

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146706.7 + phase: 0 /pseudo
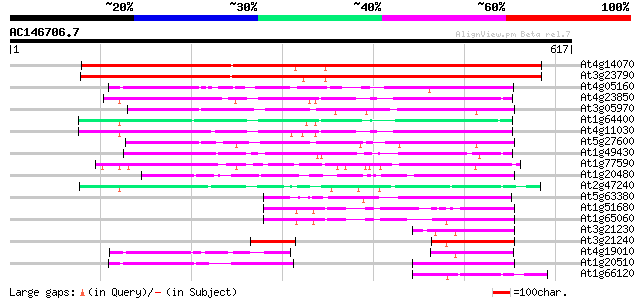
(617 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g14070 A6 anther-specific protein (Z97335.16) 677 0.0
At3g23790 AMP-binding protein, putative 667 0.0
At4g05160 4-coumarate--CoA ligase - like protein 139 5e-33
At4g23850 acyl-CoA synthetase - like protein 124 2e-28
At3g05970 AMP-binding protein 122 5e-28
At1g64400 putative acyl-CoA synthetase 122 6e-28
At4g11030 putative acyl-CoA synthetase 121 1e-27
At5g27600 long chain acyl-CoA synthetase 7 (LACS7) 120 3e-27
At1g49430 acyl CoA synthetase-like protein (At1g49430) 114 2e-25
At1g77590 long chain acyl-CoA synthetase 9 (LACS9) 108 1e-23
At1g20480 unknown protein 102 5e-22
At2g47240 putative acyl-CoA synthetase 100 4e-21
At5g63380 4-coumarate-CoA ligase-like protein 92 9e-19
At1g51680 4-coumarate--coenzyme A ligase (At4CL1) 88 2e-17
At1g65060 4-coumarate:CoA ligase 3 (4CL3) 87 4e-17
At3g21230 putative 4-coumarate:CoA ligase 2 86 6e-17
At3g21240 putative 4-coumarate:CoA ligase 2 84 3e-16
At4g19010 4-coumarate-CoA ligase - like 74 2e-13
At1g20510 4-coumarate:CoA ligase like protein 72 9e-13
At1g66120 unknown protein 69 8e-12
>At4g14070 A6 anther-specific protein (Z97335.16)
Length = 727
Score = 677 bits (1748), Expect = 0.0
Identities = 343/537 (63%), Positives = 410/537 (75%), Gaps = 32/537 (5%)
Query: 80 SPLLESSLLSGNDGVVSDEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAI 139
SP LESS SG+ + S EWK VPDIWRSSAEKYGD++AL+DPYHDPP +TYKQLE I
Sbjct: 77 SPFLESSSFSGDAALRSSEWKAVPDIWRSSAEKYGDRVALVDPYHDPPLKLTYKQLEQEI 136
Query: 140 LDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNH 199
LDFAEGLRV+GV +EKIALFADNSCRWLV+DQG+MATGA+NVVRGSRSS+EELLQIY H
Sbjct: 137 LDFAEGLRVLGVKADEKIALFADNSCRWLVSDQGIMATGAVNVVRGSRSSVEELLQIYRH 196
Query: 200 SESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPIFTFTEIMHLG 259
SES+A+ VDNPE FNRIA++F KAS+RF+ILLWGEKS LV +G ++P++++ EI++ G
Sbjct: 197 SESVAIVVDNPEFFNRIAESFTSKASLRFLILLWGEKSSLVTQGM-QIPVYSYAEIINQG 255
Query: 260 RGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQI---SKYA 316
+ SR + + + Y + I SDD A ++YTSGTTGNPKGVMLTH+NLLHQI SKY
Sbjct: 256 QESRAKLSASNDTRSYRNQFIDSDDTAAIMYTSGTTGNPKGVMLTHRNLLHQIKHLSKYV 315
Query: 317 ASLAC--I*KSL*VFHFFMRC*PS-IYNCEKLE-------------------------VY 348
+ A L +H + R I+ C + VY
Sbjct: 316 PAQAGDKFLSMLPSWHAYERASEYFIFTCGVEQMYTSIRYLKDDLKRYQPNYIVSVPLVY 375
Query: 349 ESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAMLDW 408
E+LYSGIQ+QIS SS RK +ALT I+VS+ YME KRIYEG CLTK QK P Y+ A +DW
Sbjct: 376 ETLYSGIQKQISASSAGRKFLALTLIKVSMAYMEMKRIYEGMCLTKEQKPPMYIVAFVDW 435
Query: 409 LGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAIGVTLQNG 468
L AR+IA +L+P+HMLAKKL+Y KIHS+IG SKAGISGGGSLP HVD+FFEAIGV LQNG
Sbjct: 436 LWARVIAALLWPLHMLAKKLIYKKIHSSIGISKAGISGGGSLPIHVDKFFEAIGVILQNG 495
Query: 469 YGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMK 528
YGLTETSPV+ AR LSCNV+GS GHP+ TEFK+VD ET VLPPG KGI+KVRGPQ+MK
Sbjct: 496 YGLTETSPVVCARTLSCNVLGSAGHPMHGTEFKIVDPETNNVLPPGSKGIIKVRGPQVMK 555
Query: 529 GYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSGRSRNCGGVIVVEGRAKDTIVLSS 585
GYYKNPS T Q +++ GW NTGD GWIA +HS GRSR+CGGVIV+EGRAKDTIVLS+
Sbjct: 556 GYYKNPSTTKQVLNESGWFNTGDTGWIAPHHSKGRSRHCGGVIVLEGRAKDTIVLST 612
>At3g23790 AMP-binding protein, putative
Length = 722
Score = 667 bits (1720), Expect = 0.0
Identities = 337/539 (62%), Positives = 403/539 (74%), Gaps = 32/539 (5%)
Query: 78 RFSPLLESSLLSGNDGVVSDEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLED 137
R SP LE L + S+EWK+VPDIWRSS EKYGD++A++DPYHDPPST TY+QLE
Sbjct: 59 RCSPFLERLSLPREAALSSNEWKSVPDIWRSSVEKYGDRVAVVDPYHDPPSTFTYRQLEQ 118
Query: 138 AILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIY 197
ILDF EGLRV+GV +EKIALFADNSCRWLVADQG+MATGA+NVVRGSRSS+EELLQIY
Sbjct: 119 EILDFVEGLRVVGVKADEKIALFADNSCRWLVADQGIMATGAVNVVRGSRSSVEELLQIY 178
Query: 198 NHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPIFTFTEIMH 257
HSES+AL VDNPE FNRIA++F KA+ +FVILLWGEKS LV G + P++++ EI
