+WEIGIPDR +EF VP+P P +NKLY R +QYGLWERYTE+YP +D
Sbjct: 438 EPPRDGPTVWEIGIPDRSAAEFFVPDPNPKYINKLYIGHPDRFRQYGLWERYTELYPKED 497
Query: 467 LIYTLGVNK-NKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGNYTLQLALASAADA 525
L++T+GV+ KDWF+AHVTR ++TY+ TTWQI F+ ++ KS Y +++ALA+A A
Sbjct: 498 LVFTIGVSDYKKDWFFAHVTRKMGDDTYQKTTWQIKFKLENVQKSCTYKIRIALATANVA 557
Query: 526 YLQVRFNDRSV--YPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKGKNII 583
LQVR ND P F TG IG DN I RHGIHG+YRL+++DVPS L++G N +
Sbjct: 558 ELQVRMNDDDTEKTTPIFTTGVIGH---DNAIARHGIHGIYRLYNVDVPSEKLVEGDNTL 614
Query: 584 YLTQTNADT-PFQGVMYDYIRLEQPP 608
+LTQT T F G+MYDYIRLE PP
Sbjct: 615 FLTQTMTTTGAFNGLMYDYIRLEGPP 640
>At1g09880 hypothetical protein
Length = 631
Score = 589 bits (1518), Expect = e-168
Identities = 313/629 (49%), Positives = 404/629 (63%), Gaps = 34/629 (5%)
Query: 1 MARPEGYILGISYNGIDNVLDS-ENQELDRGYFDVVWNEPGEQ-NLSKSTFQRIHGTNFS 58
++ PEG++ GI YNGIDNVL N+E DRGY+DVVWN PG++ +K T RI T
Sbjct: 10 LSNPEGFVTGIQYNGIDNVLAYYTNKEYDRGYWDVVWNFPGKKAKKTKGTLDRIEATKME 69
Query: 59 VIAADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMPGIEV 118
VI ++ +E+SF+R W S S VP+NID R++ SGFYSYAIF R +G P +E+
Sbjct: 70 VITQNDEKIELSFTRTWNTS-STTAVPVNIDKRFVMLQNSSGFYSYAIFERLQGWPAVEL 128
Query: 119 DQIRFVFKLDKDRFKYMAISDTRQRNMPAMKDR--NTGQVLAYPEAVLLTNPINPQFRGE 176
D +R VFKL+K +F YMAISD RQR MP DR GQ LAYPEAV L +PI P+F+GE
Sbjct: 129 DNMRLVFKLNKKKFHYMAISDDRQRYMPVPDDRVPPRGQPLAYPEAVQLLDPIEPEFKGE 188
Query: 177 VDDKYQYSCENKDTQFMDGLVLILIHQWVS-----G**HRVMS--FAMVDPLSKILLLML 229
VDDKY+YS E+KD I +H W+S G S F PL + L +
Sbjct: 189 VDDKYEYSMESKD---------IKVHGWISTNDSVGFWQITPSNEFRSAGPLKQFLGSHV 239
Query: 230 DQS----LFRTHYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSENDNATLWSDAVQQLS 285
+ THY G ++ M+F+ GE +KKVFGPVF+YLN+ D LW +A Q
Sbjct: 240 GPTNLAVFHSTHYVGADLIMSFKNGEAWKKVFGPVFIYLNSFPKGVDPLLLWHEAKNQTK 299
Query: 286 KEVQSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYANNAYVGLALPGDAGSW 345
E + WPY+F S D+ +QRG+V GRLLV+D + AN +YVGLA PGD GSW
Sbjct: 300 IEEEKWPYNFTASDDFPASDQRGSVSGRLLVRDRFISSEDIP-ANGSYVGLAAPGDVGSW 358
Query: 346 QRESKGYQFWTQTDAKGYFKITNVVPGHYNLFGWVSGFIGDYKYNSTITITPGGVIKLNS 405
QRE KGYQFW++ D G F I NV G YNL+ + GFIGDY ++ I+PG I L
Sbjct: 359 QRECKGYQFWSKADENGSFSINNVRSGRYNLYAFAPGFIGDYHNDTVFDISPGSKISLGD 418
Query: 406 LVYNPPRNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYTEGRDK--QYGLWERYTEMYP 463
LVY PPR+G T+WEIG+PDR +EF++P+P P+ +NKLY DK QYGLWERY+E+YP
