

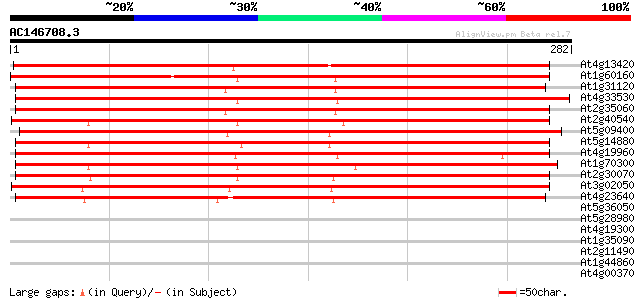
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.3 - phase: 0
(282 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g13420 K+ transporter HAK5 (HAK5) 367 e-102
At1g60160 unknown protein 288 2e-78
At1g31120 putative potassium transporter 288 3e-78
At4g33530 putative potassium transporter AtKT5p (AtKT5) 285 3e-77
At2g35060 putative potassium transporter 284 4e-77
At2g40540 putative potassium transporter AtKT2p (AtKT2) 282 1e-76
At5g09400 potassium transport protein-like 280 5e-76
At5g14880 HAK8 279 1e-75
At4g19960 potassium transporter-like protein 278 2e-75
At1g70300 high affinity potassium transporter AtHAK6 (At1g70300) 266 1e-71
At2g30070 high affinity K+ transporter (AtKUP1/AtKT1p) 256 1e-68
At3g02050 putative potassium transporter 247 4e-66
At4g23640 potassium transport like protein 236 1e-62
At5g36050 replication protein A1-like 35 0.059
At5g28980 replication protein - like 34 0.077
At4g19300 hypothtetical protein 34 0.077
At1g35090 unknown protein 33 0.17
At2g11490 pseudogene; similar to MURA transposase of maize Muta... 32 0.50
At1g44860 hypothetical protein 32 0.50
At4g00370 unknown protein 31 0.86
>At4g13420 K+ transporter HAK5 (HAK5)
Length = 785
Score = 367 bits (943), Expect = e-102
Identities = 177/273 (64%), Positives = 226/273 (81%), Gaps = 5/273 (1%)
Query: 3 TLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFI 62
T++LAFQSLGVVYGDIGTSPLYV ASTF GI+ DD++GVLSLI YTI + LLKYVFI
Sbjct: 58 TMSLAFQSLGVVYGDIGTSPLYVYASTFTDGINDKDDVVGVLSLIIYTITLVALLKYVFI 117
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSN----QQLKKKL 118
VL+ANDNG GGTFALYSL+CR+A + LIPNQ+PED+ELSNY LE P++ +K+KL
Sbjct: 118 VLQANDNGEGGTFALYSLICRYAKMGLIPNQEPEDVELSNYTLELPTTQLRRAHMIKEKL 177
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDYVVSITIAILVIL 178
ENS FA+++L +TI+GT+MVIGDG+ TP +SV+SAV+GI S LGQ+ VV +++AIL++L
Sbjct: 178 ENSKFAKIILFLVTIMGTSMVIGDGILTPSISVLSAVSGIKS-LGQNTVVGVSVAILIVL 236
Query: 179 FCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGKN 238
F QRFGT KVGFSFAPI+ +WF + G++N+FK+D+ VL A+NP YI+ YF+R G+
Sbjct: 237 FAFQRFGTDKVGFSFAPIILVWFTFLIGIGLFNLFKHDITVLKALNPLYIIYYFRRTGRQ 296
Query: 239 AWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
W+SLGGVFLCI+G EAMFADLGHF+VRA+Q+S
Sbjct: 297 GWISLGGVFLCITGTEAMFADLGHFSVRAVQIS 329
>At1g60160 unknown protein
Length = 826
Score = 288 bits (738), Expect = 2e-78
Identities = 145/279 (51%), Positives = 200/279 (70%), Gaps = 9/279 (3%)
Query: 1 MATLALAFQSLGVVYGDIGTSPLYVLASTFPK-GIDHTDDILGVLSLIYYTILALPLLKY 59