Sbjct: 179 CHSESVALVVDNPEFFNRIAESFSYKAAPKFVILLWGEKSSLVTAG-RHTPVYSYNEIKK 237
Query: 258 LGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAA 317
G+ R F + Y +E I DDIAT++YTSGTTGNPKGVMLTHQNLLHQI +
Sbjct: 238 FGQERRAKFARSNDSGKYEYEYIDPDDIATIMYTSGTTGNPKGVMLTHQNLLHQIRNLSD 297
Query: 318 SLAC-----I*KSL*VFHFFMR-C*PSIYNCEKLE------------------------- 346
+ L +H + R C I+ C +
Sbjct: 298 FVPAEAGERFLSMLPSWHAYERACEYFIFTCGVEQKYTSIRFLKDDLKRYQPHYLISVPL 357
Query: 347 VYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAML 406
VYE+LYSGIQ+QIS SS RK +ALT I+VSL Y E KR+YEG CLTKNQK P Y+ +++
Sbjct: 358 VYETLYSGIQKQISASSPARKFLALTLIKVSLAYTEMKRVYEGLCLTKNQKPPMYIVSLV 417
Query: 407 DWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAIGVTLQ 466
DWL AR++A L+P+HMLA+KLV+ KI S+IG +KAG+SGGGSLP HVD+FFEAIGV +Q
Sbjct: 418 DWLWARVVAFFLWPLHMLAEKLVHRKIRSSIGITKAGVSGGGSLPMHVDKFFEAIGVNVQ 477
Query: 467 NGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQL 526
NGYGLTETSPV++ARRL CNV+GSVGHP+K TEFK+VD ETG VLPPG KGI+KVRGP +
Sbjct: 478 NGYGLTETSPVVSARRLRCNVLGSVGHPIKDTEFKIVDHETGTVLPPGSKGIVKVRGPPV 537
Query: 527 MKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSGRSRNCGGVIVVEGRAKDTIVLSS 585
MKGYYKNP AT Q ID DGW NTGD+GWI HS+GRSR+CGGVIV+EGRAKDTIVLS+
Sbjct: 538 MKGYYKNPLATKQVIDDDGWFNTGDMGWITPQHSTGRSRSCGGVIVLEGRAKDTIVLST 596
>At4g05160 4-coumarate--CoA ligase - like protein
Length = 544
Score = 139 bits (350), Expect = 5e-33
Identities = 127/455 (27%), Positives = 200/455 (43%), Gaps = 72/455 (15%)
Query: 109 SAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWL 168
++ Y K+A+ D D ++T+ QL+ A+ A G +G+ N+ + +FA NS ++
Sbjct: 36 NSSSYPSKLAIADS--DTGDSLTFSQLKSAVARLAHGFHRLGIRKNDVVLIFAPNSYQFP 93
Query: 169 VADQGMMATGAINVVRGSRSSIEELL-QIYNHSESIALAVDNPEMFNRIAKAFDLKASMR 227
+ + A G + ++ E+ QI + + I ++V+ ++F++I K FDL
Sbjct: 94 LCFLAVTAIGGVFTTANPLYTVNEVSKQIKDSNPKIIISVN--QLFDKI-KGFDLP---- 146
Query: 228 FVILLWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIAT 287
V+LL + + + GS I +F +M L + Y F IK D A
Sbjct: 147 -VVLLGSKDTVEIPPGSNS-KILSFDNVMELS----------EPVSEYPFVEIKQSDTAA 194
Query: 288 LVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KS--L*VFHFFMRC*PSIYNCEKL 345
L+Y+SGTTG KGV LTH N + AASL + +H C +++ L
Sbjct: 195 LLYSSGTTGTSKGVELTHGNFI------AASLMVTMDQDLMGEYHGVFLCFLPMFHVFGL 248
Query: 346 EVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAM 405
V YS +QR + S+ R +E + + KN +
Sbjct: 249 AVIT--YSQLQRGNALVSMAR--------------------FELELVLKNIE-------- 278
Query: 406 LDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEA----I 461
+ T L+ + + L I S G G+ P D E
Sbjct: 279 ------KFRVTHLWVVPPVFLALSKQSIVKKFDLSSLKYIGSGAAPLGKDLMEECGRNIP 332
Query: 462 GVTLQNGYGLTETSPVIAAR--RLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
V L GYG+TET +++ RL GS G E ++V ETG+ PP +G +
Sbjct: 333 NVLLMQGYGMTETCGIVSVEDPRLGKRNSGSAGMLAPGVEAQIVSVETGKSQPPNQQGEI 392
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
VRGP +MKGY NP AT + IDK W++TGD+G+
Sbjct: 393 WVRGPNMMKGYLNNPQATKETIDKKSWVHTGDLGY 427
>At4g23850 acyl-CoA synthetase - like protein
Length = 666
Score = 124 bits (311), Expect = 2e-28
Identities = 122/505 (24%), Positives = 209/505 (41%), Gaps = 104/505 (20%)
Query: 104 DIWRSSAEKYGDKIAL-----IDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIA 158
D++R S EKY + L +D TY+++ D ++ LR +GV K
Sbjct: 49 DVFRMSVEKYPNNPMLGRREIVDGKPGKYVWQTYQEVYDIVMKLGNSLRSVGVKDEAKCG 108
Query: 159 LFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAK 218
++ NS W+++ + A G V + + I +HSE + V+ ++
Sbjct: 109 IYGANSPEWIISMEACNAHGLYCVPLYDTLGADAVEFIISHSEVSIVFVEEKKISELFKT 168
Query: 219 AFDLKASMRFVILLWGEKSCLVNEGSKE------VPIFTFTEIMHLGRGSRRLFESHDAR 272
+ M+ V+ G V+ KE + I+ + E + LG G
Sbjct: 169 CPNSTEYMKTVVSFGG-----VSREQKEEAETFGLVIYAWDEFLKLGEG----------- 212
Query: 273 KHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*V---- 328
K Y K DI T++YTSGTTG+PKGVM+++++++ I+ L ++L V
Sbjct: 213 KQYDLPIKKKSDICTIMYTSGTTGDPKGVMISNESIVTLIAGVIRLLKSANEALTVKDVY 272
Query: 329 ------FHFFMR-----------------------------C*PSIYNCEKLEVYESLYS 353
H F R P+I+ C V + +YS
Sbjct: 273 LSYLPLAHIFDRVIEECFIQHGAAIGFWRGDVKLLIEDLAELKPTIF-CAVPRVLDRVYS 331