Sbjct: 419 LVYEPPRDGSTLWEIGVPDRSAAEFYIPDPNPSFVNKLYLNHSDKYRQYGLWERYSELYP 478
Query: 464 TDDLIYTLGVNK-NKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLK--SGNYTLQLALA 520
+D++Y + ++ +K+WF+ VTR N Y+ TTWQI F+ +K +GN+ L++ALA
Sbjct: 479 DEDMVYNVDIDDYSKNWFFMQVTRKQANGGYKGTTWQIRFQFDDKMKNVTGNFKLRIALA 538
Query: 521 SAADAYLQVRFNDRSVYPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKGK 580
++ A LQVR ND S PP F T IGR DN I RHGIHGLY L+S++VP+ L G
Sbjct: 539 TSNVAELQVRVNDLSADPPLFRTEQIGR---DNTIARHGIHGLYWLYSVNVPAASLHVGN 595
Query: 581 NIIYLTQTNADTPFQGVMYDYIRLEQPPS 609
N IYLTQ A +PFQG+MYDYIRLE P S
Sbjct: 596 NTIYLTQALATSPFQGLMYDYIRLEYPDS 624
>At1g09910 hypothetical protein
Length = 675
Score = 587 bits (1514), Expect = e-168
Identities = 304/625 (48%), Positives = 396/625 (62%), Gaps = 34/625 (5%)
Query: 1 MARPEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVI 60
+++P G I GI YNGIDNVL+ N+E +RGY+D+ WNEPG K F I G F VI
Sbjct: 69 LSKPGGIITGIEYNGIDNVLEVRNKETNRGYWDLHWNEPG----GKGIFDVISGVTFRVI 124
Query: 61 AADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMPGIEVDQ 120
E VE+SF R W S+ GK +P+NID R+I G SG YSY I+ K PG E+ +
Sbjct: 125 VETEEQVEISFLRTWDPSLEGKYIPLNIDKRFIMLRGSSGVYSYGIYEHLKDWPGFELGE 184
Query: 121 IRFVFKLDKDRFKYMAISDTRQRNMPAMKDRNTG--QVLAYPEAVLLTNPINPQFRGEVD 178
R FKL KD+F YMA++D R+R MP D G Q L Y EA LLT P +P+ +GEVD
Sbjct: 185 TRIAFKLRKDKFHYMAVADDRKRIMPFPDDLCKGRCQTLDYQEASLLTAPCDPRLQGEVD 244
Query: 179 DKYQYSCENKDTQFMDGLVLILIHQWVS-----G**HRVMS--FAMVDPLSKILLLMLDQ 231
DKYQYSCENKD + +H W+S G S F PL + L +
Sbjct: 245 DKYQYSCENKD---------LRVHGWISFDPPVGFWQITPSNEFRSGGPLKQNLTSHVGP 295
Query: 232 S----LFRTHYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSENDNATLWSDAVQQLSKE 287
+ THYAGK + F+ GE +KKV+GPVF+YLN+ ++ +D LW DA ++ E
Sbjct: 296 TTLAVFHSTHYAGKTMMPRFEHGEPWKKVYGPVFIYLNSTANGDDPLCLWDDAKIKMMAE 355
Query: 288 VQSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYANNAYVGLALPGDAGSWQR 347
V+ WPY F S DY +RG GRLL++D + A AYVGLA PGD+GSWQ
Sbjct: 356 VERWPYSFVASDDYPKSEERGTARGRLLIRDRFINNDLIS-ARGAYVGLAPPGDSGSWQI 414
Query: 348 ESKGYQFWTQTDAKGYFKITNVVPGHYNLFGWVSGFIGDYKYNSTITITPGGVIKLNSLV 407
E KGYQFW D GYF I NV PG YNL+ WV FIGDY + + +T G +I++ +V
Sbjct: 415 ECKGYQFWAIADEAGYFSIGNVRPGEYNLYAWVPSFIGDYHNGTIVRVTSGCMIEMGDIV 474
Query: 408 YNPPRNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYTEGRD--KQYGLWERYTEMYPTD 465
Y PPR+GPT+WEIGIPDR SEF +P+P PTL+N++ +D +QYGLW++YT+MYP D
Sbjct: 475 YEPPRDGPTLWEIGIPDRKASEFFIPDPDPTLVNRVLVHHQDRFRQYGLWKKYTDMYPND 534