+ TL +AFQ+LGVVYGD+GTSPLYV + F K I D+LG LSL+ YTI +PL KY
Sbjct: 86 LTTLGIAFQTLGVVYGDMGTSPLYVFSDVFSKVPIRSEVDVLGALSLVIYTIAVIPLAKY 145
Query: 60 VFIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LK 115
VF+VLKANDNG G A L+ ++A V+ +PNQQP D ++S+++L+ P+ + +K
Sbjct: 146 VFVVLKANDNGEGNASAPMCLV-KYAKVNKLPNQQPADEQISSFRLKLPTPELERALGIK 204
Query: 116 KKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKL---GQDYVVSITI 172
+ LE + + LLL + ++GT+M+IGDG+ TP MSV+SA++G+ ++ G + +V +I
Sbjct: 205 EALETKGYLKTLLLLLVLMGTSMIIGDGILTPAMSVMSAMSGLQGEVKGFGTNALVMSSI 264
Query: 173 AILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYF 232
ILV LF QRFGT KVGF FAP+L +WF +GA GIYN+ KYD V+ A+NP YIV +F
Sbjct: 265 VILVALFSIQRFGTGKVGFLFAPVLALWFFSLGAIGIYNLLKYDFTVIRALNPFYIVLFF 324
Query: 233 QRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
+N K AW +LGG LCI+G EAMFADLGHF+VR+IQ++
Sbjct: 325 NKNSKQAWSALGGCVLCITGAEAMFADLGHFSVRSIQMA 363
>At1g31120 putative potassium transporter
Length = 796
Score = 288 bits (736), Expect = 3e-78
Identities = 146/272 (53%), Positives = 189/272 (68%), Gaps = 6/272 (2%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFIV 63
L L+FQSLGVVYGD+GTSPLYV +TFP+GI +DI+G LSLI Y++ +PLLKYVF+V
Sbjct: 57 LQLSFQSLGVVYGDLGTSPLYVFYNTFPRGIKDPEDIIGALSLIIYSLTLIPLLKYVFVV 116
Query: 64 LKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLET---PSSNQQLKKKLEN 120
KANDNG GGTFALYSLLCRHA VS IPNQ D EL+ Y T S + K+ LEN
Sbjct: 117 CKANDNGQGGTFALYSLLCRHAKVSTIPNQHRTDEELTTYSRTTFHERSFAAKTKRWLEN 176
Query: 121 SHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKL---GQDYVVSITIAILVI 177
+ LL + ++GT MVIGDG+ TP +SV+SA G+ L VV + + ILV
Sbjct: 177 GTSRKNALLILVLVGTCMVIGDGILTPAISVLSAAGGLRVNLPHINNGIVVVVAVVILVS 236
Query: 178 LFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGK 237
LF Q +GT +VG+ FAPI+ +WF+ I + G++N++K+D VL A +P YI YF+R G+
Sbjct: 237 LFSVQHYGTDRVGWLFAPIVFLWFLFIASIGMFNIWKHDPSVLKAFSPVYIFRYFKRGGQ 296
Query: 238 NAWMSLGGVFLCISGCEAMFADLGHFNVRAIQ 269
+ W SLGG+ L I+G EA+FADL HF V A+Q
Sbjct: 297 DRWTSLGGIMLSITGIEALFADLSHFPVSAVQ 328
>At4g33530 putative potassium transporter AtKT5p (AtKT5)
Length = 855
Score = 285 bits (728), Expect = 3e-77
Identities = 148/286 (51%), Positives = 194/286 (67%), Gaps = 8/286 (2%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKG-IDHTDDILGVLSLIYYTILALPLLKYVFI 62
L LA Q+LGVV+GDIGTSPLY F + I+ +DI+G LSL+ YT++ +PL+KYV
Sbjct: 106 LILALQTLGVVFGDIGTSPLYTFTVMFRRSPINDKEDIIGALSLVIYTLILIPLVKYVHF 165
Query: 63 VLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ----LKKKL 118
VL AND+G GGTFALYSL+CRHANVSLIPNQ P D +S + L+ PS + +K++L
