

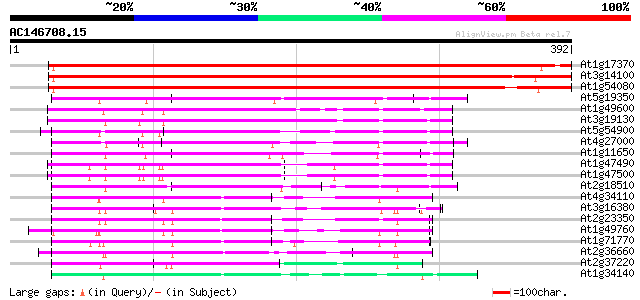
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.15 - phase: 2
(392 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g17370 oligouridylate binding protein, putative 611 e-175
At3g14100 oligouridylate binding protein, putative 599 e-172
At1g54080 unknown protein 568 e-162
At5g19350 DNA binding protein ACBF - like 134 9e-32
At1g49600 DNA binding protein ACBF, putative 131 7e-31
At3g19130 nuclear acid binding protein, putative 128 6e-30
At5g54900 unknown protein 123 2e-28
At4g27000 putative DNA binding protein 123 2e-28
At1g11650 putative DNA binding protein 117 8e-27
At1g47490 unknown protein 115 5e-26
At1g47500 unknown protein 108 4e-24
At2g18510 putative spliceosome associated protein 105 6e-23
At4g34110 poly(A)-binding protein 98 9e-21
At3g16380 putative poly(A) binding protein 98 9e-21
At2g23350 putative poly(A) binding protein 93 3e-19
At1g49760 Putative Poly-A Binding Protein (F14J22.3) 92 6e-19
At1g71770 polyadenylate-binding protein 5 88 7e-18
At2g36660 putative poly(A) binding protein 82 7e-16
At2g37220 putative RNA-binding protein 77 1e-14
At1g34140 putative protein 77 2e-14
>At1g17370 oligouridylate binding protein, putative
Length = 419
Score = 611 bits (1576), Expect = e-175
Identities = 296/370 (80%), Positives = 328/370 (88%), Gaps = 8/370 (2%)
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+H QVTEPLLQEVFAGTGPVE CKL RKEKSSYGF+HYFDRRSA LAIL+LN
Sbjct: 53 CRSVYVGNIHIQVTEPLLQEVFAGTGPVESCKLIRKEKSSYGFVHYFDRRSAGLAILSLN 112
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
GRHLFGQPIKVNWAYASGQREDTS H+NIFVGDLSPEVTDA LF CFSVY +CSDARVMW
Sbjct: 113 GRHLFGQPIKVNWAYASGQREDTSSHFNIFVGDLSPEVTDAMLFTCFSVYPTCSDARVMW 172
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
DQKTGRSRGFGFVSFR+QQDAQ+AI+++TGKWLGSRQIRCNWATKGA E+KQ+SDSKS
Sbjct: 173 DQKTGRSRGFGFVSFRNQQDAQTAIDEITGKWLGSRQIRCNWATKGATSGEDKQSSDSKS 232
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
VVELT+G SEDGK+ ++ + PENN QYTTVYVGNL E +Q+DLHRHFH+LGAGVIEEVR
Sbjct: 233 VVELTSGVSEDGKDTTNGEAPENNAQYTTVYVGNLAPEVSQVDLHRHFHSLGAGVIEEVR 292
Query: 266 VQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPPPAAA 325
VQRDKGFGFVRYSTH EAALAIQMGN SYL G+ +KCSWGSKPTP GTASNPLPPPA A
Sbjct: 293 VQRDKGFGFVRYSTHVEAALAIQMGNTHSYLSGRQMKCSWGSKPTPAGTASNPLPPPAPA 352
Query: 326 PLPGLSATDILAYERQLAMSKMGGVHAALMHPQAQHPLKQAAIGA---SQAIYDGGFQNV 382
P+PG SA+D+LAYERQLAMSKM G++ + HPQ QH KQAA+GA +QAIYDGG+QN
Sbjct: 353 PIPGFSASDLLAYERQLAMSKMAGMNPMMHHPQGQHAFKQAAMGATGSNQAIYDGGYQN- 411
Query: 383 AAAQQMMYYQ 392
AQQ+MYYQ
Sbjct: 412 --AQQLMYYQ 419
>At3g14100 oligouridylate binding protein, putative
Length = 427
Score = 599 bits (1544), Expect = e-172
Identities = 293/371 (78%), Positives = 324/371 (86%), Gaps = 7/371 (1%)
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+HTQVTEPLLQE+F TGPVE KL RK+KSSYGF+HYFDRRSAALAIL+LN
Sbjct: 58 CRSVYVGNIHTQVTEPLLQEIFTSTGPVESSKLIRKDKSSYGFVHYFDRRSAALAILSLN 117
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
GRHLFGQPIKVNWAYA+GQREDTS H+NIFVGDLSPEVTDATL+ FSV+ SCSDARVMW
Sbjct: 118 GRHLFGQPIKVNWAYATGQREDTSSHFNIFVGDLSPEVTDATLYQSFSVFSSCSDARVMW 177
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
DQKTGRSRGFGFVSFR+QQDAQ+AIN++ GKWL SRQIRCNWATKGA ++K +SD KS
Sbjct: 178 DQKTGRSRGFGFVSFRNQQDAQTAINEMNGKWLSSRQIRCNWATKGATSGDDKLSSDGKS 237
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
VVELT GSSEDGKE + + PENN Q+TTVYVGNL E TQLDLHR+FHALGAGVIEEVR
Sbjct: 238 VVELTTGSSEDGKETLNEETPENNSQFTTVYVGNLAPEVTQLDLHRYFHALGAGVIEEVR 297
Query: 266 VQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPPPAAA 325
VQRDKGFGFVRY+TH EAALAIQMGN Q YL + IKCSWG+KPTPPGTASNPLPPPA A
Sbjct: 298 VQRDKGFGFVRYNTHPEAALAIQMGNTQPYLFNRQIKCSWGNKPTPPGTASNPLPPPAPA 357
Query: 326 PLPGLSATDILAYERQLAMSKMGGVHAALMHPQAQHPLKQA----AIGASQAIYDGGFQN 381
P+PGLSA D+L YERQLA+SKM V+ ALMH Q QHPL+QA A GA+ A+YDGGFQN
Sbjct: 358 PVPGLSAADLLNYERQLALSKMASVN-ALMHQQGQHPLRQAHGINAAGATAAMYDGGFQN 416
Query: 382 VAAAQQMMYYQ 392
VAAAQQ+MYYQ
Sbjct: 417 VAAAQQLMYYQ 427
>At1g54080 unknown protein
Length = 426
Score = 568 bits (1465), Expect = e-162
Identities = 277/373 (74%), Positives = 317/373 (84%), Gaps = 16/373 (4%)
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR Y GN+HTQVTE LLQE+FA TGP+E CKL RK+KSSYGF+HYFDRR A++AI+TLN
Sbjct: 62 CRSVYAGNIHTQVTEILLQEIFASTGPIESCKLIRKDKSSYGFVHYFDRRCASMAIMTLN 121
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
GRH+FGQP+KVNWAYA+GQREDTS H+NIFVGDLSPEVTDA LF FS + SCSDARVMW
Sbjct: 122 GRHIFGQPMKVNWAYATGQREDTSSHFNIFVGDLSPEVTDAALFDSFSAFNSCSDARVMW 181
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
DQKTGRSRGFGFVSFR+QQDAQ+AIN++ GKW+ SRQIRCNWATKGA E+K +SD KS
Sbjct: 182 DQKTGRSRGFGFVSFRNQQDAQTAINEMNGKWVSSRQIRCNWATKGATFGEDKHSSDGKS 241
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
VVELTNGSSEDG+E+S+ D PENNPQ+TTVYVGNL E TQLDLHR F+ LGAGVIEEVR
Sbjct: 242 VVELTNGSSEDGRELSNEDAPENNPQFTTVYVGNLSPEVTQLDLHRLFYTLGAGVIEEVR 301
Query: 266 VQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPPPAAA 325
VQRDKGFGFVRY+TH EAALAIQMGNAQ +L + I+CSWG+KPTP GTASNPLPPPA A
Sbjct: 302 VQRDKGFGFVRYNTHDEAALAIQMGNAQPFLFSRQIRCSWGNKPTPSGTASNPLPPPAPA 361
Query: 326 PLPGLSATDILAYERQLAMSKMGGVHAALMHPQAQHPLKQAAI-----GASQAIYDGGFQ 380
+P LSA D+LAYERQLA++K MHPQAQH L+QA + G + A+YDGG+Q
Sbjct: 362 SVPSLSAMDLLAYERQLALAK--------MHPQAQHSLRQAGLGVNVAGGTAAMYDGGYQ 413
Query: 381 NVAAA-QQMMYYQ 392
NVAAA QQ+MYYQ
Sbjct: 414 NVAAAHQQLMYYQ 426
>At5g19350 DNA binding protein ACBF - like
