

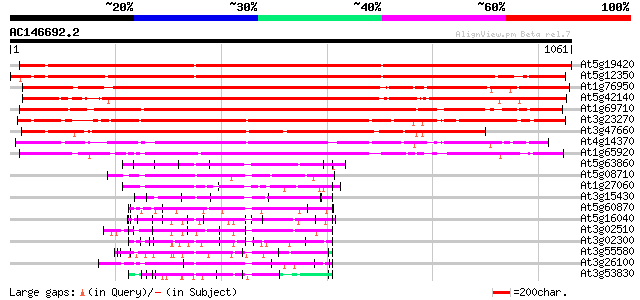
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146692.2 - phase: 0
(1061 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g19420 putative protein 1684 0.0
At5g12350 putative protein 1624 0.0
At1g76950 zinc finger protein (PRAF1) 1036 0.0
At5g42140 TMV resistance protein-like 1020 0.0
At1g69710 putative regulator of chromosome condensation 947 0.0
At3g23270 hypothetical protein 870 0.0
At3g47660 putative protein 769 0.0
At4g14370 disease resistance N like protein 750 0.0
At1g65920 634 0.0
At5g63860 UVB-resistance protein UVR8 (gb|AAD43920.1) 205 1e-52
At5g08710 unknown protein 171 2e-42
At1g27060 hypothetical protein 162 1e-39
At3g15430 unknown protein 148 1e-35
At5g60870 UVB-resistance protein UVR8 - like 148 2e-35
At5g16040 UVB-resistance protein-like 135 9e-32
At3g02510 unknown protein 134 2e-31
At3g02300 unknown protein 126 7e-29
At3g55580 regulator of chromosome condensation-like protein 123 5e-28
At3g26100 unknown protein (MPE11.29) 122 1e-27
At3g53830 putative protein 119 9e-27
>At5g19420 putative protein
Length = 1121
Score = 1684 bits (4360), Expect = 0.0
Identities = 839/1049 (79%), Positives = 926/1049 (87%), Gaps = 13/1049 (1%)
Query: 19 FEWKEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEV 78
F KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIY++RSLDL DKDEAEV
Sbjct: 80 FSGKEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYDERSLDL---DKDEAEV 136
Query: 79 WFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDH 138
WFSGLKALISR H RKWRTESRSDG PSEANSPRTYTRRSSPLHSPF SNES QK+ +H
Sbjct: 137 WFSGLKALISRCHQRKWRTESRSDGTPSEANSPRTYTRRSSPLHSPFSSNESFQKEGSNH 196
Query: 139 LRLHSPYESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTM 197
LRLHSPYESPPKNG+DKA D+ LYAVP K FFP SA+ SVHS+SSGGSD++HGHMK M
Sbjct: 197 LRLHSPYESPPKNGVDKAFSDMSLYAVPPKGFFPPGSATMSVHSLSSGGSDTLHGHMKGM 256
Query: 198 GMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDS 257
GMDAFRVSLSSA+SSSS GSGHDDGD LGDVF+WGEG G+GV+GGGNHRVGS L +K+DS
Sbjct: 257 GMDAFRVSLSSAISSSSHGSGHDDGDTLGDVFMWGEGIGEGVLGGGNHRVGSSLEIKMDS 316
Query: 258 LFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDA 317
L PKALES +VLDVQNIACGG+HA LVTKQGE FSWGEES GRLGHGVDS+V HPKLIDA
Sbjct: 317 LLPKALESTIVLDVQNIACGGQHAVLVTKQGESFSWGEESEGRLGHGVDSNVQHPKLIDA 376
Query: 318 LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIH 377
L+ TNIELVACGEYH+CAVTLSGDLYTWG G ++G+LGHGN+VSHWVPKRVN +EGIH
Sbjct: 377 LNTTNIELVACGEYHSCAVTLSGDLYTWGKG--DFGILGHGNEVSHWVPKRVNFLMEGIH 434
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
VS I+CGP+HTAVVTSAGQLFTFGDGTFG LGHGDRKSV +PREV+SLKGLRT+RA+CGV
Sbjct: 435 VSSIACGPYHTAVVTSAGQLFTFGDGTFGVLGHGDRKSVFIPREVDSLKGLRTVRAACGV 494
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQV 496
WHTAAVVEVMVG+SSSSNCSSGKLFTWGDGDK RLGHGDKE KLVPTCVA LVE NFCQV
Sbjct: 495 WHTAAVVEVMVGSSSSSNCSSGKLFTWGDGDKSRLGHGDKEPKLVPTCVAALVEPNFCQV 554
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVA 556
ACGHSLTVALTTSGHVY MGSPVYGQLGNP ADGK+PTRV+GKL KSFVEEIACGAYHVA
Sbjct: 555 ACGHSLTVALTTSGHVYTMGSPVYGQLGNPHADGKVPTRVDGKLHKSFVEEIACGAYHVA 614
Query: 557 VLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWV 616
VLT R EVYTWGKG+NGRLGHGD DDRN+PTLV++LKDK VKSIACG+NFTAA+CLHKW
Sbjct: 615 VLTSRTEVYTWGKGSNGRLGHGDADDRNSPTLVESLKDKQVKSIACGSNFTAAVCLHKWA 674