Sbjct: 166 VLWANDDGEGGTFALYSLICRHANVSLIPNQLPSDARISGFGLKVPSPELERSLIIKERL 225
Query: 119 ENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLG---QDYVVSITIAIL 175
E S + LLL + + GT MVI D V TP MSV+SA+ G+ +G QD VV I+++ L
Sbjct: 226 EASMALKKLLLILVLAGTAMVIADAVVTPAMSVMSAIGGLKVGVGVIEQDQVVVISVSFL 285
Query: 176 VILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRN 235
VILF Q++GTSK+G P L +WF + GIYN+ KYD V A NP YI +F+RN
Sbjct: 286 VILFSVQKYGTSKLGLVLGPALLLWFFCLAGIGIYNLVKYDSSVFKAFNPAYIYFFFKRN 345
Query: 236 GKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSIIKLELTFVL 281
NAW +LGG LC +G EAMFADL +F+V +IQ++ I L L +L
Sbjct: 346 SVNAWYALGGCVLCATGSEAMFADLSYFSVHSIQLTFILLVLPCLL 391
>At2g35060 putative potassium transporter
Length = 792
Score = 284 bits (726), Expect = 4e-77
Identities = 144/274 (52%), Positives = 187/274 (67%), Gaps = 6/274 (2%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFIV 63
L L+FQSLGVVYGD+GTSPLYV +TFP GI +DI+G LSLI Y++ +PLLKYVF+V
Sbjct: 58 LQLSFQSLGVVYGDLGTSPLYVFYNTFPHGIKDPEDIIGALSLIIYSLTLIPLLKYVFVV 117
Query: 64 LKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLET---PSSNQQLKKKLEN 120
KANDNG GGTFALYSLLCRHA V I NQ D EL+ Y T S + K+ LE
Sbjct: 118 CKANDNGQGGTFALYSLLCRHAKVKTIQNQHRTDEELTTYSRTTFHEHSFAAKTKRWLEK 177
Query: 121 SHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKL---GQDYVVSITIAILVI 177
+ LL + ++GT MVIGDG+ TP +SV+SA G+ L VV + + ILV
Sbjct: 178 RTSRKTALLILVLVGTCMVIGDGILTPAISVLSAAGGLRVNLPHISNGVVVFVAVVILVS 237
Query: 178 LFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGK 237
LF Q +GT +VG+ FAPI+ +WF+ I + G+YN++K+D VL A +P YI YF+R G+
Sbjct: 238 LFSVQHYGTDRVGWLFAPIVFLWFLSIASIGMYNIWKHDTSVLKAFSPVYIYRYFKRGGR 297
Query: 238 NAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
+ W SLGG+ L I+G EA+FADL HF V A+Q++
Sbjct: 298 DRWTSLGGIMLSITGIEALFADLSHFPVSAVQIA 331
>At2g40540 putative potassium transporter AtKT2p (AtKT2)
Length = 794
Score = 282 bits (722), Expect = 1e-76
Identities = 140/280 (50%), Positives = 192/280 (68%), Gaps = 10/280 (3%)
Query: 2 ATLALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTD---DILGVLSLIYYTILALPLLK 58
+ L LA+QSLGVVYGD+ SPLYV STF + I H++ +I GV+S +++T+ +PLLK
Sbjct: 21 SVLLLAYQSLGVVYGDLSISPLYVFKSTFAEDIQHSETNEEIYGVMSFVFWTLTLVPLLK 80
Query: 59 YVFIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ---LK 115
YVFIVL+A+DNG GGTFALYSL+CRH VSL+PN+Q D LS YKLE P +K
Sbjct: 81 YVFIVLRADDNGEGGTFALYSLICRHVKVSLLPNRQVSDEALSTYKLEHPPEKNHDSCVK 140
Query: 116 KKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDY----VVSIT 171