Length = 425
Score = 134 bits (337), Expect = 9e-32
Identities = 90/299 (30%), Positives = 150/299 (50%), Gaps = 12/299 (4%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK----EKSSYGFIHYFDRRSAALAILTLN 85
++G++ V E L F+ TG + K+ R + YGFI + +A + T N
Sbjct: 27 WIGDLQYWVDENYLTSCFSQTGELVSVKVIRNKITGQPEGYGFIEFISHAAAERTLQTYN 86
Query: 86 GRHLFGQPI--KVNWA-YASGQREDTSGHYNIFVGDLSPEVTDATLFACFSV-YQSCSDA 141
G + G + ++NWA + SGQ+ D ++IFVGDL+P+VTD L F V Y S A
Sbjct: 87 GTQMPGTELTFRLNWASFGSGQKVDAGPDHSIFVGDLAPDVTDYLLQETFRVHYSSVRGA 146
Query: 142 RVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNS 201
+V+ D TGRS+G+GFV F + + A+ ++ G + +R +R + AT + +Q
Sbjct: 147 KVVTDPSTGRSKGYGFVKFAEESERNRAMAEMNGLYCSTRPMRISAATP-KKNVGVQQQY 205
Query: 202 DSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVI 261
+K+V +T S+ + PE++ TT+ V NL T+ +L + F LG +
Sbjct: 206 VTKAVYPVTVPSAVAAPVQAYVAPPESDVTCTTISVANLDQNVTEEELKKAFSQLGEVIY 265
Query: 262 EEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLP 320
V++ KG+G+V++ T A A+Q Q + + ++ SW P G + P
Sbjct: 266 --VKIPATKGYGYVQFKTRPSAEEAVQRMQGQ-VIGQQAVRISWSKNPGQDGWVTQADP 321
Score = 55.1 bits (131), Expect = 7e-08
Identities = 43/176 (24%), Positives = 76/176 (42%), Gaps = 38/176 (21%)
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+++GDL V + L +CFS +V+ ++ TG+ G+GF+ F S A+ +
Sbjct: 26 LWIGDLQYWVDENYLTSCFSQTGELVSVKVIRNKITGQPEGYGFIEFISHAAAERTLQTY 85
Query: 174 TGKWLGSRQI--RCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQ 231
G + ++ R NWA+ G+G Q D+ P
Sbjct: 86 NGTQMPGTELTFRLNWASFGSG-----QKVDA-------------------------GPD 115
Query: 232 YTTVYVGNLGSEATQLDLHRHFH-----ALGAGVIEEVRVQRDKGFGFVRYSTHAE 282
+ +++VG+L + T L F GA V+ + R KG+GFV+++ +E
Sbjct: 116 H-SIFVGDLAPDVTDYLLQETFRVHYSSVRGAKVVTDPSTGRSKGYGFVKFAEESE 170
>At1g49600 DNA binding protein ACBF, putative
Length = 445
Score = 131 bits (329), Expect = 7e-31
Identities = 92/294 (31%), Positives = 152/294 (51%), Gaps = 23/294 (7%)
Query: 27 KCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS----SYGFIHYFDRRSAALAIL 82
K +VG++ + E L F+ T V K+ R +++ YGF+ + R +A A+
Sbjct: 119 KTLWVGDLLHWMDETYLHTCFSHTNEVSSVKVIRNKQTCQSEGYGFVEFLSRSAAEEALQ 178
Query: 83 TLNGRHLFG--QPIKVNWA-YASGQRE--DTSGHYNIFVGDLSPEVTDATLFACFSV-YQ 136
+ +G + QP ++NWA +++G++ + +IFVGDL+P+V+DA L F+ Y
Sbjct: 179 SFSGVTMPNAEQPFRLNWASFSTGEKRASENGPDLSIFVGDLAPDVSDAVLLETFAGRYP 238
Query: 137 SCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIE 196
S A+V+ D TGRS+G+GFV F + + A+ ++ G + SRQ+R AT
Sbjct: 239 SVKGAKVVIDSNTGRSKGYGFVRFGDENERSRAMTEMNGAFCSSRQMRVGIATPKRAAAY 298
Query: 197 EKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHAL 256
+QN + L G +G S +D NN +T++VG L ++ T+ DL + F
Sbjct: 299 GQQN--GSQALTLAGGHGGNG---SMSDGESNN---STIFVGGLDADVTEEDLMQPFSDF 350
Query: 257 GAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGK-IIKCSWGSKP 309
G + V++ KG GFV+++ A AI GN + GK ++ SWG P
Sbjct: 351 GE--VVSVKIPVGKGCGFVQFANRQSAEEAI--GNLNGTVIGKNTVRLSWGRSP 400
>At3g19130 nuclear acid binding protein, putative
Length = 435
Score = 128 bits (321), Expect = 6e-30
Identities = 86/293 (29%), Positives = 147/293 (49%), Gaps = 16/293 (5%)
Query: 27 KCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAIL 82
K +VG++ + E L F+ TG V K+ R + +S YGF+ + R +A +
Sbjct: 108 KTLWVGDLLHWMDETYLHSCFSHTGEVSSVKVIRNKLTSQSEGYGFVEFLSRAAAEEVLQ 167
Query: 83 TLNGRHL--FGQPIKVNWA-YASGQRE--DTSGHYNIFVGDLSPEVTDATLFACFSV-YQ 136
+G + QP ++NWA +++G++ + ++FVGDLSP+VTD L FS Y
Sbjct: 168 NYSGSVMPNSDQPFRINWASFSTGEKRAVENGPDLSVFVGDLSPDVTDVLLHETFSDRYP 227
Query: 137 SCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIE 196
S A+V+ D TGRS+G+GFV F + + A+ ++ G + +RQ+R AT
Sbjct: 228 SVKSAKVVIDSNTGRSKGYGFVRFGDENERSRALTEMNGAYCSNRQMRVGIATPKRAIAN 287
Query: 197 EKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHAL 256
++Q+S ++ +GS+ S +D N T++VG + + DL + F
Sbjct: 288 QQQHSSQAVILAGGHGSNGSMGYGSQSDGESTN---ATIFVGGIDPDVIDEDLRQPFSQF 344
Query: 257 GAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKP 309
G + V++ KG GFV+++ A AI+ N + + ++ SWG P
Sbjct: 345 GE--VVSVKIPVGKGCGFVQFADRKSAEDAIESLNG-TVIGKNTVRLSWGRSP 394
>At5g54900 unknown protein
Length = 387
Score = 123 bits (308), Expect = 2e-28
Identities = 90/298 (30%), Positives = 146/298 (48%), Gaps = 29/298 (9%)
Query: 22 SLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK----EKSSYGFIHYFDRRSA 77
S S K ++G++ + E + VFA +G K+ R + YGFI + A
Sbjct: 55 SASDVKSLWIGDLQQWMDENYIMSVFAQSGEATSAKVIRNKLTGQSEGYGFIEFVSHSVA 114
Query: 78 ALAILTLNGRHLFG--QPIKVNWAYASG--QREDTSG-HYNIFVGDLSPEVTDATLFACF 132
+ T NG + Q ++NWA A +R T G + IFVGDL+PEVTD L F
Sbjct: 115 ERVLQTYNGAPMPSTEQTFRLNWAQAGAGEKRFQTEGPDHTIFVGDLAPEVTDYMLSDTF 174
Query: 133 -SVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKG 191
+VY S A+V+ D+ TGRS+G+GFV F + + A+ ++ G++ +R +R A
Sbjct: 175 KNVYGSVKGAKVVLDRTTGRSKGYGFVRFADENEQMRAMTEMNGQYCSTRPMRIGPAA-- 232
Query: 192 AGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHR 251
+K+ + + ++ + ++ D N+P TT++VG L + T +L
Sbjct: 233 -----------NKNALPMQPAMYQNTQGANAGD---NDPNNTTIFVGGLDANVTDDELKS 278
Query: 252 HFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKP 309
F G + V++ K GFV+Y+ A A A+ + N + L G+ I+ SWG P
Sbjct: 279 IFGQFGE--LLHVKIPPGKRCGFVQYANKASAEHALSVLNG-TQLGGQSIRLSWGRSP 333
Score = 42.7 bits (99), Expect = 3e-04
Identities = 23/78 (29%), Positives = 42/78 (53%), Gaps = 2/78 (2%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGRHL 89
+VG + VT+ L+ +F G + K+ ++ GF+ Y ++ SA A+ LNG L