Query: 617 SGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCF 676
SG+DQSMCSGCR PFNFKRKRHNCYNCGLVFCHSCS+KKSLKA MAPNPNKPYRVCD CF
Sbjct: 675 SGMDQSMCSGCRQPFNFKRKRHNCYNCGLVFCHSCSNKKSLKACMAPNPNKPYRVCDRCF 734
Query: 677 NKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQVESR 736
NKL+K +ETD SSHSS+SRR S+NQGS + ID+D+KLDTRS QLARFS +E +QV+SR
Sbjct: 735 NKLKKAMETDPSSHSSLSRRESVNQGS-DAIDRDEKLDTRSDGQLARFSLLEPMRQVDSR 793
Query: 737 SSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRATS 796
SKKNKK EFNSSRVSP+P+GGS RG+LNI+KSFNP FGSSKKFFSASVPGSRI SRATS
Sbjct: 794 -SKKNKKYEFNSSRVSPIPSGGSHRGSLNITKSFNPTFGSSKKFFSASVPGSRIASRATS 852
Query: 797 PISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQE 856
PISRRPSPPRSTTPTPTL GLTTPKIVVDD K++ND+LSQEV+ LRSQVE+LTRKAQLQE
Sbjct: 853 PISRRPSPPRSTTPTPTLSGLTTPKIVVDDTKRSNDNLSQEVVMLRSQVENLTRKAQLQE 912
Query: 857 IELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVKSPSIAS 916
+ELERT+KQLK+A+AIA EE+A+CKAAKEVIKSLTAQLKDMAERLPVG+A++VKSPS+ S
Sbjct: 913 VELERTTKQLKEALAIASEESARCKAAKEVIKSLTAQLKDMAERLPVGSARTVKSPSLNS 972
Query: 917 FGSNELSFAAIDRLNIQATSPEADLTGSNT-QLLSNGSST--VSNRSTGQNKQSQSDSTN 973
FGS+ A + S E D T + SNG+ST + S Q +++ N
Sbjct: 973 FGSSPDYAAPSSNTLNRPNSRETDSDSLTTVPMFSNGTSTPVFDSGSYRQQANHAAEAIN 1032
Query: 974 RNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAE 1033
R +R+K+SE R+E EWVEQDEPGVYITLT+L GG DLKRVRFSRKRFSEKQAE WWAE
Sbjct: 1033 RISTRSKESEPRNENEWVEQDEPGVYITLTALAGGARDLKRVRFSRKRFSEKQAEEWWAE 1092
Query: 1034 NRVRVYEQYNVR-MVDKSSVGVGSEDLAN 1061
NR RVYEQYNVR +VDKSSVGVGSEDL +
Sbjct: 1093 NRGRVYEQYNVRIVVDKSSVGVGSEDLGH 1121
>At5g12350 putative protein
Length = 1062
Score = 1624 bits (4206), Expect = 0.0
Identities = 832/1091 (76%), Positives = 912/1091 (83%), Gaps = 74/1091 (6%)
Query: 1 MNSDLSRTGAVERDIEQT-----------------------------------FEWKEEK 25
M SDLSR G VERDIEQ F EEK
Sbjct: 1 MASDLSRAGPVERDIEQAIIALKKGAYLLKYGRRGKPKFCPFRLSNDETVLIWFSGNEEK 60
Query: 26 HLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEVWFSGLKA 85
HLKLSHVSRIISGQRT RYPRPEKEYQSFSLIY++RSLD+ DKDEAEVWF+GLKA
Sbjct: 61 HLKLSHVSRIISGQRT----RYPRPEKEYQSFSLIYSERSLDV---DKDEAEVWFTGLKA 113
Query: 86 LISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRLHSPY 145
LIS H R RTESRSDG PSEANSPRTYTRRSSPLHSPF SN+S QKD +HLR+HSP+
Sbjct: 114 LISHCHQRNRRTESRSDGTPSEANSPRTYTRRSSPLHSPFSSNDSLQKDGSNHLRIHSPF 173
Query: 146 ESPPKNGLDKAL-DVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMDAFRV 204
ESPPKNGLDKA D+ LYAVP K F+P DSA+ SVHS GGSDSMHGHM+ MGMDAFRV
Sbjct: 174 ESPPKNGLDKAFSDMALYAVPPKGFYPSDSATISVHS---GGSDSMHGHMRGMGMDAFRV 230
Query: 205 SLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALE 264
S+SSAVSSSS GSGHDDGDALGDVFIWGEG G+GV+GGGN RVGS +K+DSL PKALE
Sbjct: 231 SMSSAVSSSSHGSGHDDGDALGDVFIWGEGIGEGVLGGGNRRVGSSFDIKMDSLLPKALE 290
Query: 265 SAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIE 324
S +VLDVQNIACGG+HA LVTKQGE FSWGEES GRLGHGVDS++ PKLIDAL+ TNIE
Sbjct: 291 STIVLDVQNIACGGQHAVLVTKQGESFSWGEESEGRLGHGVDSNIQQPKLIDALNTTNIE 350
Query: 325 LVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCG 384
LVACGE+H+CAVTLSGDLYTWG G ++G+LGHGN+VSHWVPKRVN LEGIHVS I+CG
Sbjct: 351 LVACGEFHSCAVTLSGDLYTWGKG--DFGVLGHGNEVSHWVPKRVNFLLEGIHVSSIACG 408
Query: 385 PWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVV 444
P+HTAVVTSAGQLFTFGDGTFG LGHGD+KSV +PREV+SLKGLRT+RA+CGVWHTAAVV