Query: 466 DLIYTLGVNK-NKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGNYTLQLALASAAD 524
DL+YT+GV+ +DWF+AHV R + + +E TTWQIIF ++ + NY L++A+ASA
Sbjct: 535 DLVYTVGVSDYRRDWFFAHVPRK-KGDVHEGTTWQIIFNLENIDQKANYKLRVAIASATL 593
Query: 525 AYLQVRFNDRSVYPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKGKNIIY 584
A LQ+R ND P F TG IGR DN I RHGIHG+Y L+++++P N L++G N I+
Sbjct: 594 AELQIRINDAEAIRPLFTTGLIGR---DNSIARHGIHGVYMLYAVNIPGNRLVQGDNTIF 650
Query: 585 LTQTNADTPFQGVMYDYIRLEQPPS 609
L Q + PFQG+MYDYIRLE PPS
Sbjct: 651 LKQPRCNGPFQGIMYDYIRLEGPPS 675
>At4g38030 LG127/30 like gene
Length = 649
Score = 565 bits (1455), Expect = e-161
Identities = 301/628 (47%), Positives = 388/628 (60%), Gaps = 69/628 (10%)
Query: 2 ARPEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVIA 61
+ P+G I GI Y GIDNVL + DRG I GTNF +I
Sbjct: 66 SNPQGLITGIKYKGIDNVLHPHLR--DRG---------------------IEGTNFRIIT 102
Query: 62 ADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMPGIEVDQI 121
+ VE+SFSR W ++P+N+D RYI R SG Y Y IF R P +++ I
Sbjct: 103 QTQEQVEISFSRTWEDG----HIPLNVDKRYIIRRNTSGIYMYGIFERLPEWPELDMGLI 158
Query: 122 RFVFKLDKDRFKYMAISDTRQRNMPAMKDRNT----GQVLAYPEAVLLTNPINPQFRGEV 177
R FKL+ +RF YMA++D RQR MP DR+ + L Y EAV LT+P N F+ +V
Sbjct: 159 RIAFKLNPERFHYMAVADNRQREMPTEDDRDIKRGHAKALGYKEAVQLTHPHNSMFKNQV 218
Query: 178 DDKYQYSCENKDTQFMDGLVLILIHQWVSG**HRVMSFAMVDP---------LSKILLLM 228
DDKYQY+CE KD + +H W+S + F ++ P + + L
Sbjct: 219 DDKYQYTCEIKDNK---------VHGWIST--KSRVGFWIISPSGEYRSGGPIKQELTSH 267
Query: 229 LDQSLFRT----HYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSENDNAT--LWSDAVQ 282
+ + T HY G ++ ++ GE +KKV GPVF+YLN+ S+ N+ LW DA Q
Sbjct: 268 VGPTAITTFISGHYVGADMEAHYRPGEAWKKVLGPVFIYLNSDSTSNNKPQDLLWEDAKQ 327
Query: 283 QLSKEVQSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYANNAYVGLALPGDA 342
Q KEV++WPYDF S DY +RG+V GRLLV D + G+ +AYVGLA PG+A
Sbjct: 328 QSEKEVKAWPYDFVASSDYLSRRERGSVTGRLLVNDRFLTPGK-----SAYVGLAPPGEA 382
Query: 343 GSWQRESKGYQFWTQTDAKGYFKITNVVPGHYNLFGWVSGFIGDYKYNSTITITPGGVIK 402
GSWQ +KGYQFWT+T+ GYF I NV PG YNL+GWV GFIGD++Y + + + G VI
Sbjct: 383 GSWQTNTKGYQFWTKTNETGYFTIENVRPGTYNLYGWVPGFIGDFRYQNLVNVAAGSVIS 442
Query: 403 LNSLVYNPPRNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYTEGRDK--QYGLWERYTE 460
L +VY PPRNGPT+WEIG+PDR E+ +PEPY MN LY DK QYGLW+RYTE
Sbjct: 443 LGRVVYKPPRNGPTLWEIGVPDRTAREYFIPEPYKDTMNPLYLNHTDKFRQYGLWQRYTE 502
Query: 461 MYPTDDLIYTLGV-NKNKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGNYTLQLAL 519