+ LE + LL + +LGT MVIGDG+ TP +SV SAV+G+ + +++ V+ IT
Sbjct: 141 RYLEKHKWLHTALLLLVLLGTCMVIGDGLLTPAISVFSAVSGLELNMSKEHHQYAVIPIT 200
Query: 172 IAILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDY 231
ILV LF Q FGT +VGF FAPI+ W + I G+YN+ +++ + A++P Y+ +
Sbjct: 201 CFILVCLFSLQHFGTHRVGFVFAPIVLTWLLCISGIGLYNIIQWNPHIYKALSPTYMFMF 260
Query: 232 FQRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
++ + WMSLGG+ LCI+G EAMFADLGHFN AIQ++
Sbjct: 261 LRKTRVSGWMSLGGILLCITGAEAMFADLGHFNYAAIQIA 300
>At5g09400 potassium transport protein-like
Length = 858
Score = 280 bits (717), Expect = 5e-76
Identities = 143/280 (51%), Positives = 195/280 (69%), Gaps = 8/280 (2%)
Query: 6 LAFQSLGVVYGDIGTSPLYVLASTFPKG-IDHTDDILGVLSLIYYTILALPLLKYVFIVL 64
LAFQ+LGVV+GD+GTSPLY + F K + +D++G LSL+ YT+L +PL+KYV +VL
Sbjct: 107 LAFQTLGVVFGDVGTSPLYTFSVMFSKSPVQEKEDVIGALSLVLYTLLLVPLIKYVLVVL 166
Query: 65 KANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETP----SSNQQLKKKLEN 120
AND+G GGTFALYSL+ RHA +SLIPNQ D +S+++L+ P + +LK+KLEN
Sbjct: 167 WANDDGEGGTFALYSLISRHAKISLIPNQLRSDTRISSFRLKVPCPELERSLKLKEKLEN 226
Query: 121 SHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGIS---SKLGQDYVVSITIAILVI 177
S + +LL + + GT+MVI DGV TP MSV+SAV G+ + QD VV I++A LVI
Sbjct: 227 SLILKKILLVLVLAGTSMVIADGVVTPAMSVMSAVGGLKVGVDVVEQDQVVMISVAFLVI 286
Query: 178 LFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQRNGK 237
LF Q++GTSK+G P L IWF + GIYN+ KYD V A NP +I +F+RN
Sbjct: 287 LFSLQKYGTSKMGLVVGPALLIWFCSLAGIGIYNLIKYDSSVYRAFNPVHIYYFFKRNSI 346
Query: 238 NAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSIIKLEL 277
NAW +LGG LC +G EA+FADL +F+VR++Q++ + L L
Sbjct: 347 NAWYALGGCILCATGSEALFADLCYFSVRSVQLTFVCLVL 386
>At5g14880 HAK8
Length = 781
Score = 279 bits (713), Expect = 1e-75
Identities = 144/282 (51%), Positives = 193/282 (68%), Gaps = 14/282 (4%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTD---DILGVLSLIYYTILALPLLKYV 60
L LA+QSLGVVYGD+ TSPLYV STF + I H++ +I GVLSLI++T+ +PL+KYV
Sbjct: 21 LTLAYQSLGVVYGDLATSPLYVYKSTFAEDITHSETNEEIFGVLSLIFWTLTLIPLVKYV 80
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQLK----- 115
FIVL+A+DNG GGTFALYSLLCRHA +S +PN Q D +LS YK + + +LK
Sbjct: 81 FIVLRADDNGEGGTFALYSLLCRHARISSLPNFQLADEDLSEYKKNSGENPMRLKVPGWS 140
Query: 116 --KKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGIS---SKLGQDYV-VS 169
LE F + +LL + ++GT MVIGDGV TP +SV SAV+G+ SK YV V
Sbjct: 141 LKNTLEKHKFLQNMLLVLALIGTCMVIGDGVLTPAISVFSAVSGLELSMSKQQHQYVEVP 200
Query: 170 ITIAILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIV 229