Query: 354 GIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARI 413
G+Q+++S ++K + + GYM +K S++ A
Sbjct: 332 GLQKKLSDGGFLKKFIFDSAFSYKFGYM--------------KKGQSHVEA--------- 368
Query: 414 IATILFPIHMLAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFEAIGVT-LQNGYGL 471
L KLV+SK+ +G + + +SG L SHV+ F + + GYGL
Sbjct: 369 --------SPLFDKLVFSKVKQGLGGNVRIILSGAAPLASHVESFLRVVACCHVLQGYGL 420
Query: 472 TET-SPVIAARRLSCNVIGSVGHPLKHTEFKV--VDSETGEVLPPGYKGILKVRGPQLMK 528
TE+ + + ++G+VG P+ + + ++ V + L +G + +RG L
Sbjct: 421 TESCAGTFVSLPDELGMLGTVGPPVPNVDIRLESVPEMEYDALASTARGEICIRGKTLFS 480
Query: 529 GYYKNPSATNQAIDKDGWLNTGDIG 553
GYYK T + + DGWL+TGD+G
Sbjct: 481 GYYKREDLTKEVL-IDGWLHTGDVG 504
>At3g05970 AMP-binding protein
Length = 701
Score = 122 bits (307), Expect = 5e-28
Identities = 120/444 (27%), Positives = 190/444 (42%), Gaps = 42/444 (9%)
Query: 130 MTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSS 189
MTY + A GL G+ + ++ N WL+ D + ++V
Sbjct: 119 MTYGEAGTARTALGSGLVHHGIPMGSSVGIYFINRPEWLIVDHACSSYSYVSVPLYDTLG 178
Query: 190 IEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVN-EGSKEVP 248
+ + I NH+ A+ E N + S+R V+++ G L + S V
Sbjct: 179 PDAVKFIVNHATVQAIFCV-AETLNSLLSCLSEMPSVRLVVVVGGLIESLPSLPSSSGVK 237
Query: 249 IFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNL 308
+ +++ +++ GR + + F K DD+AT+ YTSGTTG PKGV+LTH NL
Sbjct: 238 VVSYSVLLNQGRSNPQRFFPP-----------KPDDVATICYTSGTTGTPKGVVLTHANL 286
Query: 309 LHQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQ---RQISTSSLV 365
+ ++ + S+ + + + P + E+ ++Y G+ Q L+
Sbjct: 287 IANVAGSSFSVKFFSSDVYISYL-----PLAHIYERANQILTVYFGVAVGFYQGDNMKLL 341
Query: 366 RKLVALTFIRVSLGYMECKRIYEGKC--------LTKNQKSPSYLYAMLDWLGARIIATI 417
L AL S RIY G L + + +Y L + + I
Sbjct: 342 DDLAALRPTVFSSVPRLYNRIYAGIINAVKTSGGLKERLFNAAYNAKKQALLNGKSASPI 401
Query: 418 LFPIHMLAKKLVYSKIHSAIGFSKAGISGGGS-LPSHVDRFFEA-IGVTLQNGYGLTETS 475
+LV++KI +G ++ G S L V F + G + GYG+TETS
Sbjct: 402 W-------DRLVFNKIKDRLGGRVRFMTSGASPLSPEVMEFLKVCFGGRVTEGYGMTETS 454
Query: 476 PVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLP---PGYKGILKVRGPQLMKGYYK 532
VI+ N+ G VG P E K+VD P +G + VRGP + GYYK
Sbjct: 455 CVISGMDEGDNLTGHVGSPNPACEVKLVDVPEMNYTSADQPHPRGEICVRGPIIFTGYYK 514
Query: 533 NPSATNQAIDKDGWLNTGDIG-WI 555
+ T + ID+DGWL+TGDIG W+
Sbjct: 515 DEIQTKEVIDEDGWLHTGDIGLWL 538
>At1g64400 putative acyl-CoA synthetase
Length = 665
Score = 122 bits (306), Expect = 6e-28
Identities = 121/529 (22%), Positives = 215/529 (39%), Gaps = 96/529 (18%)
Query: 76 NPRFSPLLESSLLSGNDGVVSDEWKTVPDIWRSSAEKYGDKIAL-----IDPYHDPPSTM 130
+P P+ S D+ + DI+R S EK + L +D
Sbjct: 21 SPSVGPVYRSIYAKDGFPEPPDDLVSAWDIFRLSVEKSPNNPMLGRREIVDGKAGKYVWQ 80
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSI 190
TYK++ + ++ +R IGV +K ++ NS W+++ + A G V
Sbjct: 81 TYKEVHNVVIKLGNSIRTIGVGKGDKCGIYGANSPEWIISMEACNAHGLYCVPLYDTLGA 140
Query: 191 EELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGE--KSCLVNEGSKEVP 248
+ I H+E ++LA + + K I+ +GE + V +
Sbjct: 141 GAIEFIICHAE-VSLAFAEENKISELLKTAPKSTKYLKYIVSFGEVTNNQRVEAERHRLT 199
Query: 249 IFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNL 308
I+++ + + LG G KHY + D+ T++YTSGTTG+PKGV+LT++++
Sbjct: 200 IYSWDQFLKLGEG-----------KHYELPEKRRSDVCTIMYTSGTTGDPKGVLLTNESI 248
Query: 309 LHQISKYAASLACI*KS----------L*VFHFFMR------------------------ 334
+H + L I + L + H F R
Sbjct: 249 IHLLEGVKKLLKTIDEELTSKDVYLSYLPLAHIFDRVIEELCIYEAASIGFWRGDVKILI 308
Query: 335 -----C*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEG 389
P+++ C V E +Y+G+Q+++S V+K + + ME + +E
Sbjct: 309 EDIAALKPTVF-CAVPRVLERIYTGLQQKLSDGGFVKKKLFNFAFKYKHKNMEKGQPHE- 366
Query: 390 KCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS-KAGISGGG 448
Q SP +A K+V+ K+ +G + + +SG
Sbjct: 367 ------QASP------------------------IADKIVFKKVKEGLGGNVRLILSGAA 396
Query: 449 SLPSHVDRFFEAIGVT-LQNGYGLTET-SPVIAARRLSCNVIGSVGHPLKHTEFKV--VD 504
L +H++ F + + GYGLTE+ + +++G+VG P+ + + ++ V
Sbjct: 397 PLAAHIESFLRVVACAHVLQGYGLTESCGGTFVSIPNELSMLGTVGPPVPNVDIRLESVP 456
Query: 505 SETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIG 553