+YPT DL+YT+GV N ++DWFYA VTR T + TY PT WQI+F + G+YTLQ+AL
Sbjct: 503 LYPTHDLVYTVGVSNYSQDWFYAQVTRKTGDLTYVPTIWQIVFHLPYVNSRGSYTLQVAL 562
Query: 520 ASAADAYLQVRFNDRSVYPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKG 579
ASAA A LQVR N+++ + P F+TG IG+ DN I RHGIHG+YRL++IDVP LL G
Sbjct: 563 ASAAWANLQVRINNQNSW-PFFSTGTIGK---DNAIARHGIHGMYRLYTIDVPGRLLRTG 618
Query: 580 KNIIYLTQTNADTPFQGVMYDYIRLEQP 607
N IYL Q A PF+G+MYDYIRLE+P
Sbjct: 619 TNTIYLRQPKAQGPFEGLMYDYIRLEEP 646
>At1g09890 hypothetical protein
Length = 477
Score = 448 bits (1152), Expect = e-126
Identities = 229/463 (49%), Positives = 298/463 (63%), Gaps = 25/463 (5%)
Query: 1 MARPEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVI 60
+++P+G + GI YNGIDN+L+ N+E++RGY+D+VW G + F I G+NF VI
Sbjct: 26 LSKPDGIVTGIEYNGIDNLLEVLNEEVNRGYWDLVWGGSG----TAGGFDVIKGSNFEVI 81
Query: 61 AADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMPGIEVDQ 120
+E +E+SF+R W S GK VP+NID R++ SG SGFY+YAI+ K P + +
Sbjct: 82 VKNEEQIELSFTRKWDPSQEGKAVPLNIDKRFVMLSGSSGFYTYAIYEHLKEWPAFSLAE 141
Query: 121 IRFVFKLDKDRFKYMAISDTRQRNMPAMKDR--NTGQVLAYPEAVLLTNPINPQFRGEVD 178
R FKL K++F YMA++D RQR MP DR + GQ LAYPEAVLL NP+ QF+GEVD
Sbjct: 142 TRIAFKLRKEKFHYMAVTDDRQRFMPLPDDRLPDRGQALAYPEAVLLVNPLESQFKGEVD 201
Query: 179 DKYQYSCENKDT---------QFMDGLVLIL-IHQWVSG**HRVMSFAMVDPLSKILLLM 228
DKYQYSCENKD Q G LI H++ +G + + V P + + +
Sbjct: 202 DKYQYSCENKDITVHGWICTEQPSVGFWLITPSHEYRTGGPQKQNLTSHVGPTALAVFIS 261
Query: 229 LDQSLFRTHYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSE-NDNATLWSDAVQQLSKE 287
HY G+++ F EGE +KKVFGPVFVYLN+++ + ND LW DA Q++ E
Sbjct: 262 -------AHYTGEDLVPKFSEGEAWKKVFGPVFVYLNSSTDDDNDPLWLWQDAKSQMNVE 314
Query: 288 VQSWPYDFPQSQDYFPPNQRGAVFGRLLVQDWYFEGGRYQYANNAYVGLALPGDAGSWQR 347
+SWPY FP S DY QRG V GRLLVQD Y + + AN YVGLA+PG AGSWQR
Sbjct: 315 AESWPYSFPASDDYVKTEQRGNVVGRLLVQDRYVDKD-FIAANRGYVGLAVPGAAGSWQR 373
Query: 348 ESKGYQFWTQTDAKGYFKITNVVPGHYNLFGWVSGFIGDYKYNSTITITPGGVIKLNSLV 407
E K YQFWT+TD +G+F I+ + PG YNL+ W+ GFIGDYKY+ ITIT G I + LV
Sbjct: 374 ECKEYQFWTRTDEEGFFYISGIRPGQYNLYAWIPGFIGDYKYDDVITITSGCYIYVEDLV 433
Query: 408 YNPPRNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLYTEGRDK 450
Y PPRNG T+WEIG PDR +EF+VP+P P +N LY D+
Sbjct: 434 YQPPRNGATLWEIGFPDRSAAEFYVPDPNPKYINNLYQNHPDR 476
>At4g37960 putative protein
Length = 289
Score = 283 bits (723), Expect = 3e-76
Identities = 144/265 (54%), Positives = 178/265 (66%), Gaps = 10/265 (3%)
Query: 349 SKGYQFWTQTDAKGYFKITNVVPGHYNLFGWVSGFIGDYKYNSTITITPGGVIKLNSLVY 408