+ AIL++LF Q +GT ++GF FAPI+ W + I G+YN+F ++ V A++P YI
Sbjct: 201 VVCAILILLFSLQHYGTHRLGFVFAPIVLAWLLCISTIGVYNIFHWNPHVYKALSPYYIY 260
Query: 230 DYFQRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
+ ++ K WMSLGG+ LCI+G EAMFADLGHF +IQ++
Sbjct: 261 KFLKKTRKRGWMSLGGILLCITGSEAMFADLGHFTQLSIQIA 302
>At4g19960 potassium transporter-like protein
Length = 842
Score = 278 bits (711), Expect = 2e-75
Identities = 143/293 (48%), Positives = 192/293 (64%), Gaps = 25/293 (8%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTDDILGVLSLIYYTILALPLLKYVFIV 63
L L+FQSLG+VYGD+GTSPLYV +TFP GID ++D++G LSLI Y++L +PL+KYVFIV
Sbjct: 58 LRLSFQSLGIVYGDLGTSPLYVFYNTFPDGIDDSEDVIGALSLIIYSLLLIPLIKYVFIV 117
Query: 64 LKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQ---QLKKKLEN 120
KANDNG GGT A+YSLLCRHA V LIPNQ D +L+ Y + + KK LE
Sbjct: 118 CKANDNGQGGTLAIYSLLCRHAKVKLIPNQHRSDEDLTTYSRTVSAEGSFAAKTKKWLEG 177
Query: 121 SHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLG-----------QDYVVS 169
+ + LL + +LGT M+IGDG+ TP +S + + K+ D VV
Sbjct: 178 KEWRKRALLVVVLLGTCMMIGDGILTPAISATGGIKVNNPKMSGGNIHFTLFSTSDIVVL 237
Query: 170 ITIAILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIV 229
+ I IL+ LF Q +GT KVG+ FAPI+ IWF+ IGATG+YN+ KYD VL A +P YI
Sbjct: 238 VAIVILIGLFSMQHYGTDKVGWLFAPIVLIWFLFIGATGMYNICKYDTSVLKAFSPTYIY 297
Query: 230 DYFQRNGKNAWMSLGGV-----------FLCISGCEAMFADLGHFNVRAIQVS 271
YF+R G++ W+SLGG+ FL I+G EA++AD+ +F + AIQ++
Sbjct: 298 LYFKRRGRDGWISLGGILLSITAISKCYFLNIAGTEALYADIAYFPLLAIQLA 350
>At1g70300 high affinity potassium transporter AtHAK6 (At1g70300)
Length = 782
Score = 266 bits (679), Expect = 1e-71
Identities = 136/287 (47%), Positives = 191/287 (66%), Gaps = 15/287 (5%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDHTD---DILGVLSLIYYTILALPLLKYV 60
L LA+QSLGVVYGD+ SPLYV STF + I H++ +I GVLS I++TI +PLLKYV
Sbjct: 20 LTLAYQSLGVVYGDLSISPLYVYKSTFAEDIHHSESNEEIFGVLSFIFWTITLVPLLKYV 79
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ------- 113
FIVL+A+DNG GGTFALYSLLCRHA V+ +P+ Q D +L YK ++ S+
Sbjct: 80 FIVLRADDNGEGGTFALYSLLCRHARVNSLPSCQLADEQLIEYKTDSIGSSSMPQSGFAA 139
Query: 114 -LKKKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSKLGQDYVVSITI 172
LK LE + +LL + ++GT MVIGDGV TP +SV SAV+G+ + +++ I +
Sbjct: 140 SLKSTLEKHGVLQKILLVLALIGTCMVIGDGVLTPAISVFSAVSGVELSMSKEHHKYIEL 199
Query: 173 ----AILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYI 228
IL+ LF Q +GT +VGF FAP++ +W + I A G+YN+F ++ V A++P Y+
Sbjct: 200 PAACVILIGLFALQHYGTHRVGFLFAPVILLWLMCISAIGVYNIFHWNPHVYQALSPYYM 259
Query: 229 VDYFQRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSIIKL 275