+ L +G + +RG L GYYK T Q + DGWL+TGD+G
Sbjct: 457 EMGYDALASNPRGEICIRGKTLFSGYYKREDLT-QEVFIDGWLHTGDVG 504
>At4g11030 putative acyl-CoA synthetase
Length = 666
Score = 121 bits (303), Expect = 1e-27
Identities = 130/531 (24%), Positives = 217/531 (40%), Gaps = 100/531 (18%)
Query: 76 NPRFSPLLESSLLSGNDGVVSDEWKTVPDIWRSSAEKYGDKIAL-----IDPYHDPPSTM 130
NP P+ S+ D ++ DI+R++ EKY + L +
Sbjct: 21 NPSVGPVYRSTFAQNGFPNPIDGIQSCWDIFRTAVEKYPNNRMLGRREISNGKAGKYVWK 80
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSI 190
TYK++ D ++ LR G+ EK ++ N C W+++ + A G V
Sbjct: 81 TYKEVYDIVIKLGNSLRSCGIKEGEKCGIYGINCCEWIISMEACNAHGLYCVPLYDTLGA 140
Query: 191 EELLQIYNHSE-SIALAVDN--PEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSK-E 246
+ I +H+E SIA + PE+F + M+ V+ G K E K
Sbjct: 141 GAVEFIISHAEVSIAFVEEKKIPELFKTCPNSTKY---MKTVVSFGGVKPEQKEEAEKLG 197
Query: 247 VPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQ 306
+ I ++ E + LG G K Y K DI T++YTSGTTG+PKGVM++++
Sbjct: 198 LVIHSWDEFLKLGEG-----------KQYELPIKKPSDICTIMYTSGTTGDPKGVMISNE 246
Query: 307 NL-------LHQISKYAASLA---CI*KSL*VFHFFMRC--------------------- 335
++ +H + ASL+ L + H F R
Sbjct: 247 SIVTITTGVMHFLGNVNASLSEKDVYISYLPLAHVFDRAIEECIIQVGGSIGFWRGDVKL 306
Query: 336 --------*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIY 387
PSI+ C V + +Y+G+Q+++S +K V G M
Sbjct: 307 LIEDLGELKPSIF-CAVPRVLDRVYTGLQQKLSGGGFFKKKVFDVAFSYKFGNM------ 359
Query: 388 EGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS-KAGISG 446
+K S++ A KLV++K+ +G + + +SG
Sbjct: 360 --------KKGQSHVAA-----------------SPFCDKLVFNKVKQGLGGNVRIILSG 394
Query: 447 GGSLPSHVDRFFEAIG-VTLQNGYGLTET-SPVIAARRLSCNVIGSVGHPLKHTEFKV-- 502
L SH++ F + + GYGLTE+ + A +++G+VG P+ + + ++
Sbjct: 395 AAPLASHIESFLRVVACCNVLQGYGLTESCAGTFATFPDELDMLGTVGPPVPNVDIRLES 454
Query: 503 VDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIG 553
V + L +G + +RG L GYYK T + + DGWL+TGD+G
Sbjct: 455 VPEMNYDALGSTPRGEICIRGKTLFSGYYKREDLTKE-VFIDGWLHTGDVG 504
>At5g27600 long chain acyl-CoA synthetase 7 (LACS7)
Length = 698
Score = 120 bits (300), Expect = 3e-27
Identities = 119/458 (25%), Positives = 188/458 (40%), Gaps = 68/458 (14%)
Query: 128 STMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSR 187
S MTY + GL GV+ + + L+ N WLV D A ++V
Sbjct: 117 SWMTYGEAASERQAIGSGLLFHGVNQGDCVGLYFINRPEWLVVDHACAAYSFVSVPLYDT 176
Query: 188 SSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEV 247
+ + + NH+ A+ P+ N + + C +E +
Sbjct: 177 LGPDAVKFVVNHANLQAIFCV-PQTLNIVIAKLPSGNPIH-------SSHCGADEHLPSL 228
Query: 248 P------IFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGV 301
P I ++ +++ GR S + F K +DIAT+ YTSGTTG PKGV
Sbjct: 229 PRGTGVTIVSYQKLLSQGRSSL-----------HPFSPPKPEDIATICYTSGTTGTPKGV 277
Query: 302 MLTHQNLLHQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQRQIST 361
+LTH NL+ ++ + + + + P + E+ +Y G+
Sbjct: 278 VLTHGNLIANVAGSSVEAEFFPSDVYISYL-----PLAHIYERANQIMGVYGGVAVGFYQ 332
Query: 362 SSLVRKLVALTFIRVSLGYMECK---RIYEGKCLTKNQKSPSYLYAMLDWLGARIIATIL 418
+ + + +R ++ + RIY+G +T KS ++ L
Sbjct: 333 GDVFKLMDDFAVLRPTIFCSVPRLYNRIYDG--ITSAVKSSG------------VVKKRL 378
Query: 419 FPIHMLAKK---------------LVYSKIHSAIGFSKAGISGGGS--LPSHVDRFFEAI 461
F I +KK LV++KI +G + G S P +D
Sbjct: 379 FEIAYNSKKQAIINGRTPSAFWDKLVFNKIKEKLGGRVRFMGSGASPLSPDVMDFLRICF 438
Query: 462 GVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLP---PGYKGI 518
G +++ GYG+TETS VI+A N+ G VG P E K+VD P +G
Sbjct: 439 GCSVREGYGMTETSCVISAMDDGDNLSGHVGSPNPACEVKLVDVPEMNYTSDDQPYPRGE 498
Query: 519 LKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIG-WI 555
+ VRGP + KGYYK+ T + +D DGWL+TGDIG W+
Sbjct: 499 ICVRGPIIFKGYYKDEEQTREILDGDGWLHTGDIGLWL 536
>At1g49430 acyl CoA synthetase-like protein (At1g49430)
Length = 665
Score = 114 bits (285), Expect = 2e-25
Identities = 123/479 (25%), Positives = 196/479 (40%), Gaps = 100/479 (20%)
Query: 126 PPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRG 185
P + +TYK+ DA + +R GV P ++ N W++A + M+ G V
Sbjct: 76 PYTWITYKEAHDAAIRIGSAIRSRGVDPGHCCGIYGANCPEWIIAMEACMSQGITYVPLY 135