Sbjct: 409 PYHTAVVTSAGQLFTFGDGTFGVLGHGDKKSVFIPREVDSLKGLRTVRAACGVWHTAAVV 468
Query: 445 EVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQVACGHSLT 503
EVMVG+SSSSNCSSGKLFTWGDGDKGRLGHG+KE KLVPTCVA LVE NFCQVACGHSLT
Sbjct: 469 EVMVGSSSSSNCSSGKLFTWGDGDKGRLGHGNKEPKLVPTCVAALVEPNFCQVACGHSLT 528
Query: 504 VALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNE 563
VALTTSGHVY MGSPVYGQLGN ADGK P RVEGKL KSFVEEIACGAYHVAVLT R E
Sbjct: 529 VALTTSGHVYTMGSPVYGQLGNSHADGKTPNRVEGKLHKSFVEEIACGAYHVAVLTSRTE 588
Query: 564 VYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGVDQSM 623
VYTWGKG+NGRLGHGD DDRN+PTLV++LKDK VKSIACGTNFTAA+C+H+W SG+DQSM
Sbjct: 589 VYTWGKGSNGRLGHGDVDDRNSPTLVESLKDKQVKSIACGTNFTAAVCIHRWASGMDQSM 648
Query: 624 CSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNKLRKTL 683
CSGCR PF+FKRKRHNCYNCGLVFCHSC+SKKSLKA MAPNPNKPYRVCD CFNKL+KT+
Sbjct: 649 CSGCRQPFSFKRKRHNCYNCGLVFCHSCTSKKSLKACMAPNPNKPYRVCDKCFNKLKKTM 708
Query: 684 ETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQVESRSSKKNKK 743
ETD SSHSS+SRRGSINQGS + IDKDDK D+RS QLARFS MES +QV+SR KKNKK
Sbjct: 709 ETDPSSHSSLSRRGSINQGS-DPIDKDDKFDSRSDGQLARFSLMESMRQVDSR-HKKNKK 766
Query: 744 LEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRATSPISRRPS 803
EFNSSRVSP+P+G SQRGALNI+KSFNPVFG+SKKFFSASVPGSRIVSRATSPISRRPS
Sbjct: 767 YEFNSSRVSPIPSGSSQRGALNIAKSFNPVFGASKKFFSASVPGSRIVSRATSPISRRPS 826
Query: 804 PPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQEIELERTS 863
PPRSTTPTPTL GL TPK VVDD K+TND+LSQEV+KLRSQVESLTRKAQLQE+ELERT+
Sbjct: 827 PPRSTTPTPTLSGLATPKFVVDDTKRTNDNLSQEVVKLRSQVESLTRKAQLQEVELERTT 886
Query: 864 KQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVKS-PSIASFGSNEL 922
KQLK+A+AI EET +CKAAKEVIKSLTAQLKDMAERLPVG+A++VKS PS+ SFGS
Sbjct: 887 KQLKEALAITNEETTRCKAAKEVIKSLTAQLKDMAERLPVGSARTVKSPPSLNSFGS--- 943
Query: 923 SFAAIDRLNI--QATSPEADLTGSNTQLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTK 980
S ID NI QA S E++ G T + SNG+ T NG T
Sbjct: 944 SPGRIDPFNILNQANSQESEPNGITTPMFSNGTMT---------------PAFGNGEAT- 987
Query: 981 DSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYE 1040
+E+R+E EWVEQDEPGVYITLT+L GG DLKRVRFSRKRFSE QAE WWA+NR RVYE
Sbjct: 988 -NEARNEKEWVEQDEPGVYITLTALAGGARDLKRVRFSRKRFSEIQAEQWWADNRGRVYE 1046
Query: 1041 QYNVRMVDKSS 1051
QYNVRMVDK+S
Sbjct: 1047 QYNVRMVDKAS 1057
>At1g76950 zinc finger protein (PRAF1)
Length = 1103
Score = 1036 bits (2679), Expect = 0.0
Identities = 564/1079 (52%), Positives = 725/1079 (66%), Gaps = 87/1079 (8%)
Query: 24 EKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDR--SLDLICKDKDEAEVWFS 81
EK LKL+ VS+I+ GQRT +FQRY RPEK+Y SFSL+YN + SLDLICKDK EAE+W
Sbjct: 58 EKRLKLASVSKIVPGQRTAVFQRYLRPEKDYLSFSLLYNGKKKSLDLICKDKVEAEIWIG 117
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRL 141
GLK LIS + + + S G S ++ R T SSP + SS S H
Sbjct: 118 GLKTLISTGQGGRSKIDGWSGGGLS-VDASRELTS-SSP-------SSSSASASRGHSSP 168
Query: 142 HSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMGMDA 201
+P+ P A V +KS LD+ + K G D
Sbjct: 169 GTPFNIDPITSPKSAEPEVPPTDSEKSHVALDNKNMQT---------------KVSGSDG 213
Query: 202 FRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPK 261
FRVS+SSA SSSS GS DD DALGDV+IWGE D VV G + S L + D L PK
Sbjct: 214 FRVSVSSAQSSSSHGSAADDSDALGDVYIWGEVICDNVVKVGIDKNASYLTTRTDVLVPK 273
Query: 262 ALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNT 321
LES +VLDV IACG RHAA VT+QGEIF+WGEESGGRLGHG+ DV HP+L+++L+ T