Sbjct: 263 FVGGLDANVTDDELKSIFGQFGELLHVKIPPGKRC--GFVQYANKASAEHALSVLNGTQL 320
Query: 90 FGQPIKVNWAYASGQRED 107
GQ I+++W + ++ D
Sbjct: 321 GGQSIRLSWGRSPNKQSD 338
>At4g27000 putative DNA binding protein
Length = 415
Score = 123 bits (308), Expect = 2e-28
Identities = 88/290 (30%), Positives = 146/290 (50%), Gaps = 29/290 (10%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSY----GFIHYFDRRSAALAILTLN 85
++G++ + E L VF TG K+ R +++ Y GFI + + +A + T N
Sbjct: 83 WIGDLQPWMDENYLMNVFGLTGEATAAKVIRNKQNGYSEGYGFIEFVNHATAERNLQTYN 142
Query: 86 GRHLFG--QPIKVNWAY-ASGQREDTSG-HYNIFVGDLSPEVTDATLFACF-SVYQSCSD 140
G + Q ++NWA +G+R G + +FVGDL+P+VTD L F +VY S
Sbjct: 143 GAPMPSSEQAFRLNWAQLGAGERRQAEGPEHTVFVGDLAPDVTDHMLTETFKAVYSSVKG 202
Query: 141 ARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQN 200
A+V+ D+ TGRS+G+GFV F + + A+ ++ G++ SR +R A +K
Sbjct: 203 AKVVNDRTTGRSKGYGFVRFADESEQIRAMTEMNGQYCSSRPMRTGPAA------NKKPL 256
Query: 201 SDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGV 260
+ + + T G+S E++P TT++VG + T+ DL F G
Sbjct: 257 TMQPASYQNTQGNS-----------GESDPTNTTIFVGAVDQSVTEDDLKSVFGQFGE-- 303
Query: 261 IEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPT 310
+ V++ K GFV+Y+ A A A+ + N + L G+ I+ SWG P+
Sbjct: 304 LVHVKIPAGKRCGFVQYANRACAEQALSVLNG-TQLGGQSIRLSWGRSPS 352
Score = 71.6 bits (174), Expect = 7e-13
Identities = 63/238 (26%), Positives = 105/238 (43%), Gaps = 48/238 (20%)
Query: 91 GQPIKVNWAYASGQREDTSGHY-NIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKT 149
GQP + + Q ++G ++++GDL P + + L F + + A+V+ +++
Sbjct: 58 GQPQQQQYGGGGSQNPGSAGEIRSLWIGDLQPWMDENYLMNVFGLTGEATAAKVIRNKQN 117
Query: 150 GRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQ--IRCNWATKGAGGIEEKQNSDSKSVV 207
G S G+GF+ F + A+ + G + S + R NWA GAG E++ ++
Sbjct: 118 GYSEGYGFIEFVNHATAERNLQTYNGAPMPSSEQAFRLNWAQLGAG---ERRQAEG---- 170
Query: 208 ELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHAL-----GAGVIE 262
PE+ TV+VG+L + T L F A+ GA V+
Sbjct: 171 ------------------PEH-----TVFVGDLAPDVTDHMLTETFKAVYSSVKGAKVVN 207
Query: 263 EVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLP 320
+ R KG+GFVR++ +E A+ N Q CS S+P G A+N P
Sbjct: 208 DRTTGRSKGYGFVRFADESEQIRAMTEMNGQ--------YCS--SRPMRTGPAANKKP 255
Score = 42.7 bits (99), Expect = 3e-04
Identities = 25/77 (32%), Positives = 40/77 (51%), Gaps = 2/77 (2%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGRHL 89
+VG V VTE L+ VF G + K+ ++ GF+ Y +R A A+ LNG L
Sbjct: 281 FVGAVDQSVTEDDLKSVFGQFGELVHVKIPAGKRC--GFVQYANRACAEQALSVLNGTQL 338
Query: 90 FGQPIKVNWAYASGQRE 106
GQ I+++W + ++
Sbjct: 339 GGQSIRLSWGRSPSNKQ 355
>At1g11650 putative DNA binding protein
Length = 405
Score = 117 bits (294), Expect = 8e-27
Identities = 88/292 (30%), Positives = 137/292 (46%), Gaps = 32/292 (10%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
++G++ + E L FA TG + K+ R +++ YGFI + +A + T N
Sbjct: 65 WIGDLQYWMDENFLYGCFAHTGEMVSAKVIRNKQTGQVEGYGFIEFASHAAAERVLQTFN 124
Query: 86 GRHLFGQP---IKVNWA-YASGQREDTSGHYNIFVGDLSPEVTDATLFACFSV-YQSCSD 140
+ P ++NWA +SG + D S Y IFVGDL+ +VTD L F Y S
Sbjct: 125 NAPIPSFPDQLFRLNWASLSSGDKRDDSPDYTIFVGDLAADVTDYILLETFRASYPSVKG 184
Query: 141 ARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWAT--KGAGGIEEK 198
A+V+ D+ TGR++G+GFV F + + A+ ++ G +R +R A KG G +
Sbjct: 185 AKVVIDRVTGRTKGYGFVRFSDESEQIRAMTEMNGVPCSTRPMRIGPAASKKGVTGQRDS 244
Query: 199 QNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGA 258
S + V +N+P TTV+VG L + T L F G
Sbjct: 245 YQSSAAGV------------------TTDNDPNNTTVFVGGLDASVTDDHLKNVFSQYGE 286
Query: 259 GVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPT 310
I V++ K GFV++S + A A++M N L G ++ SWG P+
Sbjct: 287 --IVHVKIPAGKRCGFVQFSEKSCAEEALRMLNGVQ-LGGTTVRLSWGRSPS 335
Score = 62.4 bits (150), Expect = 4e-10
Identities = 47/182 (25%), Positives = 82/182 (44%), Gaps = 39/182 (21%)
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+++GDL + + L+ CF+ A+V+ +++TG+ G+GF+ F S A+ +
Sbjct: 64 LWIGDLQYWMDENFLYGCFAHTGEMVSAKVIRNKQTGQVEGYGFIEFASHAAAERVLQTF 123
Query: 174 TGKWLGS---RQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNP 230
+ S + R NWA+ +SS D +++P
Sbjct: 124 NNAPIPSFPDQLFRLNWAS------------------------------LSSGDKRDDSP 153
Query: 231 QYTTVYVGNLGSEATQLDLHRHFHA-----LGAGVIEEVRVQRDKGFGFVRYSTHAEAAL 285
Y T++VG+L ++ T L F A GA V+ + R KG+GFVR+S +E
Sbjct: 154 DY-TIFVGDLAADVTDYILLETFRASYPSVKGAKVVIDRVTGRTKGYGFVRFSDESEQIR 212
Query: 286 AI 287
A+
Sbjct: 213 AM 214
Score = 36.6 bits (83), Expect = 0.024
Identities = 19/77 (24%), Positives = 40/77 (51%), Gaps = 2/77 (2%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGRHL 89
+VG + VT+ L+ VF+ G + K+ ++ GF+ + ++ A A+ LNG L
Sbjct: 264 FVGGLDASVTDDHLKNVFSQYGEIVHVKIPAGKRC--GFVQFSEKSCAEEALRMLNGVQL 321
Query: 90 FGQPIKVNWAYASGQRE 106
G ++++W + ++
Sbjct: 322 GGTTVRLSWGRSPSNKQ 338
>At1g47490 unknown protein
Length = 432
Score = 115 bits (287), Expect = 5e-26
Identities = 89/297 (29%), Positives = 141/297 (46%), Gaps = 34/297 (11%)
Query: 27 KCRYVGNVHTQVTEPLLQEVFAGTGPVE--GCKLFRKEKSS----YGFIHYFDRRSAALA 80
K +VG++H + E L FA E K+ R + + YGF+ + A
Sbjct: 101 KTIWVGDLHHWMDEAYLNSSFASGDEREIVSVKVIRNKNNGLSEGYGFVEFESHDVADKV 160
Query: 81 ILTLNGRHL--FGQPIKVNWA-YASGQR--EDTSGHYNIFVGDLSPEVTDATLFACFSV- 134
+ NG + QP ++NWA +++G++ E+ +IFVGDLSP+V+D L FS
Sbjct: 161 LREFNGTTMPNTDQPFRLNWASFSTGEKRLENNGPDLSIFVGDLSPDVSDNLLHETFSEK 220
Query: 135 YQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGG 194
Y S A+V+ D TGRS+G+GFV F + + A+ ++ G SR +R AT