+ ++ WMSLGG+ LCI+G EAMFADLGHF+ +I+++ L
Sbjct: 260 YKFLKKTQSRGWMSLGGILLCITGSEAMFADLGHFSQLSIKIAFTSL 306
>At2g30070 high affinity K+ transporter (AtKUP1/AtKT1p)
Length = 712
Score = 256 bits (654), Expect = 1e-68
Identities = 135/277 (48%), Positives = 182/277 (64%), Gaps = 9/277 (3%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGID-HTDD--ILGVLSLIYYTILALPLLKYV 60
L LA+QSLGV+YGD+ TSPLYV +TF + H DD I GV S I++T + L KYV
Sbjct: 26 LTLAYQSLGVIYGDLSTSPLYVYKTTFSGKLSLHEDDEEIFGVFSFIFWTFTLIALFKYV 85
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPSSNQQ---LKKK 117
FIVL A+DNG GGTFALYSLLCR+A +S++PN Q D +LS Y +P +Q +K
Sbjct: 86 FIVLSADDNGEGGTFALYSLLCRYAKLSILPNHQEMDEKLSTYATGSPGETRQSAAVKSF 145
Query: 118 LENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK---LGQDYVVSITIAI 174
E ++ LL +LGT M IGD V TP +SV+SAV+G+ K L ++YVV I I
Sbjct: 146 FEKHPKSQKCLLLFVLLGTCMAIGDSVLTPTISVLSAVSGVKLKIPNLHENYVVIIACII 205
Query: 175 LVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYFQR 234
LV +F QR+GT +V F FAPI T W + I + G+YN K++ R++ A++P Y+ + +
Sbjct: 206 LVAIFSVQRYGTHRVAFIFAPISTAWLLSISSIGVYNTIKWNPRIVSALSPVYMYKFLRS 265
Query: 235 NGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
G W+SLGGV L I+G E MFADLGHF+ +I+V+
Sbjct: 266 TGVEGWVSLGGVVLSITGVETMFADLGHFSSLSIKVA 302
>At3g02050 putative potassium transporter
Length = 789
Score = 247 bits (631), Expect = 4e-66
Identities = 128/279 (45%), Positives = 185/279 (65%), Gaps = 9/279 (3%)
Query: 2 ATLALAFQSLGVVYGDIGTSPLYVLASTFPKGID--HTDD-ILGVLSLIYYTILALPLLK 58
+ L LA+QS GVVYGD+ TSPLYV STF + H +D + G SLI++T+ +PLLK
Sbjct: 24 SNLILAYQSFGVVYGDLSTSPLYVFPSTFIGKLHKHHNEDAVFGAFSLIFWTLTLIPLLK 83
Query: 59 YVFIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNYKLETPS---SNQQLK 115
Y+ ++L A+DNG GGTFALYSLLCRHA +SL+PNQQ D ELS YK + ++ +
Sbjct: 84 YLLVLLSADDNGEGGTFALYSLLCRHAKLSLLPNQQAADEELSAYKFGPSTDTVTSSPFR 143
Query: 116 KKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISS---KLGQDYVVSITI 172
LE R LL + + G MVIGDGV TP +SV+S+++G+ + + ++ +
Sbjct: 144 TFLEKHKRLRTALLLVVLFGAAMVIGDGVLTPALSVLSSLSGLQATEKNVTDGELLVLAC 203
Query: 173 AILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYF 232
ILV LF Q GT +V F FAPI+ IW I I G+YN+ +++ +++ A++P YI+ +F
Sbjct: 204 VILVGLFALQHCGTHRVAFMFAPIVIIWLISIFFIGLYNIIRWNPKIIHAVSPLYIIKFF 263
Query: 233 QRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVS 271
+ G++ W+SLGGV L ++G EAMFA+LGHF +I+V+
Sbjct: 264 RVTGQDGWISLGGVLLSVTGTEAMFANLGHFTSVSIRVA 302
>At4g23640 potassium transport like protein
Length = 775