Sbjct: 274 PLESNIVLDVHQIACGVRHAAFVTRQGEIFTWGEESGGRLGHGIGKDVFHPRLVESLTAT 333
Query: 322 N-IELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSY 380
+ ++ VACGE+HTCAVTL+G+LYTWG+G +N GLLGHG+ +SHW+PKR+ G LEG+HV+
Sbjct: 334 SSVDFVACGEFHTCAVTLAGELYTWGDGTHNVGLLGHGSDISHWIPKRIAGSLEGLHVAS 393
Query: 381 ISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHT 440
+SCGPWHTA++TS G+LFTFGDGTFG LGHGD+++V PREVESL GLRT+ SCGVWHT
Sbjct: 394 VSCGPWHTALITSYGRLFTFGDGTFGVLGHGDKETVQYPREVESLSGLRTIAVSCGVWHT 453
Query: 441 AAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACG 499
AAVVE++V S+SS+ SSGKLFTWGDGDK RLGHGDK+ +L PTCV AL+++NF ++ACG
Sbjct: 454 AAVVEIIVTQSNSSSVSSGKLFTWGDGDKNRLGHGDKDPRLKPTCVPALIDYNFHKIACG 513
Query: 500 HSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLT 559
HSLTV LTTSG V+ MGS VYGQLGN Q DGKLP VE KL FVEEI+CGAYHVA LT
Sbjct: 514 HSLTVGLTTSGQVFTMGSTVYGQLGNLQTDGKLPCLVEDKLASEFVEEISCGAYHVAALT 573
Query: 560 LRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGV 619
RNEVYTWGKGANGRLGHGD +DR PT+V+ALKD+HVK IACG+N+TAAICLHKWVSG
Sbjct: 574 SRNEVYTWGKGANGRLGHGDLEDRKVPTIVEALKDRHVKYIACGSNYTAAICLHKWVSGA 633
Query: 620 DQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCFNKL 679
+QS CS CRL F F RKRHNCYNCGLV CHSCSSKK+ +A++AP+ + YRVCD C+ KL
Sbjct: 634 EQSQCSTCRLAFGFTRKRHNCYNCGLVHCHSCSSKKAFRAALAPSAGRLYRVCDSCYVKL 693
Query: 680 RKTLETDSSS--HSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARF--SSMESFKQVES 735
K E + ++ +S+V R N+ D+LD +S +LA+F S+M+ KQ++S
Sbjct: 694 SKVSEINDTNRRNSAVPRLSGENR---------DRLD-KSEIRLAKFGTSNMDLIKQLDS 743
Query: 736 RSSKKNKKLE-FNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRA 794
+++K+ KK + F+ R S +P+ + A + + + ++ K A S I SR+
Sbjct: 744 KAAKQGKKTDTFSLGRNSQLPSLLQLKDA--VQSNIGDMRRATPKLAQAP---SGISSRS 798
Query: 795 TSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQL 854
SP SRR SPPRS TP P+ GL P + D+ KKTN+ L+QE++KLR+QV+SLT+K +
Sbjct: 799 VSPFSRRSSPPRSATPMPSTSGLYFPVGIADNMKKTNEILNQEIVKLRTQVDSLTQKCEF 858
Query: 855 QEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVK---- 910
QE+EL+ + K+ ++A+A+A EE+AK +AAKE IKSL AQLKD+AE+LP G +SVK
Sbjct: 859 QEVELQNSVKKTQEALALAEEESAKSRAAKEAIKSLIAQLKDVAEKLPPG--ESVKLACL 916
Query: 911 -------------------------SPSIASFGSNELSFAAIDRLNIQA--TSPEADLTG 943
+ SI+S + +FA N+Q+ +P A
Sbjct: 917 QNGLDQNGFHFPEENGFHPSRSESMTSSISSVAPFDFAFANASWSNLQSPKQTPRASERN 976
Query: 944 SNT----QLLSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVY 999
SN LS+ S +S R + Q Q++S N + ++ ++ E EW+EQ EPGVY
Sbjct: 977 SNAYPADPRLSSSGSVISERI--EPFQFQNNSDNGSSQTGVNNTNQVEAEWIEQYEPGVY 1034
Query: 1000 ITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSSVGVGSED 1058
ITL +L G DL+RVRFSR+RF E QAE WW+ENR +VYE+YNVR+ +KS+ D
Sbjct: 1035 ITLVALHDGTRDLRRVRFSRRRFGEHQAETWWSENREKVYEKYNVRVSEKSTASQTHRD 1093
>At5g42140 TMV resistance protein-like
Length = 1073
Score = 1020 bits (2638), Expect = 0.0
Identities = 565/1062 (53%), Positives = 719/1062 (67%), Gaps = 84/1062 (7%)
Query: 24 EKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDR--SLDLICKDKDEAEVWFS 81
EK LKL+ VS+I+ GQRT +FQRY RP+K+Y SFSLIY++R +LDLICKDK EAEVW +
Sbjct: 53 EKRLKLATVSKIVPGQRTAVFQRYLRPDKDYLSFSLIYSNRKRTLDLICKDKVEAEVWIA 112
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDHLRL 141
GLKALIS R + + SDG S A+S R L SP S+ + +D
Sbjct: 113 GLKALISGQAGRS-KIDGWSDGGLSIADS------RDLTLSSPTNSSVCASRDFNI---A 162