Sbjct: 221 YPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEMNGVKCSSRAMRIGPATP---- 276
Query: 195 IEEKQNSDSKSVVELTNGSSEDGKEISSNDV--PENNPQYTTVYVGNLGSEATQLDLHRH 252
TNG + G + + + PE + TT++VG L S T DL +
Sbjct: 277 -------------RKTNGYQQQGGYMPNGTLTRPEGDIMNTTIFVGGLDSSVTDEDLKQP 323
Query: 253 FHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKP 309
F+ G I V++ KG GFV++ A A++ N + + + ++ SWG P
Sbjct: 324 FNEFGE--IVSVKIPVGKGCGFVQFVNRPNAEEALEKLNG-TVIGKQTVRLSWGRNP 377
Score = 49.7 bits (117), Expect = 3e-06
Identities = 45/185 (24%), Positives = 75/185 (40%), Gaps = 28/185 (15%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGP-VEGCKLFRKEKSS----YGFIHYFDRRSAALAILTL 84
+VG++ V++ LL E F+ P V+ K+ + YGF+ + D A+ +
Sbjct: 200 FVGDLSPDVSDNLLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEM 259
Query: 85 NGRHLFGQ--------PIKVNWAYASG---------QREDTSGHYNIFVGDLSPEVTDAT 127
NG + P K N G + E + IFVG L VTD
Sbjct: 260 NGVKCSSRAMRIGPATPRKTNGYQQQGGYMPNGTLTRPEGDIMNTTIFVGGLDSSVTDED 319
Query: 128 LFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNW 187
L F+ + ++ +G GFV F ++ +A+ A+ L G +G + +R +W
Sbjct: 320 LKQPFNEFGEIVSVKIPV------GKGCGFVQFVNRPNAEEALEKLNGTVIGKQTVRLSW 373
Query: 188 ATKGA 192
A
Sbjct: 374 GRNPA 378
Score = 33.1 bits (74), Expect = 0.27
Identities = 20/87 (22%), Positives = 42/87 (47%), Gaps = 2/87 (2%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGRHL 89
+VG + + VT+ L++ F G + K+ GF+ + +R +A A+ LNG +
Sbjct: 307 FVGGLDSSVTDEDLKQPFNEFGEIVSVKI--PVGKGCGFVQFVNRPNAEEALEKLNGTVI 364
Query: 90 FGQPIKVNWAYASGQREDTSGHYNIFV 116
Q ++++W ++ + N +V
Sbjct: 365 GKQTVRLSWGRNPANKQPRDKYGNQWV 391
>At1g47500 unknown protein
Length = 434
Score = 108 bits (271), Expect = 4e-24
Identities = 87/297 (29%), Positives = 137/297 (45%), Gaps = 34/297 (11%)
Query: 27 KCRYVGNVHTQVTEPLLQEVFAGTGPVE--GCKLFRKEKSS----YGFIHYFDRRSAALA 80
K +VG++ + E L F E K+ R + + YGF+ + A
Sbjct: 103 KTIWVGDLQNWMDEAYLNSAFTSAEEREIVSLKVIRNKHNGSSEGYGFVEFESHDVADKV 162
Query: 81 ILTLNGRHL--FGQPIKVNWA-YASGQR--EDTSGHYNIFVGDLSPEVTDATLFACFSV- 134
+ NG + QP ++NWA +++G++ E+ +IFVGDL+P+V+DA L FS
Sbjct: 163 LQEFNGAPMPNTDQPFRLNWASFSTGEKRLENNGPDLSIFVGDLAPDVSDALLHETFSEK 222
Query: 135 YQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGG 194
Y S A+V+ D TGRS+G+GFV F + + A+ ++ G SR +R AT
Sbjct: 223 YPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEMNGVKCSSRAMRIGPATP---- 278
Query: 195 IEEKQNSDSKSVVELTNGSSEDGKEISSNDV--PENNPQYTTVYVGNLGSEATQLDLHRH 252
TNG + G + S E + TT++VG L S T DL +
Sbjct: 279 -------------RKTNGYQQQGGYMPSGAFTRSEGDTINTTIFVGGLDSSVTDEDLKQP 325
Query: 253 FHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKP 309
F G I V++ KG GFV++ A A++ N + + + ++ SWG P
Sbjct: 326 FSEFGE--IVSVKIPVGKGCGFVQFVNRPNAEEALEKLNG-TVIGKQTVRLSWGRNP 379
Score = 52.0 bits (123), Expect = 6e-07
Identities = 46/185 (24%), Positives = 76/185 (40%), Gaps = 28/185 (15%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGP-VEGCKLFRKEKSS----YGFIHYFDRRSAALAILTL 84
+VG++ V++ LL E F+ P V+ K+ + YGF+ + D A+ +
Sbjct: 202 FVGDLAPDVSDALLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEM 261
Query: 85 NGRHLFGQ--------PIKVNWAYASG---------QREDTSGHYNIFVGDLSPEVTDAT 127
NG + P K N G + E + + IFVG L VTD
Sbjct: 262 NGVKCSSRAMRIGPATPRKTNGYQQQGGYMPSGAFTRSEGDTINTTIFVGGLDSSVTDED 321
Query: 128 LFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNW 187
L FS + ++ +G GFV F ++ +A+ A+ L G +G + +R +W
Sbjct: 322 LKQPFSEFGEIVSVKIPV------GKGCGFVQFVNRPNAEEALEKLNGTVIGKQTVRLSW 375
Query: 188 ATKGA 192
A
Sbjct: 376 GRNPA 380
Score = 34.3 bits (77), Expect = 0.12
Identities = 20/87 (22%), Positives = 43/87 (48%), Gaps = 2/87 (2%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGRHL 89
+VG + + VT+ L++ F+ G + K+ GF+ + +R +A A+ LNG +
Sbjct: 309 FVGGLDSSVTDEDLKQPFSEFGEIVSVKI--PVGKGCGFVQFVNRPNAEEALEKLNGTVI 366
Query: 90 FGQPIKVNWAYASGQREDTSGHYNIFV 116
Q ++++W ++ + N +V
Sbjct: 367 GKQTVRLSWGRNPANKQPRDKYGNQWV 393
>At2g18510 putative spliceosome associated protein
Length = 363
Score = 105 bits (261), Expect = 6e-23
Identities = 67/198 (33%), Positives = 109/198 (54%), Gaps = 16/198 (8%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
YVG + Q++E LL E+F GPV + + ++ YGFI Y A AI LN
Sbjct: 28 YVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAIKVLN 87
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQS-CSDARVM 144
L G+PI+VN A + D N+F+G+L P+V + L+ FS + S+ ++M
Sbjct: 88 MIKLHGKPIRVNKASQDKKSLDVGA--NLFIGNLDPDVDEKLLYDTFSAFGVIASNPKIM 145
Query: 145 WDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWA----TKGAGGIEEKQN 200
D TG SRGFGF+S+ S + + +AI +TG++L +RQI ++A TKG E+
Sbjct: 146 RDPDTGNSRGFGFISYDSFEASDAAIESMTGQYLSNRQITVSYAYKKDTKG-----ERHG 200
Query: 201 SDSKSVVELTNGSSEDGK 218
+ ++ ++ TN +++ +
Sbjct: 201 TPAERLLAATNPTAQKSR 218
Score = 52.8 bits (125), Expect = 3e-07
Identities = 51/206 (24%), Positives = 85/206 (40%), Gaps = 42/206 (20%)
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
++VG L ++++ L+ F + V D+ T + +GF+ +RS++DA AI L
Sbjct: 27 VYVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAIKVL 86
Query: 174 TGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYT 233
L + IR N K + D KS+ DV N
Sbjct: 87 NMIKLHGKPIRVN-----------KASQDKKSL-----------------DVGAN----- 113
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRD------KGFGFVRYSTHAEAALAI 287
+++GNL + + L+ F A G + ++ RD +GFGF+ Y + + AI
Sbjct: 114 -LFIGNLDPDVDEKLLYDTFSAFGV-IASNPKIMRDPDTGNSRGFGFISYDSFEASDAAI 171