S GYQFWT+ D G F I NV PG Y+L+ WVSGFIGDYKY ITITPG I + +VY
Sbjct: 26 SSGYQFWTRADKMGMFTIANVRPGTYSLYAWVSGFIGDYKYVRDITITPGREIDVGHIVY 85
Query: 409 NPPRNGPTIWEIGIPDRLTSEFHVPEPYPTLMNKLY-----TEGRDKQYGLWERYTEMYP 463
PPRNGPT+WEIG PDR +EF++P+P PTL KLY + R +QYGLW+RY+ +YP
Sbjct: 86 VPPRNGPTLWEIGQPDRTAAEFYIPDPDPTLFTKLYLNYSNPQDRFRQYGLWDRYSVLYP 145
Query: 464 TDDLIYTLGVNK-NKDWFYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGNYTLQLALASA 522
+DL++T GV+ KDWFYAHV R N TY+ TTWQI F + +++ YTL++ALA+A
Sbjct: 146 RNDLVFTAGVSDYKKDWFYAHVNRKAGNGTYKATTWQIKFNLKAVIQTRIYTLRIALAAA 205
Query: 523 ADAYLQVRFNDRSVYPPHFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKGKNI 582
+ L V N+ P F TG IGR DN I RHGIHGLY+L++IDV LL G N
Sbjct: 206 STIDLLVWVNEVD-SKPLFITGLIGR---DNAIARHGIHGLYKLYNIDVHGKLLRVGNNT 261
Query: 583 IYLTQTNADTPFQGVMYDYIRLEQP 607
I+LT F GVMYDY+RLE P
Sbjct: 262 IFLTHGRNSDSFSGVMYDYLRLEGP 286
>At1g65210 hypothetical protein
Length = 248
Score = 214 bits (546), Expect = 9e-56
Identities = 112/188 (59%), Positives = 131/188 (69%), Gaps = 7/188 (3%)
Query: 423 PDRLTSEFHVPEPYPTLMNKLYTEGRDK--QYGLWERYTEMYPTDDLIYTLGV-NKNKDW 479
P + E+ VPEPY MN LY DK QYGLW+RYTE+YP DLIYT+GV N +KDW
Sbjct: 62 PHQRAREYFVPEPYKNTMNPLYLNHTDKFRQYGLWQRYTELYPNHDLIYTIGVSNYSKDW 121
Query: 480 FYAHVTRNTENNTYEPTTWQIIFEHQHDLKSGNYTLQLALASAADAYLQVRFNDRSVYPP 539
FY+ VTR ++TY PTTWQ +F + G+YTLQLALASAA A LQVRFN+ P
Sbjct: 122 FYSQVTRKIGDSTYTPTTWQTVFHLPYVNMRGSYTLQLALASAAWANLQVRFNNEYTRP- 180
Query: 540 HFATGHIGRIRGDNCIQRHGIHGLYRLFSIDVPSNLLLKGKNIIYLTQTNADTPFQGVMY 599
F+TG+IGR DN I RHGIHGLYRL+SI+VP LL G N IYL Q A P +GVMY
Sbjct: 181 FFSTGYIGR---DNAIARHGIHGLYRLYSINVPGRLLRTGTNTIYLRQAKASGPLEGVMY 237
Query: 600 DYIRLEQP 607
DYIRLE+P
Sbjct: 238 DYIRLEEP 245
>At4g37950 hypothetical protein
Length = 296
Score = 128 bits (322), Expect = 8e-30
Identities = 89/242 (36%), Positives = 126/242 (51%), Gaps = 43/242 (17%)
Query: 60 IAADENMVEVSFSRLWTHSMSGKNVPINIDIRYIFRSGDSGFYSYAIFNRPKGMPGIEVD 119
+ D +++V+FS S G I I +G + + I NR P +++D
Sbjct: 69 VVVDNGIIDVTFS-----SPQGL-------ITRIKYNGLNNVLNDQIENRGYSWPDVDMD 116
Query: 120 QIRFVFKLDKDRFKYMAISDTRQRNMPAMKDRNT----GQVLAYPEAVLLTNPINPQFRG 175
QIR VFKL+ +F +MA+SD RQ+ MP DR+ LAY EAV L NP N +G
Sbjct: 117 QIRIVFKLNTTKFDFMAVSDNRQKIMPFDTDRDITKGRASPLAYKEAVHLINPQNHMLKG 176
Query: 176 EVDDKYQYSCENKDTQFMDGLVLILIHQWVSG**HRVMSFAMVDPLSKILLL-------- 227
+VDDKY YS ENKD + +H W+S + + F M+ P +
Sbjct: 177 QVDDKYMYSVENKDNK---------VHGWISS--DQRIGFWMITPSDEFHACGPIKQDLT 225