Query: 142 HSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSG----GSDSMHGHMKTM 197
SPY S + FP S + +S+SS SDS + ++
Sbjct: 163 DSPYNS--------------------TNFP--RTSRTENSVSSERSHVASDSPNMLVRGT 200
Query: 198 GMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDS 257
G DAFRVS+SS SSSS GS DD DALGDV+IWGE + V G + LG + D
Sbjct: 201 GSDAFRVSVSSVQSSSSHGSAPDDCDALGDVYIWGEVLCENVTKFGADKNIGYLGSRSDV 260
Query: 258 LFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDA 317
L PK LES VVLDV +IACG +HAALV++QGE+F+WGE SGGRLGHG+ DV P+LI++
Sbjct: 261 LIPKPLESNVVLDVHHIACGVKHAALVSRQGEVFTWGEASGGRLGHGMGKDVTGPQLIES 320
Query: 318 LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIH 377
L+ T+I+ VACGE+HTCAVT++G++YTWG+G +N GLLGHG VSHW+PKR++GPLEG+
Sbjct: 321 LAATSIDFVACGEFHTCAVTMTGEIYTWGDGTHNAGLLGHGTDVSHWIPKRISGPLEGLQ 380
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
++ +SCGPWHTA++TS GQLFTFGDGTFG LGHGD+++V PREVESL GLRT+ +CGV
Sbjct: 381 IASVSCGPWHTALITSTGQLFTFGDGTFGVLGHGDKETVFYPREVESLSGLRTIAVACGV 440
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQV 496
WH AA+VEV+V +SSSS SSGKLFTWGDGDK RLGHGDKE +L PTCV AL++H F +V
Sbjct: 441 WHAAAIVEVIVTHSSSS-VSSGKLFTWGDGDKSRLGHGDKEPRLKPTCVSALIDHTFHRV 499
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVA 556
ACGHSLTV LTTSG VY MGS VYGQLGNP ADGKLP VE KL K VEEIACGAYHVA
Sbjct: 500 ACGHSLTVGLTTSGKVYTMGSTVYGQLGNPNADGKLPCLVEDKLTKDCVEEIACGAYHVA 559
Query: 557 VLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWV 616
VLT RNEV+TWGKGANGRLGHGD +DR PTLVDALK++HVK+IACG+NFTAAICLHKWV
Sbjct: 560 VLTSRNEVFTWGKGANGRLGHGDVEDRKAPTLVDALKERHVKNIACGSNFTAAICLHKWV 619
Query: 617 SGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCF 676
SG +QS CS CR F F RKRHNCYNCGLV CHSCSSKKSLKA++APNP KPYRVCD C
Sbjct: 620 SGTEQSQCSACRQAFGFTRKRHNCYNCGLVHCHSCSSKKSLKAALAPNPGKPYRVCDSCH 679
Query: 677 NKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARF---SSMESFKQV 733
+KL K E + S +V R S + D+LD ++ +LA+ S+++ KQ+
Sbjct: 680 SKLSKVSEANIDSRKNVMPRLS--------GENKDRLD-KTEIRLAKSGIPSNIDLIKQL 730
Query: 734 ESRSSKKNKKLE-FNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVS 792
++R++++ KK + F+ R S P + N++ G K A P S S
Sbjct: 731 DNRAARQGKKADTFSLVRTSQTPLTQLKDALTNVADLRR---GPPK---PAVTPSS---S 781
Query: 793 RATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKA 852
R SP SRR SPPRS TP P GL + + KKTN+ L+QEV++LR+Q ESL +
Sbjct: 782 RPVSPFSRRSSPPRSVTPIPLNVGLGFSTSIAESLKKTNELLNQEVVRLRAQAESLRHRC 841
Query: 853 QLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGT--AKSVK 910
++QE E++++ K++++A+++A EE+AK +AAKEVIKSLTAQ+KD+A LP G A++ +
Sbjct: 842 EVQEFEVQKSVKKVQEAMSLAAEESAKSEAAKEVIKSLTAQVKDIAALLPPGAYEAETTR 901
Query: 911 SPSIAS-FGSNELSFA---------AIDRLNIQATSPEADLTGSNTQLLSNGSS-TVSNR 959
+ ++ + F N F + + SP A S L N S ++
Sbjct: 902 TANLLNGFEQNGFHFTNANGQRQSRSDSMSDTSLASPLAMPARSMNGLWRNSQSPRNTDA 961
Query: 960 STGQ---------NKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVI 1010
S G+ N S+ +R+ + + + S+ E EW+EQ EPGVYITL +L G
Sbjct: 962 SMGELLSEGVRISNGFSEDGRNSRSSAASASNASQVEAEWIEQYEPGVYITLLALGDGTR 1021
Query: 1011 DLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSSV 1052
DLKRVRFSR+RF E+QAE WW+ENR RVYE+YN+R D+SSV
Sbjct: 1022 DLKRVRFSRRRFREQQAETWWSENRERVYEKYNIRGTDRSSV 1063
>At1g69710 putative regulator of chromosome condensation
Length = 1028
Score = 947 bits (2447), Expect = 0.0
