

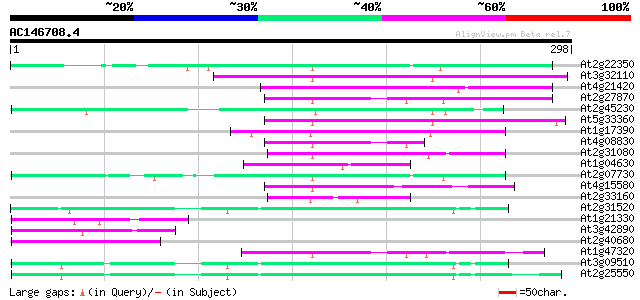
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.4 + phase: 0
(298 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g22350 putative non-LTR retroelement reverse transcriptase 67 9e-12
At3g32110 non-LTR reverse transcriptase, putative 66 2e-11
At4g21420 hypothetical protein 65 4e-11
At2g27870 putative non-LTR retroelement reverse transcriptase 60 2e-09
At2g45230 putative non-LTR retroelement reverse transcriptase 58 5e-09
At5g33360 putative protein 57 1e-08
At1g17390 hypothetical protein 56 3e-08
At4g08830 putative protein 54 1e-07
At2g31080 putative non-LTR retroelement reverse transcriptase 54 1e-07
At1g04630 unknown protein 50 1e-06
At2g07730 putative non-LTR retroelement reverse transcriptase 49 3e-06
At4g15580 splicing factor like protein 47 1e-05
At2g33160 putative polygalacturonase 47 1e-05
At2g31520 putative non-LTR retroelement reverse transcriptase 46 3e-05
At1g21330 hypothetical protein 46 3e-05
At3g42890 putative protein 45 4e-05
At2g40680 putative non-LTR retroelement reverse transcriptase 45 5e-05
At1g47320 hypothetical protein 45 5e-05
At3g09510 putative non-LTR reverse transcriptase 44 8e-05
At2g25550 putative non-LTR retroelement reverse transcriptase 44 8e-05
>At2g22350 putative non-LTR retroelement reverse transcriptase
Length = 321
Score = 67.4 bits (163), Expect = 9e-12
Identities = 80/305 (26%), Positives = 121/305 (39%), Gaps = 46/305 (15%)
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDL 60
+C C ET H+ C + +WS L +PR Q+R
Sbjct: 34 VCQICQGGEETILHVLRDCPAMAGIWSRL-------------------VPR---DQIRQF 71
Query: 61 YVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQV------RVHAFIGLSGA----VSTGKC 110
+ A+++ W N+R + + + V R G +G V K
Sbjct: 72 FTASLLE------WIYKNLRERGSWPTVFVMAVWWGWKWRCGNIFGGNGKCRDRVKFIKD 125
Query: 111 IAADAAILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDH 168
+A + AI +AF R R V W P WVK+NTDG+ N G GG+ RDH
Sbjct: 126 LAEEVAIANAFVKGNEVRVSRVERLVSWVSPEDGWVKLNTDGASRGNPGFATAGGVLRDH 185
Query: 169 LGTFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPSLV- 227
G ++G F N+G CS E+ G + A G + LE D S ++ F +
Sbjct: 186 NGAWIGGFAVNIGVCSAPLAELWGVYYGLFIAWGRGARRVELEVD-SKMVVGFLTTGIAD 244
Query: 228 --PI-LLRNRWHNARTLNVQVISSHIFREGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQ 283
P+ L ++ + V SH++RE N D LAN S+ +G L + P
Sbjct: 245 SHPLSFLLRLCYDFLSKGWIVRISHVYREANRLADGLANYAFSLSLGLHLLESRPDVVSS 304
Query: 284 VFYED 288
+ +D
Sbjct: 305 ILLDD 309
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 66.2 bits (160), Expect = 2e-11
Identities = 54/194 (27%), Positives = 84/194 (42%), Gaps = 6/194 (3%)
Query: 109 KCIAADAAILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFR 166
K +A + +I + + +R + W PP W K+NTDG+ N G + GG+ R
Sbjct: 1714 KDLAVEVSIAYSREVELRLSGLRVNKPIRWTPPMEGWYKINTDGASRGNPGLASAGGVLR 1773
Query: 167 DHLGTFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKN--P 224