Score = 236 bits (601), Expect = 1e-62
Identities = 127/277 (45%), Positives = 176/277 (62%), Gaps = 13/277 (4%)
Query: 4 LALAFQSLGVVYGDIGTSPLYVLASTFPKGIDH---TDDILGVLSLIYYTILALPLLKYV 60
L LA+QS G+V+GD+ SPLYV TF G+ H D I G SLI++TI L L+KY+
Sbjct: 12 LLLAYQSFGLVFGDLSISPLYVYKCTFYGGLRHHQTEDTIFGAFSLIFWTITLLSLIKYM 71
Query: 61 FIVLKANDNGNGGTFALYSLLCRHANVSLIPNQQPEDMELSNY------KLETPSSNQQL 114
VL A+DNG GG FALY+LLCRHA SL+PNQQ D E+S Y PSS
Sbjct: 72 VFVLSADDNGEGGIFALYALLCRHARFSLLPNQQAADEEISTYYGPGDASRNLPSS--AF 129
Query: 115 KKKLENSHFARVLLLFMTILGTTMVIGDGVFTPPMSVISAVNGISSK--LGQDYVVSITI 172
K +E + ++ LL + ++GT+MVI GV TP +SV S+++G+ +K L VV I
Sbjct: 130 KSLIERNKRSKTALLVLVLVGTSMVITIGVLTPAISVSSSIDGLVAKTSLKHSTVVMIAC 189
Query: 173 AILVILFCAQRFGTSKVGFSFAPILTIWFILIGATGIYNVFKYDVRVLLAINPKYIVDYF 232
A+LV LF Q GT+KV F FAPI+ +W ++I G+YN+ ++ V A++P YI +F
Sbjct: 190 ALLVGLFVLQHRGTNKVAFLFAPIMILWLLIIATAGVYNIVTWNPSVYKALSPYYIYVFF 249
Query: 233 QRNGKNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQ 269
+ G + W+SLGG+ LCI+G EA+FA+LG F +I+
Sbjct: 250 RDTGIDGWLSLGGILLCITGTEAIFAELGQFTATSIR 286
>At5g36050 replication protein A1-like
Length = 252
Score = 34.7 bits (78), Expect = 0.059
Identities = 24/97 (24%), Positives = 45/97 (45%), Gaps = 15/97 (15%)
Query: 191 FSFAPILTIWFILIGATGIYNVF----KYDVRVLLA----------INPKYIVDYFQRNG 236
+ +P++T W I + IY F +++R++LA I P++ YF R
Sbjct: 8 YELSPMVTGWTICVMVLRIYKKFLNPNAFELRLVLADEWGTQIEATIGPRFSAFYFDRIK 67
Query: 237 KNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSII 273
+N W ++ FL CEA+ F++ + +I+
Sbjct: 68 ENQWKTI-TTFLVRPNCEAIKTTPHQFHILFMDHTIV 103
>At5g28980 replication protein - like
Length = 354
Score = 34.3 bits (77), Expect = 0.077
Identities = 24/97 (24%), Positives = 44/97 (44%), Gaps = 15/97 (15%)
Query: 191 FSFAPILTIWFILIGATGIYNVF----KYDVRVLLA----------INPKYIVDYFQRNG 236
+ +P++T W I + IY F +++R++LA I P++ YF R
Sbjct: 8 YELSPMVTGWTICVMVLRIYKKFLNLNAFELRLVLADEWGTQIEATIGPRFSAFYFDRIK 67
Query: 237 KNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSII 273
+N W ++ FL CEA+ F + + +I+
Sbjct: 68 ENQWKTI-TTFLVRPNCEAIKTTPHQFRILFMDHTIV 103
>At4g19300 hypothtetical protein
Length = 252
Score = 34.3 bits (77), Expect = 0.077
Identities = 24/97 (24%), Positives = 44/97 (44%), Gaps = 15/97 (15%)
Query: 191 FSFAPILTIWFILIGATGIYNVF----KYDVRVLLA----------INPKYIVDYFQRNG 236
+ +P++T W I + IY F +++R++LA I P++ YF R
Sbjct: 8 YELSPMVTGWTICVMVLRIYKKFLNPNAFELRLVLADEWGTQIEATIGPRFSAFYFDRIK 67
Query: 237 KNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSII 273
+N W ++ FL CEA+ F + + +I+
Sbjct: 68 ENQWKTI-TTFLVRPNCEAIKTTPHQFRILFMDHTIV 103