Identities = 503/1029 (48%), Positives = 668/1029 (64%), Gaps = 59/1029 (5%)
Query: 19 FEWKEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEV 78
+ KEEK +KLS V RI+ GQRTP F+RYPRPEKEYQSFSLI DRSLDLICKDKDEAEV
Sbjct: 53 YSGKEEKQIKLSQVLRIVPGQRTPTFKRYPRPEKEYQSFSLICPDRSLDLICKDKDEAEV 112
Query: 79 WFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNESSQKDSGDH 138
W GLK+LI+R KW+T + + +E +P + RR SP + ++
Sbjct: 113 WVVGLKSLITRVKVSKWKTTIKPEITSAECPTP--HARRVSPFVTILDQVIQPSNETSTQ 170
Query: 139 LRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTMG 198
RL + D+V P + +++ S ++ + +++
Sbjct: 171 TRLGKVFS-----------DIVAITAPPSNNNQTEASGNLFCPFSPTPANVENSNLRFST 219
Query: 199 MDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSL 258
D FR+SLSSAVS+SS GS H+D DALGDVF+WGE DGV+ G + + S D+L
Sbjct: 220 NDPFRLSLSSAVSTSSHGSYHEDFDALGDVFVWGESISDGVLSGTGNSLNS---TTEDAL 276
Query: 259 FPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDAL 318
PKALES +VLD QNIACG HA LVTKQGEIFSWGE GG+LGHG+++D PK I ++
Sbjct: 277 LPKALESTIVLDAQNIACGKCHAVLVTKQGEIFSWGEGKGGKLGHGLETDAQKPKFISSV 336
Query: 319 SNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHV 378
+ +ACG++HTCA+T SGDLY+WG+G +N LLGHGN+ S W+PKRV G L+G++V
Sbjct: 337 RGLGFKSLACGDFHTCAITQSGDLYSWGDGTHNVDLLGHGNESSCWIPKRVTGDLQGLYV 396
Query: 379 SYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVW 438
S ++CGPWHTAVV S+GQLFTFGDGTFGALGHGDR+S S+PREVESL GL + +CGVW
Sbjct: 397 SDVACGPWHTAVVASSGQLFTFGDGTFGALGHGDRRSTSVPREVESLIGLIVTKVACGVW 456
Query: 439 HTAAVVEVMVGNSSSS-NCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQV 496
HTAAVVEV S + + S G++FTWGDG+KG+LGHGD + KL+P CV +L N CQV
Sbjct: 457 HTAAVVEVTNEASEAEVDSSRGQVFTWGDGEKGQLGHGDNDTKLLPECVISLTNENICQV 516
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVA 556
ACGHSLTV+ T+ GHVY MGS YGQLGNP A G P RVEG ++++ VEEIACG+YHVA
Sbjct: 517 ACGHSLTVSRTSRGHVYTMGSTAYGQLGNPTAKGNFPERVEGDIVEASVEEIACGSYHVA 576
Query: 557 VLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKWV 616
VLT ++E+YTWGKG NG+LGHG+ +++ P +V L++K VK+I CG+NFTA IC+HKWV
Sbjct: 577 VLTSKSEIYTWGKGLNGQLGHGNVENKREPAVVGFLREKQVKAITCGSNFTAVICVHKWV 636
Query: 617 SGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGCF 676
G + S+C+GCR PFNF+RKRHNCYNCGLVFC CSS+KSL+A++AP+ NKPYRVC GCF
Sbjct: 637 PGSEHSLCAGCRNPFNFRRKRHNCYNCGLVFCKVCSSRKSLRAALAPDMNKPYRVCYGCF 696
Query: 677 NKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSME-SFKQVES 735
KL+K+ E+ S+ +S +R+ +N + + D L + + AR SS + S E
Sbjct: 697 TKLKKSRESSPSTPTSRTRK-LLNMRKSTDVSERDSLTQKFLSVNARLSSADSSLHYSER 755
Query: 736 RSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFFSASVPGSRIVSRAT 795
R +++ K E N+S V P NG Q SK + K +PGS + SR T
Sbjct: 756 RHHRRDLKPEVNNSNVFPSMNGSLQPVGSPFSKG-STALPKIPKNMMVKIPGSGMSSRTT 814
Query: 796 SPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKLRSQVESLTRKAQLQ 855
SP+S + + PR + ++ ++K+ DS +Q++ L+ QVE L KA
Sbjct: 815 SPVSVKSTSPRRSY-----------EVAAAESKQLKDSFNQDMAGLKEQVEQLASKAHQL 863
Query: 856 EIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAERLPVGTAKSVKSPSIA 915
E ELE+T +QLK A+A +E + ++AKEVI+SLT QLK+MAE K + SI+
Sbjct: 864 EEELEKTKRQLKVVTAMAADEAEENRSAKEVIRSLTTQLKEMAE-------KQSQKDSIS 916
Query: 916 SFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSNGSSTVSNRSTGQNKQSQSDSTNRN 975
+ N + T E T + T ++ S VS S QN+ + + + N