+ G + G F N+GRCS E+ G + A ++ LE D+ + K
Sbjct: 1774 NSAGAWCGGFAVNIGRCSAPLAELWGVYYGLYMAWAKQLTHLELEVDSEVVVGFLKTGIG 1833
Query: 225 SLVPI-LLRNRWHNARTLNVQVISSHIFREGNVCVDRLANLGHSV-VGEVWLSTLPSDFH 282
P+ L HN + + V SH++RE N D LAN S+ +G +P
Sbjct: 1834 ETHPLSFLVRLCHNFLSKDWTVRISHVYREANSLADGLANHAFSLSLGLHVFDEIPISLV 1893
Query: 283 QVFYEDRCSMPRIR 296
+ ED R+R
Sbjct: 1894 MLLSEDNDGPARLR 1907
>At4g21420 hypothetical protein
Length = 229
Score = 65.1 bits (157), Expect = 4e-11
Identities = 47/162 (29%), Positives = 73/162 (45%), Gaps = 8/162 (4%)
Query: 134 VSVCWKPPSAPWVKVNTDGSV-LNNSGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSG 192
V V WK P +K+N DGS + GG+FR+H FL + ++G + E +
Sbjct: 59 VKVVWKKPENGRIKLNFDGSRGREGQASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAA 118
Query: 193 FILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILLRNRWHN-----ARTLNVQVIS 247
+E A +G ++WLE DA + + + N+ N LN +
Sbjct: 119 LKRGLELALENGLTDLWLEGDAKIIMDIISRRGRLRCEKTNKHVNYIKVVMPELN-NCVL 177
Query: 248 SHIFREGNVCVDRLANLGHSVVG-EVWLSTLPSDFHQVFYED 288
SH++REGN D+LA LGH +VW P + ++D
Sbjct: 178 SHVYREGNRVADKLAKLGHQFQDPKVWRVQPPEIVLPIMHDD 219
>At2g27870 putative non-LTR retroelement reverse transcriptase
Length = 314
Score = 59.7 bits (143), Expect = 2e-09
Identities = 49/167 (29%), Positives = 74/167 (43%), Gaps = 22/167 (13%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
+ W P W K+NTDG+ N G + GG+ RD G + G F N+G CS E+ G
Sbjct: 144 IAWSKPEEGWWKLNTDGASRGNPGLASAGGVLRDEEGAWRGGFALNIGVCSAPLAELWG- 202
Query: 194 ILVMEHAALHGWYNIW------LESDASSALMV-FKNPSLVPI----LLRNRWHNARTLN 242
+G Y W LE + S ++V F + + L H+ + +
Sbjct: 203 -------VYYGLYIAWERRVTRLEIEVDSEIVVGFLKIGINEVHPLSFLVRLCHDFISRD 255
Query: 243 VQVISSHIFREGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQVFYED 288
+V SH++RE N D LAN S+ +G LS +P + +D
Sbjct: 256 WRVRISHVYREANRLADGLANYAFSLPLGFHSLSLVPDSLRFILLDD 302
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 58.2 bits (139), Expect = 5e-09
Identities = 67/273 (24%), Positives = 108/273 (39%), Gaps = 32/273 (11%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDF--SSIKALMSSLPRHCSSQVRD 59
C C ET +HL +C F W+ G ++ S + + L H S
Sbjct: 1092 CVRCPSHGETVNHLLFKCPFARLTWAISPLPAPPGGEWAESLFRNMHHVLSVHKSQPEES 1151
Query: 60 LYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAILD 119
+ A + ++ +W RN++ F + ++ V ++ D +
Sbjct: 1152 DHHALIPWILWRLWKNRNDLVFKGREFTAPQVILKA----------------TEDMDAWN 1195
Query: 120 AFRIP-PHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSGAC--GGLFRDHLGTFLGAF 176
+ P P S V W+PPS WVK NTDG+ + G C G + R+H G L
Sbjct: 1196 NRKEPQPQVTSSTRDRCVKWQPPSHGWVKCNTDGAWSKDLGNCGVGWVLRNHTGRLLWLG 1255
Query: 177 VGNL-GRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKN----PSLVPIL- 230
+ L + SV +TEV + + + + ESD+ + + +N PSL P +
Sbjct: 1256 LRALPSQQSVLETEVEALRWAVLSLSRFNYRRVIFESDSQYLVSLIQNEMDIPSLAPRIQ 1315