Query: 288 QMGNAQSYLCGKIIKCSWGSKPTPPG 313
+ Q YL + I S+ K G
Sbjct: 172 ESMTGQ-YLSNRQITVSYAYKKDTKG 196
>At4g34110 poly(A)-binding protein
Length = 629
Score = 97.8 bits (242), Expect = 9e-21
Identities = 75/273 (27%), Positives = 123/273 (44%), Gaps = 38/273 (13%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFR----KEKSSYGFIHYFDRRSAALAILTLN 85
YVG++ VT+ L + F G V ++ R + YG++++ + + AA AI LN
Sbjct: 39 YVGDLDFNVTDSQLFDAFGQMGTVVTVRVCRDLVTRRSLGYGYVNFTNPQDAARAIQELN 98
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
L+G+PI+V +++ SG NIF+ +L + L FS + + +V
Sbjct: 99 YIPLYGKPIRVMYSHRDPSVR-RSGAGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVAV 157
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
D +G+S+G+GFV + +++ AQ AI L G L +Q+ G +Q DS +
Sbjct: 158 DS-SGQSKGYGFVQYANEESAQKAIEKLNGMLLNDKQVY-------VGPFLRRQERDSTA 209
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALG---AGVIE 262
N ++T VYV NL T DL F G + V+
Sbjct: 210 ----------------------NKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVM 247
Query: 263 EVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSY 295
+ + KGFGFV + +AA A++ N +
Sbjct: 248 KDGEGKSKGFGFVNFENADDAARAVESLNGHKF 280
Score = 63.5 bits (153), Expect = 2e-10
Identities = 44/172 (25%), Positives = 81/172 (46%), Gaps = 19/172 (11%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
YV N+ T+ L+ F G + + + + +GF+++ + AA A+ +LNG
Sbjct: 218 YVKNLAESTTDDDLKNAFGEYGKITSAVVMKDGEGKSKGFGFVNFENADDAARAVESLNG 277
Query: 87 RHLFGQPIKVNWAYASGQRE---------------DTSGHYNIFVGDLSPEVTDATLFAC 131
+ V A +RE D N++V +L P ++D L
Sbjct: 278 HKFDDKEWYVGRAQKKSERETELRVRYEQNLKEAADKFQSSNLYVKNLDPSISDEKLKEI 337
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
FS + + + ++VM D G S+G GFV+F + ++A A++ L+GK + S+ +
Sbjct: 338 FSPFGTVTSSKVMRDPN-GTSKGSGFVAFATPEEATEAMSQLSGKMIESKPL 388
Score = 32.7 bits (73), Expect = 0.35
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 4/60 (6%)
Query: 233 TTVYVGNLGSEATQLDLHRHFHALG----AGVIEEVRVQRDKGFGFVRYSTHAEAALAIQ 288
T++YVG+L T L F +G V ++ +R G+G+V ++ +AA AIQ
Sbjct: 36 TSLYVGDLDFNVTDSQLFDAFGQMGTVVTVRVCRDLVTRRSLGYGYVNFTNPQDAARAIQ 95
>At3g16380 putative poly(A) binding protein
Length = 537
Score = 97.8 bits (242), Expect = 9e-21
Identities = 79/280 (28%), Positives = 132/280 (46%), Gaps = 33/280 (11%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS--YGFIHYFDRRSAALAILTLNGR 87
YV N+ + +T L+ +F G + CK+ + S +GF+ + +SA A L+G
Sbjct: 115 YVKNLDSSITSSCLERMFCPFGSILSCKVVEENGQSKGFGFVQFDTEQSAVSARSALHGS 174
Query: 88 HLFGQPIKVNWAYASGQREDTSGHY---NIFVGDLSPEVTDATLFACFSVYQSCSDARVM 144
++G+ + V +R +G+ N++V +L VTD L FS Y + S VM
Sbjct: 175 MVYGKKLFVAKFINKDERAAMAGNQDSTNVYVKNLIETVTDDCLHTLFSQYGTVSSVVVM 234
Query: 145 WDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSK 204
D GRSRGFGFV+F + ++A+ A+ L G LGS+++ A K + +
Sbjct: 235 RDGM-GRSRGFGFVNFCNPENAKKAMESLCGLQLGSKKLFVGKALK---------KDERR 284
Query: 205 SVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEV 264
+++ ++ S N + + N +++ +YV NL + L F G V +V
Sbjct: 285 EMLK---------QKFSDNFIAKPNMRWSNLYVKNLSESMNETRLREIFGCYGQIVSAKV 335
Query: 265 RVQ---RDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKII 301
R KGFGFV +S E+ A+ YL G ++
Sbjct: 336 MCHENGRSKGFGFVCFSNCEESK------QAKRYLNGFLV 369
Score = 77.0 bits (188), Expect = 2e-14
Identities = 76/290 (26%), Positives = 128/290 (43%), Gaps = 51/290 (17%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK----EKSSYGFIHYFDRRSAALAILTLN 85
YVG++ VTE L + F+ PV L R + Y +I++ SA+ A+ LN
Sbjct: 24 YVGDLSPDVTEKDLIDKFSLNVPVVSVHLCRNSVTGKSMCYAYINFDSPFSASNAMTRLN 83
Query: 86 GRHLFGQPIKVNWAY--ASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARV 143
L G+ +++ W+ + +R +G N++V +L +T + L F + S +V
Sbjct: 84 HSDLKGKAMRIMWSQRDLAYRRRTRTGFANLYVKNLDSSITSSCLERMFCPFGSILSCKV 143
Query: 144 MWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDS 203
+ ++ G+S+GFGFV F ++Q A SA + L G + +++ + + N D
Sbjct: 144 V--EENGQSKGFGFVQFDTEQSAVSARSALHGSMVYGKKL----------FVAKFINKDE 191
Query: 204 KSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEE 263
++ + N T VYV NL T LH F G +
Sbjct: 192 RAAM-------------------AGNQDSTNVYVKNLIETVTDDCLHTLFSQY--GTVSS 230
Query: 264 VRVQRD-----KGFGFVRYSTHAEAALA------IQMGNAQSYLCGKIIK 302
V V RD +GFGFV + A A +Q+G+ + ++ GK +K
Sbjct: 231 VVVMRDGMGRSRGFGFVNFCNPENAKKAMESLCGLQLGSKKLFV-GKALK 279
Score = 53.9 bits (128), Expect = 1e-07
Identities = 47/190 (24%), Positives = 76/190 (39%), Gaps = 36/190 (18%)
Query: 101 ASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSF 160
A G + +S +++VGDLSP+VT+ L FS+ + + TG+S + +++F
Sbjct: 10 ALGNHQHSSRFGSLYVGDLSPDVTEKDLIDKFSLNVPVVSVHLCRNSVTGKSMCYAYINF 69
Query: 161 RSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEI 220
S A +A+ L L + +R W+ + +
Sbjct: 70 DSPFSASNAMTRLNHSDLKGKAMRIMWSQRDLAYRRRTRTG------------------- 110
Query: 221 SSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGA----GVIEEVRVQRDKGFGFVR 276
+ +YV NL S T L R F G+ V+EE + KGFGFV+
Sbjct: 111 -----------FANLYVKNLDSSITSSCLERMFCPFGSILSCKVVEE--NGQSKGFGFVQ 157
Query: 277 YSTHAEAALA 286
+ T A A
Sbjct: 158 FDTEQSAVSA 167
>At2g23350 putative poly(A) binding protein
Length = 662
Score = 92.8 bits (229), Expect = 3e-19
Identities = 83/275 (30%), Positives = 117/275 (42%), Gaps = 31/275 (11%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
+V N+ V L E F+G G + CK+ + YGF+ + SA AI LNG
Sbjct: 137 FVKNLDKSVDNKTLHEAFSGCGTIVSCKVATDHMGQSRGYGFVQFDTEDSAKNAIEKLNG 196