Query: 186 SRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSK 245
+ + I NH+E + V + + ++ ++++ I+ +GE S E +K
Sbjct: 136 DSLGVNAVEFIINHAEVSLVFVQEKTVSSILSCQKGCSSNLK-TIVSFGEVSSTQKEEAK 194
Query: 246 E--VPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVML 303
V +F++ E +G L E++ RK + DI T++YTSGTTG PKGV+L
Sbjct: 195 NQCVSLFSWNEFSLMGN----LDEANLPRK-------RKTDICTIMYTSGTTGEPKGVIL 243
Query: 304 THQNLLHQISKYAASLACI*KSL*VFHFFMRC*P---------SIYN------------- 341
+ + Q+ L +S F P IY
Sbjct: 244 NNAAISVQVLSIDKMLEVTDRSCDTSDVFFSYLPLAHCYDQVMEIYFLSRGSSVGYWRGD 303
Query: 342 ----------------CEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKR 385
C VY+ LY+GI ++IS S L+RK + LG M
Sbjct: 304 IRYLMDDVQALKPTVFCGVPRVYDKLYAGIMQKISASGLIRKKLFDFAYNYKLGNMR--- 360
Query: 386 IYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAI-GFSKAGI 444
K ++ + SP LD +L++ KI A+ G + +
Sbjct: 361 ----KGFSQEEASP-----RLD-------------------RLMFDKIKEALGGRAHMLL 392
Query: 445 SGGGSLPSHVDRFFEAIGVT-LQNGYGLTET-SPVIAARRLSCNVIGSVGHPLKHTEFKV 502
SG LP HV+ F I + L GYGLTE+ +++G+VG P+ E ++
Sbjct: 393 SGAAPLPRHVEEFLRIIPASNLSQGYGLTESCGGSFTTLAGVFSMVGTVGVPMPTVEARL 452
Query: 503 VDSETGEVLPPGY--------KGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIG 553
V V GY +G + +RG + GY+K T+Q + DGW +TGDIG
Sbjct: 453 V-----SVPEMGYDAFSADVPRGEICLRGNSMFSGYHKRQDLTDQVL-IDGWFHTGDIG 505
>At1g77590 long chain acyl-CoA synthetase 9 (LACS9)
Length = 691
Score = 108 bits (269), Expect = 1e-23
Identities = 135/529 (25%), Positives = 219/529 (40%), Gaps = 95/529 (17%)
Query: 95 VSDEWK---TVPDIWRSSAEKYGDKIAL---------IDPYHDPPST----------MTY 132
VS W+ T+P+++ S + D++ L I+ D + +T+
Sbjct: 50 VSSHWEHISTLPELFEISCNAHSDRVFLGTRKLISREIETSEDGKTFEKLHLGDYEWLTF 109
Query: 133 KQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEE 192
+ +A+ DFA GL IG E++A+FAD W ++ QG V S E
Sbjct: 110 GKTLEAVCDFASGLVQIGHKTEERVAIFADTREEWFISLQGCFRRNVTVVTIYSSLGEEA 169
Query: 193 LLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVP---- 248
L N +E + + E+ + + L+ R + C+ +E +V
Sbjct: 170 LCHSLNETEVTTVICGSKELKKLMDISQQLETVKRVI--------CMDDEFPSDVNSNWM 221
Query: 249 IFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNL 308
+FT++ LGR + ++ A D+A ++YTSG+TG PKGVM+TH N+
Sbjct: 222 ATSFTDVQKLGR-------ENPVDPNFPLSA----DVAVIMYTSGSTGLPKGVMMTHGNV 270
Query: 309 LHQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQRQI--------- 359
L +S ++ I L +M P + E ES+ + I I
Sbjct: 271 LATVS----AVMTIVPDLGKRDIYMAYLPLAHILEL--AAESVMATIGSAIGYGSPLTLT 324
Query: 360 STSSLVRK-----LVALTFIRVSLGYMECKRIYEG---KCLTK---NQKSPSYLYAML-- 406
TS+ ++K + AL ++ R+ +G K K ++K + YA
Sbjct: 325 DTSNKIKKGTKGDVTALKPTIMTAVPAILDRVRDGVRKKVDAKGGLSKKLFDFAYARRLS 384
Query: 407 ----DWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFE-A 460
W GA + +L+ + LV+ KI + +G + +SGG L RF
Sbjct: 385 AINGSWFGAWGLEKLLWDV------LVFRKIRAVLGGQIRYLLSGGAPLSGDTQRFINIC 438
Query: 461 IGVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVL---PPGYKG 517
+G + GYGLTET +G VG PL + K+VD G L P +G
Sbjct: 439 VGAPIGQGYGLTETCAGGTFSEFEDTSVGRVGAPLPCSFVKLVDWAEGGYLTSDKPMPRG 498
Query: 518 ILKVRGPQLMKGYYKNPSATNQA--IDKDG--WLNTGDIGWIAAYHSSG 562
+ + G + GY+KN T + +D+ G W TGDIG +H G
Sbjct: 499 EIVIGGSNITLGYFKNEEKTKEVYKVDEKGMRWFYTGDIG---RFHPDG 544
>At1g20480 unknown protein
Length = 565
Score = 102 bits (255), Expect = 5e-22
Identities = 105/414 (25%), Positives = 177/414 (42%), Gaps = 59/414 (14%)
Query: 146 LRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIAL 205
L +GV + + + NS + + +M+ GAI ++ +E+ + S + L
Sbjct: 91 LYALGVRKGNVVIILSPNSILFPIVSLSVMSLGAIITTANPINTSDEISKQIGDSRPV-L 149
Query: 206 AVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRL 265
A ++ +++A A + V+L+ VP ++ + + L +
Sbjct: 150 AFTTCKLVSKLAAASNFNLP---VVLM----------DDYHVPSQSYGDRVKLVGRLETM 196
Query: 266 FESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KS 325
E+ + V + + DD A L+Y+SGTTG KGVML+H+NL+ + Y A
Sbjct: 197 IETEPSESR-VKQRVNQDDTAALLYSSGTTGTSKGVMLSHRNLIALVQAYRAR------- 248
Query: 326 L*VFHFFMRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKR 385
F R +I C L+AL + V L + +
Sbjct: 249 ---FGLEQRTICTIPMCHIF--------------GFGGFATGLIALGWTIVVLPKFDMAK 291