Query: 231 -LRNRWHNARTLNVQVISSHIFREGNVCVDRLA 262
+RN + + Q REGN DR A
Sbjct: 1316 DIRNLLRHFEEVKFQFTR----REGNNVADRTA 1344
>At5g33360 putative protein
Length = 306
Score = 57.0 bits (136), Expect = 1e-08
Identities = 49/168 (29%), Positives = 73/168 (43%), Gaps = 8/168 (4%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
V W PS W K+NTDG+ N G GG R+ G + F N+GRCS E+ G
Sbjct: 136 VRWSKPSLGWCKLNTDGASHGNPGLAIAGGALRNEYGEWCFGFALNIGRCSAPLAELWGV 195
Query: 194 ILVMEHAALHGWYNIWLESDASSALMVFKN--PSLVPI-LLRNRWHNARTLNVQVISSHI 250
+ A G + LE D+ + + S P+ L H + + V H+
Sbjct: 196 YYGLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLSRDWIVRIGHV 255
Query: 251 FREGNVCVDRLANLGHSV-VGEVWLSTLPSDFHQVFYEDR--CSMPRI 295
+RE N D LAN + +G ++ PS + +D S+PR+
Sbjct: 256 YREANRLADELANYAFDLPLGYHGFASPPSSLDSILRDDELGTSVPRL 303
>At1g17390 hypothetical protein
Length = 322
Score = 55.8 bits (133), Expect = 3e-08
Identities = 45/156 (28%), Positives = 67/156 (42%), Gaps = 10/156 (6%)
Query: 118 LDAFRIPPHR-----RSMREIVSVCWKPPSAPWVKVNTDGSVLNNS--GACGGLFRDHLG 170
LD ++ H+ R+ RE + W PP W K+NTDG+ N GG+ RD G
Sbjct: 79 LDVWKAHVHKMGVTTRTAREERLIAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDG 138
Query: 171 TFLGAFVGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFK---NPSLV 227
+ F N+G CS E+ G + A G + +E D+ + + + S
Sbjct: 139 NWCYGFSLNIGICSAPLAELWGAYYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHP 198
Query: 228 PILLRNRWHNARTLNVQVISSHIFREGNVCVDRLAN 263
L H + + V SH++RE N D LAN
Sbjct: 199 LSFLVRLCHGLLSKDWSVRISHVYREANRLADGLAN 234
>At4g08830 putative protein
Length = 947
Score = 53.9 bits (128), Expect = 1e-07
Identities = 33/93 (35%), Positives = 45/93 (47%), Gaps = 16/93 (17%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
V WKPP WVK+NTDG+ N G GG+ RD +G + G F ++G CS E+ G
Sbjct: 839 VAWKPPDGEWVKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWG- 897
Query: 194 ILVMEHAALHGWYNIW------LESDASSALMV 220
+G Y W +E + S L+V
Sbjct: 898 -------VYYGLYMAWERRFTRVELEVDSELVV 923
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 53.5 bits (127), Expect = 1e-07
Identities = 42/132 (31%), Positives = 60/132 (44%), Gaps = 7/132 (5%)
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFIL 195
W+ PS WVK+ TDG+ N G A GG R+ G +LG F N+G C+ E+ G
Sbjct: 1063 WQVPSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYY 1122
Query: 196 VMEHAALHGWYNIWLESDASSALMVF----KNPSLVPILLRNRWHNARTLNVQVISSHIF 251
+ A G+ + L+ D + N + L+R T + V SH++
Sbjct: 1123 GLLIAWDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVR-LCQGFFTRDWLVRVSHVY 1181
Query: 252 REGNVCVDRLAN 263
RE N D LAN
Sbjct: 1182 REANRLADGLAN 1193
>At1g04630 unknown protein
Length = 356
Score = 50.4 bits (119), Expect = 1e-06
Identities = 29/92 (31%), Positives = 47/92 (50%), Gaps = 4/92 (4%)
Query: 125 PHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNS-GACGGLFRDHLGTFLGA--FVGNLG 181
P R + +E + W P W K N DG+ +N+ G L RD GTFLGA +G++