Query: 87 RHLFGQPIKVNWAYASGQRE---DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARV 143
+ L + I V +RE D N++V +LS TD L F Y S S A V
Sbjct: 197 KVLNDKQIFVGPFLRKEERESAADKMKFTNVYVKNLSEATTDDELKTTFGQYGSISSAVV 256
Query: 144 MWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDS 203
M D G+SR FGFV+F + +DA A+ L GK ++ W G ++K +
Sbjct: 257 MRDGD-GKSRCFGFVNFENPEDAARAVEALNGKKFDDKE----WYV---GKAQKKSEREL 308
Query: 204 KSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEE 263
+ GSS+ G + +YV NL T L F G I
Sbjct: 309 ELSRRYEQGSSDGGNKFDG----------LNLYVKNLDDTVTDEKLRELFAEFGT--ITS 356
Query: 264 VRVQRD-----KGFGFVRYSTHAEAALAIQMGNAQ 293
+V RD KG GFV +S +EA+ + N +
Sbjct: 357 CKVMRDPSGTSKGSGFVAFSAASEASRVLNEMNGK 391
Score = 83.2 bits (204), Expect = 2e-16
Identities = 74/275 (26%), Positives = 120/275 (42%), Gaps = 42/275 (15%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
YVG++ VT+ L + F V ++ R ++ YG+++Y + A A+ LN
Sbjct: 49 YVGDLDFNVTDSQLYDYFTEVCQVVSVRVCRDAATNTSLGYGYVNYSNTDDAEKAMQKLN 108
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
+L G+ I++ ++ SG N+FV +L V + TL FS + +V
Sbjct: 109 YSYLNGKMIRITYSSRDSSAR-RSGVGNLFVKNLDKSVDNKTLHEAFSGCGTIVSCKVAT 167
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
D G+SRG+GFV F ++ A++AI L GK L +QI G K+ +S +
Sbjct: 168 DHM-GQSRGYGFVQFDTEDSAKNAIEKLNGKVLNDKQI-------FVGPFLRKEERESAA 219
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
+ ++T VYV NL T +L F G+ I
Sbjct: 220 ----------------------DKMKFTNVYVKNLSEATTDDELKTTFGQYGS--ISSAV 255
Query: 266 VQRD-----KGFGFVRYSTHAEAALAIQMGNAQSY 295
V RD + FGFV + +AA A++ N + +
Sbjct: 256 VMRDGDGKSRCFGFVNFENPEDAARAVEALNGKKF 290
Score = 63.5 bits (153), Expect = 2e-10
Identities = 45/172 (26%), Positives = 80/172 (46%), Gaps = 19/172 (11%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
YV N+ T+ L+ F G + + R + +GF+++ + AA A+ LNG
Sbjct: 228 YVKNLSEATTDDELKTTFGQYGSISSAVVMRDGDGKSRCFGFVNFENPEDAARAVEALNG 287
Query: 87 RHLFGQPIKVNWAYASGQRE-DTSGHY--------------NIFVGDLSPEVTDATLFAC 131
+ + V A +RE + S Y N++V +L VTD L
Sbjct: 288 KKFDDKEWYVGKAQKKSERELELSRRYEQGSSDGGNKFDGLNLYVKNLDDTVTDEKLREL 347
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
F+ + + + +VM D +G S+G GFV+F + +A +N++ GK +G + +
Sbjct: 348 FAEFGTITSCKVMRDP-SGTSKGSGFVAFSAASEASRVLNEMNGKMVGGKPL 398
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/79 (29%), Positives = 40/79 (50%), Gaps = 3/79 (3%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSY---GFIHYFDRRSAALAILTLNG 86
YV N+ VT+ L+E+FA G + CK+ R + GF+ + A+ + +NG
Sbjct: 331 YVKNLDDTVTDEKLRELFAEFGTITSCKVMRDPSGTSKGSGFVAFSAASEASRVLNEMNG 390
Query: 87 RHLFGQPIKVNWAYASGQR 105
+ + G+P+ V A +R
Sbjct: 391 KMVGGKPLYVALAQRKEER 409
>At1g49760 Putative Poly-A Binding Protein (F14J22.3)
Length = 671
Score = 91.7 bits (226), Expect = 6e-19
Identities = 84/296 (28%), Positives = 129/296 (43%), Gaps = 37/296 (12%)
Query: 14 IQVLAAVSSLSVRKCR----YVGNVHTQVTEPLLQEVFAGTGPVEGCKLF---RKEKSSY 66
I+V+ +V S+RK ++ N+ + L E F+ GP+ CK+ + Y
Sbjct: 116 IRVMYSVRDPSLRKSGVGNIFIKNLDKSIDHKALHETFSAFGPILSCKVAVDPSGQSKGY 175
Query: 67 GFIHYFDRRSAALAILTLNGRHLFGQPIKVNWAYASGQREDTSGHY----NIFVGDLSPE 122
GF+ Y +A AI LNG L + + V + + D SG N++V +LS
Sbjct: 176 GFVQYDTDEAAQGAIDKLNGMLLNDKQVYVG-PFVHKLQRDPSGEKVKFTNVYVKNLSES 234
Query: 123 VTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQ 182
++D L F + + +M D + G+S+GFGFV+F + DA A++ L GK ++
Sbjct: 235 LSDEELNKVFGEFGVTTSCVIMRDGE-GKSKGFGFVNFENSDDAARAVDALNGKTFDDKE 293
Query: 183 IRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGS 242
W K S+ EL + KE + + Q + +YV NL
Sbjct: 294 ----W-------FVGKAQKKSERETELKQKFEQSLKEAA------DKSQGSNLYVKNLDE 336
Query: 243 EATQLDLHRHFHALGAGVIEEVRVQRD-----KGFGFVRYSTHAEAALAIQMGNAQ 293
T L HF G I +V RD +G GFV +ST EA AI N +
Sbjct: 337 SVTDDKLREHFAPF--GTITSCKVMRDPSGVSRGSGFVAFSTPEEATRAITEMNGK 390
Score = 84.3 bits (207), Expect = 1e-16
Identities = 72/273 (26%), Positives = 120/273 (43%), Gaps = 38/273 (13%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFR----KEKSSYGFIHYFDRRSAALAILTLN 85
YVG++ VT+ L E F G V ++ R + YG+++Y + A+ A+ LN
Sbjct: 48 YVGDLDATVTDSQLFEAFTQAGQVVSVRVCRDMTTRRSLGYGYVNYATPQDASRALNELN 107
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
L G+ I+V ++ SG NIF+ +L + L FS + +V
Sbjct: 108 FMALNGRAIRVMYSVRDPSLR-KSGVGNIFIKNLDKSIDHKALHETFSAFGPILSCKVAV 166
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
D +G+S+G+GFV + + + AQ AI+ L G L +Q+ G K D
Sbjct: 167 D-PSGQSKGYGFVQYDTDEAAQGAIDKLNGMLLNDKQVY-------VGPFVHKLQRD--- 215
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALG---AGVIE 262
G+++ ++T VYV NL + +L++ F G + VI
Sbjct: 216 ---------PSGEKV----------KFTNVYVKNLSESLSDEELNKVFGEFGVTTSCVIM 256
Query: 263 EVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSY 295
+ KGFGFV + +AA A+ N +++
Sbjct: 257 RDGEGKSKGFGFVNFENSDDAARAVDALNGKTF 289
Score = 64.7 bits (156), Expect = 8e-11
Identities = 47/172 (27%), Positives = 82/172 (47%), Gaps = 19/172 (11%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
YV N+ +++ L +VF G C + R + +GF+++ + AA A+ LNG
Sbjct: 227 YVKNLSESLSDEELNKVFGEFGVTTSCVIMRDGEGKSKGFGFVNFENSDDAARAVDALNG 286
Query: 87 RHLFGQPIKVNWAYASGQRE---------------DTSGHYNIFVGDLSPEVTDATLFAC 131
+ + V A +RE D S N++V +L VTD L
Sbjct: 287 KTFDDKEWFVGKAQKKSERETELKQKFEQSLKEAADKSQGSNLYVKNLDESVTDDKLREH 346
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
F+ + + + +VM D +G SRG GFV+F + ++A AI ++ GK + ++ +
Sbjct: 347 FAPFGTITSCKVMRDP-SGVSRGSGFVAFSTPEEATRAITEMNGKMIVTKPL 397
Score = 39.3 bits (90), Expect = 0.004