Sbjct: 917 T--------------NSKHTDKEKSETVTQTSNQTHIRSMVSQDS--QNENNLTSKSFAN 960
Query: 976 GSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVRFSRKRFSEKQAENWWAENR 1035
G R + + E V QDEPGVY+TL SLPGG +LKRVRFSRK+F+E+QAE WW EN
Sbjct: 961 GHR----KQNDKPEKVVQDEPGVYLTLLSLPGGGTELKRVRFSRKQFTEEQAEKWWGENG 1016
Query: 1036 VRVYEQYNV 1044
+V E++N+
Sbjct: 1017 AKVCERHNI 1025
>At3g23270 hypothetical protein
Length = 1045
Score = 870 bits (2247), Expect = 0.0
Identities = 487/1055 (46%), Positives = 658/1055 (62%), Gaps = 110/1055 (10%)
Query: 16 EQTFEW---KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYN--DRSLDLIC 70
E+T W EEK LKL VSRI+ GQRT R+ RPEK++ SFSL+YN +RSLDLIC
Sbjct: 48 EKTLIWFSRGEEKGLKLFEVSRIVPGQRT----RFLRPEKDHLSFSLLYNNRERSLDLIC 103
Query: 71 KDKDEAEVWFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSPFGSNES 130
KDK E EVWF+ LK LI +S +R+ R+E IP +S R S
Sbjct: 104 KDKAETEVWFAALKFLIEKSRNRRARSE-----IPEIHDSDTFSVGRQS----------- 147
Query: 131 SQKDSGDHLRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSM 190
+D V +P+ S + S G +
Sbjct: 148 --------------------------IDFVPSNIPRGR----TSIDLGYQNNSDVGYE-- 175
Query: 191 HGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSG 250
G+M D FR+S+SS S SS GSG DD ++LGDV++WGE +G++ G S
Sbjct: 176 RGNMLRPSTDGFRISVSSTPSCSSGGSGPDDIESLGDVYVWGEVWTEGILPDGT---ASN 232
Query: 251 LGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVL 310
VK D L P+ LES VVLDV I CG RH ALVT+QGE+F+WGEE GGRLGHG+ D+
Sbjct: 233 ETVKTDVLTPRPLESNVVLDVHQIVCGVRHVALVTRQGEVFTWGEEVGGRLGHGIQVDIS 292
Query: 311 HPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVN 370
PKL++ L+ TNI+ VACGEYHTC V+ SGDL++WG+G +N GLLGHG+ +SHW+PKRV+
Sbjct: 293 RPKLVEFLALTNIDFVACGEYHTCVVSTSGDLFSWGDGIHNVGLLGHGSDISHWIPKRVS 352
Query: 371 GPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRT 430
GPLEG+ V ++CG WH+A+ T+ G+LFTFGDG FG LGHG+R+SVS P+EV+SL GL+T
Sbjct: 353 GPLEGLQVLSVACGTWHSALATANGKLFTFGDGAFGVLGHGNRESVSYPKEVQSLNGLKT 412
Query: 431 MRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LV 489
++ +C +WHTAA+VEVM ++++ SS KLFTWGDGDK RLGHG+KE L+PTCV+ L+
Sbjct: 413 VKVACSIWHTAAIVEVM--GQTATSMSSRKLFTWGDGDKNRLGHGNKETYLLPTCVSSLI 470
Query: 490 EHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIA 549
++NF ++ACGH+ TVALTTSGHV+ MG +GQLGN +DGKLP V+ +L+ FVEEIA
Sbjct: 471 DYNFHKIACGHTFTVALTTSGHVFTMGGTAHGQLGNSISDGKLPCLVQDRLVGEFVEEIA 530
Query: 550 CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAA 609
CGA+HVAVLT R+EV+TWGKGANGRLGHGDT+D+ PTLV+AL+D+HVKS++CG+NFT++
Sbjct: 531 CGAHHVAVLTSRSEVFTWGKGANGRLGHGDTEDKRTPTLVEALRDRHVKSLSCGSNFTSS 590
Query: 610 ICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPY 669
IC+HKWVSG DQS+CSGCR F F RKRHNCYNCGLV CH+CSSKK+LKA++AP P KP+
Sbjct: 591 ICIHKWVSGADQSICSGCRQAFGFTRKRHNCYNCGLVHCHACSSKKALKAALAPTPGKPH 650
Query: 670 RVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMES 729
RVCD C++KL+ +S S+V+R + S++ + D+ TRS L + +
Sbjct: 651 RVCDACYSKLK---AAESGYSSNVNRNVATPGRSIDGSVRTDRETTRSSKVL-----LSA 702
Query: 730 FKQVESRSSKKNKKLEFNSSRVSPVPNGGSQR------GALNISKSFNPVFGSSKK---- 779
K SS+ E +++R S VP+ + I +F PV +
Sbjct: 703 NKNSVMSSSRPGFTPESSNARASQVPSLQQLKDIAFPSSLSAIQNAFKPVVAPTTTPPRT 762
Query: 780 -FFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEV 838
S P R++SP +RRPSPPR++ G + + V+D +KTN+ ++QE+
Sbjct: 763 LVIGPSSPSPPPPPRSSSPYARRPSPPRTS-------GFS--RSVIDSLRKTNEVMNQEM 813