Sbjct: 182 PDRVTSQEPTANLWTTPPIGWTKCNYDGTYHSNAPSKAGWLLRDDRGTFLGAAHAIGSI- 240
Query: 182 RCSVFDTEVSGFILVMEHAALHGWYNIWLESD 213
+ ++E+ ++ M+H G+ I+ E D
Sbjct: 241 TTNPMESELQALVMAMQHCWSRGYRKIYFEGD 272
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 49.3 bits (116), Expect = 3e-06
Identities = 68/271 (25%), Positives = 100/271 (36%), Gaps = 32/271 (11%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDLY 61
CS C E+ H+ C + +W L Q FS + L ++L L+
Sbjct: 685 CSVCNGADESILHVLRDCPAMTPIWQRLLPQRRQNEFFSQFEWLFTNLDP-AKGDWPTLF 743
Query: 62 VAAVVHMVHSIWWA---RNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAADAAIL 118
IWWA R F K+ ++ FI K IA +
Sbjct: 744 SMG-------IWWAWKWRCGDVFGERKLCRDRLK-----FI---------KDIAEEVRKA 782
Query: 119 DAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAF 176
+ H + R + WK PS WVK+ TDG+ + G A G + G +LG F
Sbjct: 783 HVGTLNNHVKRARVERMIRWKAPSDRWVKLTTDGASRGHQGLAAASGAILNLQGEWLGGF 842
Query: 177 VGNLGRCSVFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPI----LLR 232
N+G C E+ G + A G+ + L D S ++ F + + L
Sbjct: 843 ALNIGSCDAPLAELWGAYYGLLIAWDKGFRRVELNLD-SELVVGFLSTGISKAHPLSFLV 901
Query: 233 NRWHNARTLNVQVISSHIFREGNVCVDRLAN 263
T + V SH++RE N D LAN
Sbjct: 902 RLCQGFFTRDWLVRVSHVYREANRLADGLAN 932
>At4g15580 splicing factor like protein
Length = 559
Score = 47.4 bits (111), Expect = 1e-05
Identities = 42/136 (30%), Positives = 59/136 (42%), Gaps = 15/136 (11%)
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
+ W P WVKV+TDG+ N G A GG+ RD G ++G F L + G
Sbjct: 26 IAWTKPPEGWVKVSTDGASRGNPGPAAAGGVIRDEDGLWVGGFALQLAFVRLRWQSCGGR 85
Query: 194 I-LVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILLRNRWHNARTLNVQVISSHIFR 252
+ L ++ + G +L+S S A + L LL W V SH++R
Sbjct: 86 VELEVDSELVVG----FLQSGISEAHPLAFLVRLCHGLLSKDW--------LVRVSHVYR 133
Query: 253 EGNVCVDRLANLGHSV 268
E N D LAN S+
Sbjct: 134 EANRLADGLANYAFSL 149
>At2g33160 putative polygalacturonase
Length = 664
Score = 47.0 bits (110), Expect = 1e-05
Identities = 27/81 (33%), Positives = 40/81 (49%), Gaps = 7/81 (8%)
Query: 138 WKPPSAPWVKVNTDGSVLNNS--GACGGLFRDHLGTFLGAFVGN-LGRC--SVFDTEVSG 192
W+ P+ WVK N DGS +N + G G + RDH G + F G +G C + + E
Sbjct: 503 WRRPTNGWVKCNVDGSFINGNIPGKGGWIVRDHNGRY--EFAGQAIGNCIDNALECEFQA 560
Query: 193 FILVMEHAALHGWYNIWLESD 213
++ M+H G+ I E D
Sbjct: 561 LLMAMQHCWSKGYRRICFEGD 581
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 45.8 bits (107), Expect = 3e-05
Identities = 64/278 (23%), Positives = 108/278 (38%), Gaps = 31/278 (11%)
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSWLC--SQLHVGLDFSSIKALMSSLPRHCS-SQV 57
+C C + E+ +H C F W WL S + L + + +S++ + +
Sbjct: 1239 ICPRCHRENESINHALFTCPFATMAW-WLSDSSLIRNQLMSNDFEENISNILNFVQDTTM 1297
Query: 58 RDLYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAAD--A 115
D + V ++ IW ARNNV F+ + S S V + K D
Sbjct: 1298 SDFHKLLPVWLIWRIWKARNNVVFNKFRESP-------------SKTVLSAKAETHDWLN 1344