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 3/80 (3%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
YV N+ VT+ L+E FA G + CK+ R GF+ + A AI +NG
Sbjct: 330 YVKNLDESVTDDKLREHFAPFGTITSCKVMRDPSGVSRGSGFVAFSTPEEATRAITEMNG 389
Query: 87 RHLFGQPIKVNWAYASGQRE 106
+ + +P+ V A R+
Sbjct: 390 KMIVTKPLYVALAQRKEDRK 409
Score = 34.7 bits (78), Expect = 0.093
Identities = 26/76 (34%), Positives = 40/76 (52%), Gaps = 9/76 (11%)
Query: 233 TTVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDK------GFGFVRYSTHAEAALA 286
T++YVG+L + T L F AG + VRV RD G+G+V Y+T +A+ A
Sbjct: 45 TSLYVGDLDATVTDSQLFEAFTQ--AGQVVSVRVCRDMTTRRSLGYGYVNYATPQDASRA 102
Query: 287 IQMGNAQSYLCGKIIK 302
+ N + L G+ I+
Sbjct: 103 LNELNFMA-LNGRAIR 117
>At1g71770 polyadenylate-binding protein 5
Length = 668
Score = 88.2 bits (217), Expect = 7e-18
Identities = 68/271 (25%), Positives = 122/271 (44%), Gaps = 35/271 (12%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEG---CKLFRKEKSSYGFIHYFDRRSAALAILTLNG 86
YVG++ V E L ++F PV C+ Y ++++ + A+ A+ +LN
Sbjct: 48 YVGDLDPSVNESHLLDLFNQVAPVHNLRVCRDLTHRSLGYAYVNFANPEDASRAMESLNY 107
Query: 87 RHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWD 146
+ +PI++ + SG N+F+ +L + + L+ FS + + +V D
Sbjct: 108 APIRDRPIRIMLSNRDPSTR-LSGKGNVFIKNLDASIDNKALYETFSSFGTILSCKVAMD 166
Query: 147 QKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSV 206
GRS+G+GFV F ++ AQ+AI+ L G L +Q+ + +++ S+S +V
Sbjct: 167 V-VGRSKGYGFVQFEKEETAQAAIDKLNGMLLNDKQVFVGHFVRR----QDRARSESGAV 221
Query: 207 VELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALG---AGVIEE 263
P +T VYV NL E T +L + F G + V+ +
Sbjct: 222 -----------------------PSFTNVYVKNLPKEITDDELKKTFGKYGDISSAVVMK 258
Query: 264 VRVQRDKGFGFVRYSTHAEAALAIQMGNAQS 294
+ + FGFV + + AA+A++ N S
Sbjct: 259 DQSGNSRSFGFVNFVSPEAAAVAVEKMNGIS 289
Score = 77.0 bits (188), Expect = 2e-14
Identities = 77/278 (27%), Positives = 113/278 (39%), Gaps = 35/278 (12%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
++ N+ + L E F+ G + CK+ YGF+ + +A AI LNG
Sbjct: 135 FIKNLDASIDNKALYETFSSFGTILSCKVAMDVVGRSKGYGFVQFEKEETAQAAIDKLNG 194
Query: 87 RHLFGQPIKV-NWAYASGQREDTSGHY----NIFVGDLSPEVTDATLFACFSVYQSCSDA 141
L + + V ++ + SG N++V +L E+TD L F Y S A
Sbjct: 195 MLLNDKQVFVGHFVRRQDRARSESGAVPSFTNVYVKNLPKEITDDELKKTFGKYGDISSA 254
Query: 142 RVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEE---K 198
VM DQ +G SR FGFV+F S + A A+ + G LG + A K + EE K
Sbjct: 255 VVMKDQ-SGNSRSFGFVNFVSPEAAAVAVEKMNGISLGEDVLYVGRAQKKSDREEELRRK 313
Query: 199 QNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGA 258
+ S E GS+ +Y+ NL L F G
Sbjct: 314 FEQERISRFEKLQGSN--------------------LYLKNLDDSVNDEKLKEMFSEYGN 353
Query: 259 GVIEEVRVQR---DKGFGFVRYSTHAEAALAIQMGNAQ 293
+V + +GFGFV YS EA LA++ N +
Sbjct: 354 VTSCKVMMNSQGLSRGFGFVAYSNPEEALLAMKEMNGK 391
Score = 67.0 bits (162), Expect = 2e-11
Identities = 43/172 (25%), Positives = 83/172 (48%), Gaps = 19/172 (11%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
YV N+ ++T+ L++ F G + + + + S+GF+++ +AA+A+ +NG
Sbjct: 228 YVKNLPKEITDDELKKTFGKYGDISSAVVMKDQSGNSRSFGFVNFVSPEAAAVAVEKMNG 287
Query: 87 RHLFGQPIKVNWAYASGQREDTSGHY---------------NIFVGDLSPEVTDATLFAC 131
L + V A RE+ N+++ +L V D L
Sbjct: 288 ISLGEDVLYVGRAQKKSDREEELRRKFEQERISRFEKLQGSNLYLKNLDDSVNDEKLKEM 347
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
FS Y + + +VM + + G SRGFGFV++ + ++A A+ ++ GK +G + +
Sbjct: 348 FSEYGNVTSCKVMMNSQ-GLSRGFGFVAYSNPEEALLAMKEMNGKMIGRKPL 398
Score = 40.4 bits (93), Expect = 0.002
Identities = 22/80 (27%), Positives = 41/80 (50%), Gaps = 3/80 (3%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
Y+ N+ V + L+E+F+ G V CK+ + +GF+ Y + A LA+ +NG
Sbjct: 331 YLKNLDDSVNDEKLKEMFSEYGNVTSCKVMMNSQGLSRGFGFVAYSNPEEALLAMKEMNG 390
Query: 87 RHLFGQPIKVNWAYASGQRE 106
+ + +P+ V A +R+
Sbjct: 391 KMIGRKPLYVALAQRKEERQ 410
>At2g36660 putative poly(A) binding protein
Length = 609
Score = 81.6 bits (200), Expect = 7e-16
Identities = 71/284 (25%), Positives = 119/284 (41%), Gaps = 44/284 (15%)
Query: 21 SSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRS 76
SS V YVG++H VTE +L + FA + +L + S YG+ ++ R+
Sbjct: 18 SSSPVTASLYVGDLHPSVTEGILYDAFAEFKSLTSVRLCKDASSGRSLCYGYANFLSRQD 77
Query: 77 AALAILTLNGRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQ 136
A LAI N L G+ I+V W+ + +G N+FV +L VT+A L F +
Sbjct: 78 ANLAIEKKNNSLLNGKMIRVMWSVRAPDAR-RNGVGNVFVKNLPESVTNAVLQDMFKKFG 136
Query: 137 SCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIE 196
+ +V + G+SRG+GFV F + A +AI L + ++I
Sbjct: 137 NIVSCKVA-TLEDGKSRGYGFVQFEQEDAAHAAIQTLNSTIVADKEIYV----------- 184
Query: 197 EKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHAL 256
GK + D + +YT +Y+ NL ++ ++ L F
Sbjct: 185 --------------------GKFMKKTDRVKPEEKYTNLYMKNLDADVSEDLLREKFAEF 224
Query: 257 GAGVIEEVRVQRD-----KGFGFVRYSTHAEAALAIQMGNAQSY 295
G I + + +D +G+ FV + +A A + N +
Sbjct: 225 GK--IVSLAIAKDENRLCRGYAFVNFDNPEDARRAAETVNGTKF 266
Score = 75.5 bits (184), Expect = 5e-14
Identities = 68/273 (24%), Positives = 120/273 (43%), Gaps = 25/273 (9%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS---YGFIHYFDRRSAALAILTLNG 86
+V N+ VT +LQ++F G + CK+ E YGF+ + +A AI TLN
Sbjct: 115 FVKNLPESVTNAVLQDMFKKFGNIVSCKVATLEDGKSRGYGFVQFEQEDAAHAAIQTLNS 174
Query: 87 RHLFGQPIKVNWAYASGQREDTSGHY-NIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
+ + I V R Y N+++ +L +V++ L F+ + +
Sbjct: 175 TIVADKEIYVGKFMKKTDRVKPEEKYTNLYMKNLDADVSEDLLREKFAEFGKIVSLAIAK 234