Query: 228 ----MLDQSLFRT-HYAGKEVTMAFQEGETYKKVFGPVFVYLNTASSENDNATLWSDAVQ 282
S+F + HYAGK++ ++ E +KKVFGPVFVYLN+ASS N LW+DA +
Sbjct: 226 SHVGPTTLSMFTSVHYAGKDMNTNYKSKEPWKKVFGPVFVYLNSASSRN---LLWTDAKR 282
Query: 283 QL 284
Q+
Sbjct: 283 QV 284
Score = 29.3 bits (64), Expect = 6.7
Identities = 19/66 (28%), Positives = 37/66 (55%), Gaps = 7/66 (10%)
Query: 4 PEGYILGISYNGIDNVLDSENQELDRGYFDVVWNEPGEQNLSKSTFQRIHGTNFSVIAAD 63
P+G I I YNG++NVL+ + + +RGY W + + + + F +++ T F +A
Sbjct: 83 PQGLITRIKYNGLNNVLNDQIE--NRGY---SWPDV-DMDQIRIVF-KLNTTKFDFMAVS 135
Query: 64 ENMVEV 69
+N ++
Sbjct: 136 DNRQKI 141
>At2g27950 unknown protein
Length = 778
Score = 29.6 bits (65), Expect = 5.1
Identities = 16/57 (28%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query: 432 VPEPYPTLMNKLYTEGRDKQYGLWERYTEMYPTDDLIYTLGVNKNKDWFYAHVTRNT 488
VPE +P +++L + D++ + TEM P DD + +DW H ++T
Sbjct: 537 VPENFPNQLSQLSSVDEDREANILGELTEMQPDDDAASL--QSTIQDWSEEHSDQDT 591
>At4g14570 acylamino acid-releasing enzyme (aare)
Length = 764
Score = 29.3 bits (64), Expect = 6.7
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 2/44 (4%)
Query: 391 STITITPGGVIKLNSLVYNPPRNGPTIWEIGIPDRLTSEFHVPE 434
S +P G+ L ++ NP PT +EI +L EFH+P+
Sbjct: 100 SAFVPSPSGLKLL--VIRNPENESPTKFEIWNSSQLEKEFHIPQ 141
>At5g45800 receptor kinase-like protein
Length = 666
Score = 28.9 bits (63), Expect = 8.7
Identities = 25/73 (34%), Positives = 38/73 (51%), Gaps = 11/73 (15%)
Query: 11 ISYNGIDN-----VLDSENQELDRGYFDVVWNEPG--EQNLSKSTFQ---RIHGTNFSVI 60
+S+N I N VLD N LD +W+ PG NLS++ F R+ N SV+
Sbjct: 71 VSWNPIRNLTRLRVLDLSNNSLDGSLPTWLWSMPGLVSVNLSRNRFGGSIRVIPVNGSVL 130
Query: 61 AADENMVEVSFSR 73
+A + + +SF+R
Sbjct: 131 SAVKEL-NLSFNR 142
>At1g10170 hypothetical protein
Length = 1188
Score = 28.9 bits (63), Expect = 8.7
Identities = 16/44 (36%), Positives = 24/44 (54%)
Query: 137 ISDTRQRNMPAMKDRNTGQVLAYPEAVLLTNPINPQFRGEVDDK 180
+ D RQR + N G+ +A E V+LT+P PQ E+ +K
Sbjct: 172 LPDNRQRVASRTRPVNQGKRVAKEENVVLTDPNLPQLVQELQEK 215
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,983,736
Number of Sequences: 26719
Number of extensions: 688773
Number of successful extensions: 1264
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 1193
Number of HSP's gapped (non-prelim): 18
length of query: 609
length of database: 11,318,596
effective HSP length: 105
effective length of query: 504
effective length of database: 8,513,101
effective search space: 4290602904
effective search space used: 4290602904
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146709.12