>At1g35090 unknown protein
Length = 244
Score = 33.1 bits (74), Expect = 0.17
Identities = 24/97 (24%), Positives = 44/97 (44%), Gaps = 15/97 (15%)
Query: 191 FSFAPILTIWFILIGATGIY----NVFKYDVRVLLA----------INPKYIVDYFQRNG 236
+ +P++T W I + IY N +++R++LA I P++ YF R
Sbjct: 8 YELSPMVTGWTICVMVLRIYKKILNPNAFELRLVLADEWGTQIEATIGPRFSAFYFDRIK 67
Query: 237 KNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSII 273
+N W ++ FL CEA+ F + + +I+
Sbjct: 68 ENQWKTI-TTFLVRPNCEAIKTTPHQFRILFMDHTIV 103
>At2g11490 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 240
Score = 31.6 bits (70), Expect = 0.50
Identities = 20/97 (20%), Positives = 44/97 (44%), Gaps = 15/97 (15%)
Query: 191 FSFAPILTIWFILIGATGIY----NVFKYDVRVLLA----------INPKYIVDYFQRNG 236
+ +P++T W I + +Y N +++R+LL I P++ Y+ R
Sbjct: 8 YELSPLVTGWAICVMVVRVYKKILNHDAFELRLLLVDKLGTQIEATIGPRFSAFYYDRIK 67
Query: 237 KNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSII 273
+N W ++ F+ CEA+ F + ++ +++
Sbjct: 68 ENQWKTI-TTFIVRPNCEAIRTTPHEFRILFMEQTVV 103
>At1g44860 hypothetical protein
Length = 244
Score = 31.6 bits (70), Expect = 0.50
Identities = 24/97 (24%), Positives = 41/97 (41%), Gaps = 15/97 (15%)
Query: 191 FSFAPILTIWFILIGATGIYNVF----KYDVRVLLA----------INPKYIVDYFQRNG 236
+ +P++T W I + IY F +++R++LA I P + YF R
Sbjct: 8 YELSPMVTGWTICVIVLRIYKKFLNTNAFELRLVLADEWGTQIEATIGPHFSAFYFDRIQ 67
Query: 237 KNAWMSLGGVFLCISGCEAMFADLGHFNVRAIQVSII 273
+N W + FL CE + F + + +II
Sbjct: 68 ENQWKKI-STFLVRPNCEPIRTTPHEFRILFMDHTII 103
>At4g00370 unknown protein
Length = 541
Score = 30.8 bits (68), Expect = 0.86
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 11/89 (12%)
Query: 124 ARVLLLFMTILGTTMVIGDGVFTPPMS--------VISAVNGISSKLGQDYVVSIT-IAI 174
AR+ L F+ ++ M IG+GV P M+ V ++ Y+ S+T +A
Sbjct: 219 ARLGLPFLLVVRAFMGIGEGVAMPAMNNMLSKWIPVSERSRSLALVYSGMYLGSVTGLAF 278
Query: 175 LVILFCAQRFGTSKVGFSFAPILTIWFIL 203
+L +FG V +SF + +IWF+L
Sbjct: 279 SPMLIT--KFGWPSVFYSFGSLGSIWFLL 305
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.143 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,005,422
Number of Sequences: 26719
Number of extensions: 248749
Number of successful extensions: 807
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 752
Number of HSP's gapped (non-prelim): 28
length of query: 282
length of database: 11,318,596
effective HSP length: 98
effective length of query: 184
effective length of database: 8,700,134
effective search space: 1600824656
effective search space used: 1600824656
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146708.3