Query: 116 AILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTD-GSVLNNSGACGG-LFRDHLGTFL 173
A + P R + E + W+ P A +VK N D G + A GG + R+H GT +
Sbjct: 1345 ATQSHKKTPSPTRQIAE-NKIEWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNHYGTPI 1403
Query: 174 GAFVGNLGRCS-VFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILLR 232
L S + E + ++ + G+ +++E D + + + S L
Sbjct: 1404 SWGSMKLAHTSNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLINLINGISFHSSLAN 1463
Query: 233 NR-----WHNARTLNVQVISSHIFREGNVCVDRLANLG 265
+ W N + ++Q I R+GN LA G
Sbjct: 1464 HLEDISFWAN-KFASIQF--GFIRRKGNKLAHVLAKYG 1498
>At1g21330 hypothetical protein
Length = 211
Score = 45.8 bits (107), Expect = 3e-05
Identities = 32/99 (32%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWSWLCSQL---HVGLDFSSIKALM--SSLPRHCSSQ 56
C FC VET +HLF C++ +W+ L +L D+ I L+ +SL + C
Sbjct: 86 CVFCHAPVETRNHLFFACQYSADVWTGLSRKLLSQRFSTDWDMIIKLITDTSLGKGCLFL 145
Query: 57 VRDLYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRV 95
VR + VHSIW RN R A + S + R+
Sbjct: 146 VRYTFQLT----VHSIWKERNGHRHGEAPIPSSQLTRRL 180
>At3g42890 putative protein
Length = 534
Score = 45.4 bits (106), Expect = 4e-05
Identities = 28/90 (31%), Positives = 44/90 (48%), Gaps = 5/90 (5%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGL---DFSSIKALMSSLPRHCSSQVR 58
CSFC E +ET +H+F C + +WS L L L + S+ AL++ PR
Sbjct: 146 CSFCDEAMETREHIFFSCPYSSAVWSALTKDLLGQLFTTSWESMTALLTDPPRPKVHMFV 205
Query: 59 DLYVAAVVHMVHSIWWARNNVRFSSAKVSS 88
YV + +H++W RN R ++ + S
Sbjct: 206 LRYVFQL--SIHTLWRERNGRRHGASPIPS 233
>At2g40680 putative non-LTR retroelement reverse transcriptase
Length = 296
Score = 45.1 bits (105), Expect = 5e-05
Identities = 27/80 (33%), Positives = 38/80 (46%), Gaps = 1/80 (1%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWSWLCSQLHVGLDFSSIKALMSSLPRHCSSQVRDLY 61
C FC E VET +HLF +C + +W L L G SS +++ L + L
Sbjct: 170 CKFCAEPVETRNHLFFQCPYSTQVWEKLMKGLLQGHYTSSWDRIVTLLTSSSLGRKHLLL 229
Query: 62 VAAVVHM-VHSIWWARNNVR 80
+ + VH+IW RNN R
Sbjct: 230 LRYTFQIKVHTIWRERNNRR 249
>At1g47320 hypothetical protein
Length = 259
Score = 45.1 bits (105), Expect = 5e-05
Identities = 45/175 (25%), Positives = 74/175 (41%), Gaps = 32/175 (18%)
Query: 124 PPHRRSMREIVSVCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLG 181
P + ++ C + +K+NTDG+ N G GG+ +D+ G + G F N+G
Sbjct: 79 PDSHQCPKDYYYACLNNGESGCLKINTDGASRGNPGLATAGGVLQDNEGRWCGGFSLNIG 138
Query: 182 RCSVFDTEVSGFILVMEHAALHGWYNIW------LESDASSALMV------FKNPSLVPI 229
R S E+ G A +G Y W +E + S ++V + +
Sbjct: 139 RSSAPMAELWG--------AYYGLYLAWERKSSHIELEVDSEIVVGFLKTGISDHHPLSF 190
Query: 230 LLRNRWHNARTLNVQVISSHIFREGNVCVDRLANLGHSVVGEVWLSTLPSDFHQV 284
L+R H + + +V H++RE N D LAN ++ +LP FH V
Sbjct: 191 LVR-LCHGFISKDWRVRIFHVYREANRFADGLAN---------YVFSLPLGFHFV 235
>At3g09510 putative non-LTR reverse transcriptase
Length = 484
Score = 44.3 bits (103), Expect = 8e-05