Query: 386 IYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGIS 445
+ + +S SYL L I+ ++ + + K S +H+ + +
Sbjct: 292 LLSA---VETHRS-SYLS-----LVPPIVVAMVNGANEINSKYDLSSLHTVV-------A 335
Query: 446 GGGSLPSHV-DRFFEAIG-VTLQNGYGLTETSPVIAA--RRLSCNVIGSVGHPLKHTEFK 501
GG L V ++F E V + GYGLTE++ + A+ + G+ G + E K
Sbjct: 336 GGAPLSREVTEKFVENYPKVKILQGYGLTESTAIAASMFNKEETKRYGASGLLAPNVEGK 395
Query: 502 VVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+VD +TG VL G L +R P +MKGY+KN AT ID +GWL TGD+ +I
Sbjct: 396 IVDPDTGRVLGVNQTGELWIRSPTVMKGYFKNKEATASTIDSEGWLKTGDLCYI 449
>At2g47240 putative acyl-CoA synthetase
Length = 660
Score = 99.8 bits (247), Expect = 4e-21
Identities = 130/552 (23%), Positives = 223/552 (39%), Gaps = 92/552 (16%)
Query: 77 PRFSPLLESSLLSGNDGVVSDEWKTVPDIWRSSAEKYGDKIAL-----IDPYHDPPSTMT 131
P P+ + L + E T DI+ S EK+ D L +D P T
Sbjct: 19 PSVGPVYRNLLSEKGFPPIDSEITTAWDIFSKSVEKFPDNNMLGWRRIVDEKVGPYMWKT 78
Query: 132 YKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIE 191
YK++ + +L LR G P ++ ++ N +W++A + A I V
Sbjct: 79 YKEVYEEVLQIGSALRAAGAEPGSRVGIYGVNCPQWIIAMEACAAHTLICVPLYDTLGSG 138
Query: 192 ELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRF-VILLWGEKSCLVNEGSKEVPIF 250
+ I H+E + V + ++ + D K + R I+ + S ++ + E+ +
Sbjct: 139 AVDYIVEHAEIDFVFVQDTKIKGLLEP--DCKCAKRLKAIVSFTNVSDELSHKASEIGVK 196
Query: 251 TFT--EIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNL 308
T++ + +H+GR K K+ +I T++YTSGT+G+PKGV+ L
Sbjct: 197 TYSWIDFLHMGR-----------EKPEDTNPPKAFNICTIMYTSGTSGDPKGVV-----L 240
Query: 309 LHQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLY------SGIQRQISTS 362
HQ A + + L + F E ++ +Y + I +++
Sbjct: 241 THQ----AVATFVVGMDLYMDQF-----------EDKMTHDDVYLSFLPLAHILDRMNEE 285
Query: 363 SLVRKLVALTFIRVSLGYME-----------------CKRIYEGKCLTKNQKSPSYLYAM 405
RK ++ + +L + +RI+EG + +P +
Sbjct: 286 YFFRKGASVGYYHGNLNVLRDDIQELKPTYLAGVPRVFERIHEGIQKALQELNPRRRFIF 345
Query: 406 -------LDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRF 457
L WL + P +A + + KI +G + +SGG L ++ F
Sbjct: 346 NALYKHKLAWLNRGYSHSKASP---MADFIAFRKIRDKLGGRIRLLVSGGAPLSPEIEEF 402
Query: 458 FEAIGVT-LQNGYGLTET--SPVIAARRLSCNVIGSVGHPLKHTEFKVVD-SETG-EVLP 512
+ GYGLTET + C ++G+VG P + E ++ + SE G + L
Sbjct: 403 LRVTCCCFVVQGYGLTETLGGTALGFPDEMC-MLGTVGIPAVYNEIRLEEVSEMGYDPLG 461
Query: 513 PGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSGRSRNCGGVIV 572
G + +RG + GYYKNP T + + KDGW +TGDIG I GV+
Sbjct: 462 ENPAGEICIRGQCMFSGYYKNPELTEEVM-KDGWFHTGDIGEILP----------NGVLK 510
Query: 573 VEGRAKDTIVLS 584
+ R K+ I LS
Sbjct: 511 IIDRKKNLIKLS 522
>At5g63380 4-coumarate-CoA ligase-like protein
Length = 562
Score = 92.0 bits (227), Expect = 9e-19
Identities = 83/279 (29%), Positives = 127/279 (44%), Gaps = 46/279 (16%)
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*VFHFFMRC*PSI 339
+ D A ++++SGTTG KGV+LTH+NL+ AS A V H R
Sbjct: 201 VNQSDPAAILFSSGTTGRVKGVLLTHRNLI-------ASTA-------VSH--QRTLQDP 244
Query: 340 YNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIY---EGKCLTKNQ 396
N +++ L+S + ++ + ++L V LG E + ++ E +T
Sbjct: 245 VNYDRV----GLFSLPLFHVFGFMMMIRAISLGETLVLLGRFELEAMFKAVEKYKVTGMP 300
Query: 397 KSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDR 456
SP + A++ K +K + G G +R
Sbjct: 301 VSPPLIVALV--------------------KSELTKKYDLRSLRSLGCGGAPLGKDIAER 340
Query: 457 FFEAI-GVTLQNGYGLTETSPVIAARRLSCNVI--GSVGHPLKHTEFKVVDSETGEVLPP 513
F + V + GYGLTE+S A+ ++ GSVG ++ E K+VD TGE LPP
Sbjct: 341 FKQKFPDVDIVQGYGLTESSGPAASTFGPEEMVKYGSVGRISENMEAKIVDPSTGESLPP 400
Query: 514 GYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDI 552
G G L +RGP +MKGY N A+ + +DK+GWL TGD+
Sbjct: 401 GKTGELWLRGPVIMKGYVGNEKASAETVDKEGWLKTGDL 439
>At1g51680 4-coumarate--coenzyme A ligase (At4CL1)
Length = 561
Score = 87.8 bits (216), Expect = 2e-17
Identities = 82/295 (27%), Positives = 132/295 (43%), Gaps = 67/295 (22%)
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISK---------YAASLACI*KSL*VFH 330
I DD+ L Y+SGTTG PKGVMLTH+ L+ +++ Y S I L +FH
Sbjct: 199 ISPDDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYFHSDDVILCVLPMFH 258
Query: 331 FF---------MRC*PSIYNCEKLEVYESLYSGIQR-QISTSSLVRKLVALTFIRVSLGY 380
+ +R +I K E+ L IQR +++ + +V +V
Sbjct: 259 IYALNSIMLCGLRVGAAILIMPKFEI-NLLLELIQRCKVTVAPMVPPIVL---------- 307
Query: 381 MECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS 440
+ K+ ++ Y D R++ + P+ + V +K +A
Sbjct: 308 ----------AIAKSSETEKY-----DLSSIRVVKSGAAPLGKELEDAVNAKFPNAKLGQ 352
Query: 441 KAGISGGGSLPSHVDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEF 500
G++ G + + + G+ E PV + +C + +++ E
Sbjct: 353 GYGMTEAGPV------------LAMSLGFA-KEPFPVKSG---ACGTV------VRNAEM 390
Query: 501 KVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
K+VD +TG+ L G + +RG Q+MKGY NP+AT + IDKDGWL+TGDIG I
Sbjct: 391 KIVDPDTGDSLSRNQPGEICIRGHQIMKGYLNNPAATAETIDKDGWLHTGDIGLI 445
>At1g65060 4-coumarate:CoA ligase 3 (4CL3)
Length = 561
Score = 86.7 bits (213), Expect = 4e-17
Identities = 84/300 (28%), Positives = 129/300 (43%), Gaps = 77/300 (25%)
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISK---------YAASLACI*KSL*VFH 330
I DD A L ++SGTTG PKGV+LTH++L+ +++ Y S I L +FH
Sbjct: 202 IGGDDAAALPFSSGTTGLPKGVVLTHKSLITSVAQQVDGDNPNLYLKSNDVILCVLPLFH 261
Query: 331 FF---------MRC*PSIYNCEKLEVYESLYSGIQR-QISTSSLVRKLVALTFIRVSLGY 380
+ +R ++ K E+ +L IQR +++ ++LV LV
Sbjct: 262 IYSLNSVLLNSLRSGATVLLMHKFEI-GALLDLIQRHRVTIAALVPPLVI---------- 310
Query: 381 MECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS 440
L KN SY D R + + P+ + + ++ AI
Sbjct: 311 ----------ALAKNPTVNSY-----DLSSVRFVLSGAAPLGKELQDSLRRRLPQAI--- 352
Query: 441 KAGISGGGSLPSHVDRFFEAIGVTLQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPL 495
L GYG+TE PV++ A+ GS G +
Sbjct: 353 ------------------------LGQGYGMTEAGPVLSMSLGFAKEPIPTKSGSCGTVV 388
Query: 496 KHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
++ E KVV ET L G + +RG Q+MK Y +P AT+ ID++GWL+TGDIG++
Sbjct: 389 RNAELKVVHLETRLSLGYNQPGEICIRGQQIMKEYLNDPEATSATIDEEGWLHTGDIGYV 448
>At3g21230 putative 4-coumarate:CoA ligase 2
Length = 570
Score = 85.9 bits (211), Expect = 6e-17
Identities = 49/121 (40%), Positives = 71/121 (58%), Gaps = 12/121 (9%)
Query: 444 ISGGGSLPSHVDRFFEAIGVTLQN-----GYGLTETSPVIAARRLSCNVI----GSVGHP 494
+SG +L ++ +A+ + N GYG+TE+ V + + N G+ G
Sbjct: 335 LSGAATLKKELE---DAVRLKFPNAIFGQGYGMTESGTVAKSLAFAKNPFKTKSGACGTV 391
Query: 495 LKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
+++ E KVVD+ETG LP G + VRG QLMKGY +P AT + IDKDGWL+TGDIG+
Sbjct: 392 IRNAEMKVVDTETGISLPRNKSGEICVRGHQLMKGYLNDPEATARTIDKDGWLHTGDIGF 451
Query: 555 I 555
+
Sbjct: 452 V 452
Score = 40.4 bits (93), Expect = 0.003
Identities = 17/35 (48%), Positives = 25/35 (70%)
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISK 314
I +D + Y+SGTTG PKGVM+TH+ L+ I++
Sbjct: 207 ISPEDTVAMPYSSGTTGLPKGVMITHKGLVTSIAQ 241
>At3g21240 putative 4-coumarate:CoA ligase 2
Length = 556
Score = 83.6 bits (205), Expect = 3e-16
Identities = 42/96 (43%), Positives = 60/96 (61%), Gaps = 5/96 (5%)
Query: 465 LQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
L GYG+TE PV+A A+ G+ G +++ E K++D +TG+ LP G +
Sbjct: 343 LGQGYGMTEAGPVLAMSLGFAKEPFPVKSGACGTVVRNAEMKILDPDTGDSLPRNKPGEI 402
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG Q+MKGY +P AT IDKDGWL+TGD+G+I
Sbjct: 403 CIRGNQIMKGYLNDPLATASTIDKDGWLHTGDVGFI 438
Score = 46.2 bits (108), Expect = 5e-05
Identities = 21/50 (42%), Positives = 33/50 (66%)
Query: 265 LFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISK 314
L +S + R + E I +D+ L ++SGTTG PKGVMLTH+ L+ +++
Sbjct: 177 LTQSEEPRVDSIPEKISPEDVVALPFSSGTTGLPKGVMLTHKGLVTSVAQ 226
>At4g19010 4-coumarate-CoA ligase - like
Length = 566
Score = 74.3 bits (181), Expect = 2e-13
Identities = 41/95 (43%), Positives = 55/95 (57%), Gaps = 4/95 (4%)
Query: 463 VTLQNGYGLTETSPVIAARRLSCNVIG---SVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
V L GYG+TE S + R + + SVG + + KVVD +G LPPG +G L
Sbjct: 352 VDLIQGYGMTE-STAVGTRGFNSEKLSRYSSVGLLAPNMQAKVVDWSSGSFLPPGNRGEL 410
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
++GP +MKGY NP AT +I +D WL TGDI +
Sbjct: 411 WIQGPGVMKGYLNNPKATQMSIVEDSWLRTGDIAY 445
Score = 48.9 bits (115), Expect = 8e-06