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKS 205
D+ RG+ FV+F + +DA+ A + G GS+ C + G +K+ +
Sbjct: 235 DENR-LCRGYAFVNFDNPEDARRAAETVNGTKFGSK---CLYV-----GRAQKKAEREQL 285
Query: 206 VVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVR 265
+ E E+ K I+ + + +YV N+ T+ +L +HF G ++
Sbjct: 286 LREQFKEKHEEQKMIA---------KVSNIYVKNVNVAVTEEELRKHFSQCGTITSTKLM 336
Query: 266 VQ---RDKGFGFVRYSTHAEAALAIQMGNAQSY 295
+ KGFGFV +ST EA A++ + Q +
Sbjct: 337 CDEKGKSKGFGFVCFSTPEEAIDAVKTFHGQMF 369
Score = 61.6 bits (148), Expect = 7e-10
Identities = 55/228 (24%), Positives = 98/228 (42%), Gaps = 29/228 (12%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
Y+ N+ V+E LL+E FA G + + + E Y F+++ + A A T+NG
Sbjct: 204 YMKNLDADVSEDLLREKFAEFGKIVSLAIAKDENRLCRGYAFVNFDNPEDARRAAETVNG 263
Query: 87 RHLFGQPIKVNWAYASGQREDTSGHY---------------NIFVGDLSPEVTDATLFAC 131
+ + V A +RE NI+V +++ VT+ L
Sbjct: 264 TKFGSKCLYVGRAQKKAEREQLLREQFKEKHEEQKMIAKVSNIYVKNVNVAVTEEELRKH 323
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKG 191
FS + + ++M D+K G+S+GFGFV F + ++A A+ G+ + + A K
Sbjct: 324 FSQCGTITSTKLMCDEK-GKSKGFGFVCFSTPEEAIDAVKTFHGQMFHGKPLYVAIAQK- 381
Query: 192 AGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGN 239
D K +++ G+ + ++ SS+ N Y +Y N
Sbjct: 382 --------KEDRKMQLQVQFGNRVEARK-SSSSASVNPGTYAPLYYTN 420
>At2g37220 putative RNA-binding protein
Length = 289
Score = 77.4 bits (189), Expect = 1e-14
Identities = 58/187 (31%), Positives = 87/187 (46%), Gaps = 28/187 (14%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKE----KSSYGFIHYFDRRSAALAILTLN 85
+VGN+ V L ++F G VE ++ + +GF+ A N
Sbjct: 94 FVGNLPFNVDSAQLAQLFESAGNVEMVEVIYDKITGRSRGFGFVTMSSVSEVEAAAQQFN 153
Query: 86 GRHLFGQPIKVNWAYASGQREDT-------------SGH-----------YNIFVGDLSP 121
G L G+P++VN +RED SG+ ++VG+LS
Sbjct: 154 GYELDGRPLRVNAGPPPPKREDGFSRGPRSSFGSSGSGYGGGGGSGAGSGNRVYVGNLSW 213
Query: 122 EVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSR 181
V D L + FS +ARV++D+ +GRS+GFGFV++ S Q+ Q+AI L G L R
Sbjct: 214 GVDDMALESLFSEQGKVVEARVIYDRDSGRSKGFGFVTYDSSQEVQNAIKSLDGADLDGR 273
Query: 182 QIRCNWA 188
QIR + A
Sbjct: 274 QIRVSEA 280
Score = 67.0 bits (162), Expect = 2e-11
Identities = 59/192 (30%), Positives = 78/192 (39%), Gaps = 12/192 (6%)
Query: 101 ASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSF 160
A + + S +FVG+L V A L F + V++D+ TGRSRGFGFV+
Sbjct: 80 APPKEQSFSADLKLFVGNLPFNVDSAQLAQLFESAGNVEMVEVIYDKITGRSRGFGFVTM 139
Query: 161 RSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEI 220
S + ++A G L R +R N AG K+ + GSS G
Sbjct: 140 SSVSEVEAAAQQFNGYELDGRPLRVN-----AGPPPPKREDGFSRGPRSSFGSSGSGYGG 194
Query: 221 SSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRD----KGFGFVR 276
+ VYVGNL + L F G V V RD KGFGFV
Sbjct: 195 GGGSGAGSG---NRVYVGNLSWGVDDMALESLFSEQGKVVEARVIYDRDSGRSKGFGFVT 251
Query: 277 YSTHAEAALAIQ 288
Y + E AI+
Sbjct: 252 YDSSQEVQNAIK 263
Score = 35.0 bits (79), Expect = 0.071
Identities = 24/80 (30%), Positives = 39/80 (48%), Gaps = 4/80 (5%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
YVGN+ V + L+ +F+ G V ++ S +GF+ Y + AI +L+
Sbjct: 207 YVGNLSWGVDDMALESLFSEQGKVVEARVIYDRDSGRSKGFGFVTYDSSQEVQNAIKSLD 266
Query: 86 GRHLFGQPIKVNWAYASGQR 105
G L G+ I+V+ A A R
Sbjct: 267 GADLDGRQIRVSEAEARPPR 286
Score = 32.3 bits (72), Expect = 0.46
Identities = 29/84 (34%), Positives = 40/84 (47%), Gaps = 11/84 (13%)
Query: 235 VYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDK------GFGFVRYSTHAEAALAIQ 288
++VGNL L + F + AG +E V V DK GFGFV S+ +E A Q
Sbjct: 93 LFVGNLPFNVDSAQLAQLFES--AGNVEMVEVIYDKITGRSRGFGFVTMSSVSEVEAAAQ 150
Query: 289 MGNAQSYLCGKIIKCSWGSKPTPP 312
N L G+ ++ + G P PP
Sbjct: 151 QFNGYE-LDGRPLRVNAG--PPPP 171
>At1g34140 putative protein
Length = 398
Score = 76.6 bits (187), Expect = 2e-14
Identities = 79/307 (25%), Positives = 124/307 (39%), Gaps = 34/307 (11%)
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
+V N+ + L ++F+ G V CK+ R YGF+ ++ S A NG
Sbjct: 34 FVKNLDESIDNKQLCDMFSAFGKVLSCKVARDASGVSKGYGFVQFYSDLSVYTACNFHNG 93
Query: 87 RHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWD 146
+ Q I V + GQ + + N++V +L TDA L F + + A VM D
Sbjct: 94 TLIRNQHIHVCPFVSRGQWDKSRVFTNVYVKNLVETATDADLKRLFGEFGEITSAVVMKD 153
Query: 147 QKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSV 206
+ G+SR FGFV+F + A +AI + G + +++ A + + + D K+
Sbjct: 154 GE-GKSRRFGFVNFEKAEAAVTAIEKMNGVVVDEKELHVGRAQR-----KTNRTEDLKAK 207
Query: 207 VELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAGVIEEVRV 266
EL E D+ +YV NL L F G
Sbjct: 208 FEL---------EKIIRDMKTRKGM--NLYVKNLDDSVDNTKLEELFSEFG------TIT 250
Query: 267 QRDKGFGFVRYSTHAEAALAI------QMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLP 320
+KG GFV +ST EA+ A+ +GN Y+ + +C K +NP P
Sbjct: 251 SCNKGVGFVEFSTSEEASKAMLKMNGKMVGNKPIYV--SLAQCKEQHKLHLQTQFNNPPP 308
Query: 321 PPAAAPL 327
P P+
Sbjct: 309 SPHQQPI 315
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,814,142
Number of Sequences: 26719
Number of extensions: 388802
Number of successful extensions: 2758
Number of sequences better than 10.0: 201
Number of HSP's better than 10.0 without gapping: 152
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 2013
Number of HSP's gapped (non-prelim): 525
length of query: 392
length of database: 11,318,596
effective HSP length: 101
effective length of query: 291
effective length of database: 8,619,977
effective search space: 2508413307
effective search space used: 2508413307
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146708.15