Identities = 63/279 (22%), Positives = 107/279 (37%), Gaps = 35/279 (12%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLW-----SWLCSQLHVGLDFSSIKALMSSLPRHCSSQ 56
C C + E+ +H C F W S + +QL +I +++ + +
Sbjct: 200 CPRCHRENESINHALFTCPFATMAWRLSDSSLIRNQLMSNDFEENISNILNFVQ---DTT 256
Query: 57 VRDLYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAAD-- 114
+ D + V ++ IW ARNNV F+ + S S V + K D
Sbjct: 257 MSDFHKLLPVWLIWRIWKARNNVVFNKFRESP-------------SKTVLSAKAETHDWL 303
Query: 115 AAILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTD-GSVLNNSGACGG-LFRDHLGTF 172
A + P R + E + W+ P A +VK N D G + A GG + R+H GT
Sbjct: 304 NATQSHKKTPSPTRQIAE-NKIEWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNHYGTP 362
Query: 173 LGAFVGNLGRCS-VFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILL 231
+ L S + E + ++ + G+ +++E D + + + S L
Sbjct: 363 ISWGSMKLAHTSNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLINLINGISFHSSLA 422
Query: 232 RNR-----WHNARTLNVQVISSHIFREGNVCVDRLANLG 265
+ W N + ++Q I R+GN LA G
Sbjct: 423 NHLEDISFWAN-KFASIQF--GFIRRKGNKLAHVLAKYG 458
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 44.3 bits (103), Expect = 8e-05
Identities = 67/307 (21%), Positives = 115/307 (36%), Gaps = 49/307 (15%)
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLW-----SWLCSQLHVGLDFSSIKALMSSLPRHCSSQ 56
C C + E+ +H C F W S + +QL +I +++ + +
Sbjct: 1466 CPRCHRENESINHALFTCPFATMAWRLSDSSLIRNQLMSNDFEENISNILNFVQ---DTT 1522
Query: 57 VRDLYVAAVVHMVHSIWWARNNVRFSSAKVSSHAVQVRVHAFIGLSGAVSTGKCIAAD-- 114
+ D + V ++ IW ARNNV F+ + S S V + K D
Sbjct: 1523 MSDFHKLLPVWLIWRIWKARNNVVFNKFRESP-------------SKTVLSAKAETHDWL 1569
Query: 115 AAILDAFRIPPHRRSMREIVSVCWKPPSAPWVKVNTD-GSVLNNSGACGG-LFRDHLGTF 172
A + P R + E + W+ P A +VK N D G + A GG + R+H GT
Sbjct: 1570 NATQSHKKTPSPTRQIAE-NKIEWRNPPATYVKCNFDAGFDVQKLEATGGWIIRNHYGTP 1628
Query: 173 LGAFVGNLGRCS-VFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPSLVPILL 231
+ L S + E + ++ + G+ +++E D + + + S L
Sbjct: 1629 ISWGSMKLAHTSNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLINLINGISFHSSLA 1688
Query: 232 RNR-----WHNARTLNVQVISSHIFREGNVCVDRLANLGHSVVGEVWLSTLPSDFHQVFY 286
+ W N + ++Q I ++GN LA G + + FY
Sbjct: 1689 NHLEDISFWAN-KFASIQF--GFIRKKGNKLAHVLAKYGCT--------------YSTFY 1731
Query: 287 EDRCSMP 293
D S+P
Sbjct: 1732 SDSGSLP 1738
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.137 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,808,686
Number of Sequences: 26719
Number of extensions: 271316
Number of successful extensions: 843
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 733
Number of HSP's gapped (non-prelim): 99
length of query: 298
length of database: 11,318,596
effective HSP length: 99
effective length of query: 199
effective length of database: 8,673,415
effective search space: 1726009585
effective search space used: 1726009585
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146708.4