Identities = 53/201 (26%), Positives = 90/201 (44%), Gaps = 26/201 (12%)
Query: 110 AEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGL-RVIGVSPNEKIALFADNSCRWL 168
+ K+ ALID ++++ +L+ + A G+ V+GV + ++L NS +
Sbjct: 55 SHKHHGDTALIDSLTG--FSISHTELQIMVQSMAAGIYHVLGVRQGDVVSLVLPNSVYFP 112
Query: 169 VADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRF 228
+ +++ GAI SS+ E+ + + S+ LA + E + K L S+
Sbjct: 113 MIFLSLISLGAIVTTMNPSSSLGEIKKQVSEC-SVGLAFTSTE---NVEKLSSLGVSVIS 168
Query: 229 VILLWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATL 288
V + S + E P F G + L IK DD+A +
Sbjct: 169 VSESYDFDSIRI-----ENPKFYSIMKESFGFVPKPL--------------IKQDDVAAI 209
Query: 289 VYTSGTTGNPKGVMLTHQNLL 309
+Y+SGTTG KGV+LTH+NL+
Sbjct: 210 MYSSGTTGASKGVLLTHRNLI 230
>At1g20510 4-coumarate:CoA ligase like protein
Length = 546
Score = 72.0 bits (175), Expect = 9e-13
Identities = 41/116 (35%), Positives = 62/116 (53%), Gaps = 4/116 (3%)
Query: 444 ISGGGSLPSHVDRFFEAIGVTLQ--NGYGLTETSPVIAARRL--SCNVIGSVGHPLKHTE 499
+ GG L V F T++ GYGLTE++ + A+ G+ G E
Sbjct: 313 LCGGAPLSKEVTEGFAEKYPTVKILQGYGLTESTGIGASTDTVEESRRYGTAGKLSASME 372
Query: 500 FKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
++VD TG++L P G L ++GP +MKGY+ N AT+ +D +GWL TGD+ +I
Sbjct: 373 GRIVDPVTGQILGPKQTGELWLKGPSIMKGYFSNEEATSSTLDSEGWLRTGDLCYI 428
Score = 57.4 bits (137), Expect = 2e-08
Identities = 54/207 (26%), Positives = 91/207 (43%), Gaps = 30/207 (14%)
Query: 109 SAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWL 168
S++ + +IA ID +T+ +L A+ A+ L IG+ + L + NS +
Sbjct: 39 SSQAHRGRIAFIDA--STGQNLTFTELWRAVESVADCLSEIGIRKGHVVLLLSPNSILFP 96
Query: 169 VADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRF 228
V +M+ GAI ++ E+ + S + LA ++ +I+ A
Sbjct: 97 VVCLSVMSLGAIITTTNPLNTSNEIAKQIKDSNPV-LAFTTSQLLPKISAA--------- 146
Query: 229 VILLWGEKSCLVNEGSKEVPIFTFTEIMHLGRGS-RRLFE--SHDARKHYVFEAIKSDDI 285
+K++PI E G RRL E + + V E + DD
Sbjct: 147 ---------------AKKLPIVLMDEERVDSVGDVRRLVEMMKKEPSGNRVKERVDQDDT 191
Query: 286 ATLVYTSGTTGNPKGVMLTHQNLLHQI 312
ATL+Y+SGTTG KGV+ +H+NL+ +
Sbjct: 192 ATLLYSSGTTGMSKGVISSHRNLIAMV 218
>At1g66120 unknown protein
Length = 572
Score = 68.9 bits (167), Expect = 8e-12
Identities = 52/163 (31%), Positives = 83/163 (50%), Gaps = 27/163 (16%)
Query: 444 ISGGGSLPSHVDRFFEAIGVTLQNGYGLTE-TSPVIAA------------RRLSCNVIGS 490
++GG S P+ + + E +G + +GYGLTE T PV+ +++
Sbjct: 306 LTGGSSPPAVLIKKVEQLGFHVMHGYGLTEATGPVLFCEWQDEWNKLPEHQQIELQQRQG 365
Query: 491 VGHPLKHTEFKVVDSETGEVLPPGYK--GILKVRGPQLMKGYYKNPSATNQAIDKDGWLN 548
V + L + V +++T E +P K G + ++G LMKGY KNP AT++A K GWLN
Sbjct: 366 VRN-LTLADVDVKNTKTLESVPRDGKTMGEIVIKGSSLMKGYLKNPKATSEAF-KHGWLN 423
Query: 549 TGDIGWIAAYHSSGRSRNCGGVIVVEGRAKDTIVLSSEKVKML 591
TGDIG I + G + ++ R+KD I+ E + +
Sbjct: 424 TGDIGVI----------HPDGYVEIKDRSKDIIISGGENISSI 456
Score = 41.6 bits (96), Expect = 0.001
Identities = 42/181 (23%), Positives = 71/181 (39%), Gaps = 17/181 (9%)
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSI 190
T+ Q D A L + ++ N+ +++ A N + TGA+ +R
Sbjct: 41 TWPQTYDRCCRLAASLLSLNITRNDVVSILAPNVPAMYEMHFSVPMTGAVLNPINTRLDA 100
Query: 191 EELLQIYNHSESIALAVDNP-----EMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSK 245
+ + I H+E L VD + R+ + + R +++ E SK
Sbjct: 101 KTIAIILRHAEPKILFVDYEFAPLIQEVLRLIPTYQSQPHPRIILI--NEIDSTTKPFSK 158
Query: 246 EVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTH 305
E+ G R E + +F D +L YTSGTT +PKGV+++H
Sbjct: 159 ELDY----------EGLIRKGEPTPSSSASMFRVHNEHDPISLNYTSGTTADPKGVVISH 208
Query: 306 Q 306
Q
Sbjct: 209 Q 209
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,533,626
Number of Sequences: 26719
Number of extensions: 581813
Number of successful extensions: 1977
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1820
Number of HSP's gapped (non-prelim): 105
length of query: 617
length of database: 11,318,596
effective HSP length: 105
effective length of query: 512
effective length of database: 8,513,101
effective search space: 4358707712
effective search space used: 4358707712
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146706.7