Query: 839 IKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMA 898
KL SQ ++ Q E+ER K KDA +A +++K KAA E +KS+ QLK++
Sbjct: 814 TKLHSQ-----QRCNNQGTEIERFQKAAKDASELAARQSSKHKAATEALKSVAEQLKELK 868
Query: 899 ERLPVGTAKSVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNT--QLLSNGSSTV 956
E+LP ++S SI S L+ + + TS + T T Q+ SN S T
Sbjct: 869 EKLPPEVSESEAFESINSQAEAYLNANKVSETSPLTTSGQEQETYQKTEEQVPSNSSITE 928
Query: 957 SNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVIDLKRVR 1016
++ S S++ ST + SR ES+ EQ EPGVY+T G +RVR
Sbjct: 929 TSSS------SRAPSTEASSSRISGKESK------EQFEPGVYVTYEVDMNGNKIFRRVR 976
Query: 1017 FSRKRFSEKQAENWWAENRVRVYEQYNVRMVDKSS 1051
FS+KRF E QAE+WW +N+ R+ + Y+ SS
Sbjct: 977 FSKKRFDEHQAEDWWTKNKDRLLKCYSSNSSSSSS 1011
>At3g47660 putative protein
Length = 951
Score = 770 bits (1987), Expect = 0.0
Identities = 428/914 (46%), Positives = 571/914 (61%), Gaps = 65/914 (7%)
Query: 22 KEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIYNDRSLDLICKDKDEAEVWFS 81
K+EK LKLS V+RII GQRT +F+RYP+P KEYQSFSLIY +RSLDL DKDEAE W +
Sbjct: 56 KKEKRLKLSSVTRIIPGQRTAVFRRYPQPTKEYQSFSLIYGERSLDL---DKDEAEFWLT 112
Query: 82 GLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSP----LHSPFGSNESSQKDSGD 137
L+AL+SR++ SRS + E + ++ S + S E ++K SG
Sbjct: 113 TLRALLSRNNSSALVLHSRSRSLAPENGEQSSSSQNSKSNIRSVSSDTSYEEHAKKASGS 172
Query: 138 HLRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISSGGSDSMHGHMKTM 197
H +P + G K VL LD H+ ++ + +
Sbjct: 173 HCN------TPQRLG--KVFSEVLSQTAVLKALSLDELVHKPHTSPPETIENRPTNHSSP 224
Query: 198 GMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDS 257
G+D + S+SSAVSSSSQGS +D +L DVF+WGE GDG++GGG H+ GS + DS
Sbjct: 225 GVDTSKYSISSAVSSSSQGSTFEDLKSLCDVFVWGESIGDGLLGGGMHKSGSSSSLMTDS 284
Query: 258 LFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDA 317
PK L+S V LD Q+I+CG +A LVTKQG+++SWGEESGGRLGHGV S V HPKLID
Sbjct: 285 FLPKVLKSHVALDAQSISCGTNYAVLVTKQGQMYSWGEESGGRLGHGVCSYVPHPKLIDE 344
Query: 318 LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIH 377
+ + +EL CGE+HTCAVT SGDLY WG+G +N GLLG G+ SHW P R+ G +EGI+
Sbjct: 345 FNGSTVELADCGEFHTCAVTASGDLYAWGDGDHNAGLLGLGSGASHWKPVRILGQMEGIY 404
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
V ISCGPWHTA VTS G+LFTFGDGTFGALGHGDR S ++PREVE+L G RT++A+CGV
Sbjct: 405 VKAISCGPWHTAFVTSEGKLFTFGDGTFGALGHGDRISTNIPREVEALNGCRTIKAACGV 464
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQV 496
WH+AAVV V SSGKLFTWGDGD GRLGHGD E +L+P+CV L +F QV
Sbjct: 465 WHSAAVVSVF-----GEATSSGKLFTWGDGDDGRLGHGDIECRLIPSCVTELDTTSFQQV 519
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLGNPQAD-GKLPTRVEGKLLKSFVEEIACGAYHV 555
ACG S+TVAL+ SG VYAMG+ +P D + P+ +EG L KSFV+E+ACG +H+
Sbjct: 520 ACGQSITVALSMSGQVYAMGT------ADPSHDIVRAPSCIEGGLGKSFVQEVACGFHHI 573
Query: 556 AVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLHKW 615
AVL + EVYTWGKG+NG+LGHGDT+ R PTLV ALK K V+ + CG+N+TA ICLHK
Sbjct: 574 AVLNSKAEVYTWGKGSNGQLGHGDTEYRCMPTLVKALKGKQVRKVVCGSNYTATICLHKP 633
Query: 616 VSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAPNPNKPYRVCDGC 675
++G D + CSGCR PFN+ RK HNCYNCG VFC+SC+SKKSL A+MAP N+PYRVCD C
Sbjct: 634 ITGTDSTKCSGCRHPFNYMRKLHNCYNCGSVFCNSCTSKKSLAAAMAPKTNRPYRVCDDC 693
Query: 676 FNKL---RKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRSRNQLARFSSMESFKQ 732
+ KL R++L T ++S + SL + D++ + QL R S + F+Q
Sbjct: 694 YIKLEGIRESLATPANS-------ARFSNASLPSSYEMDEIGITPQRQLLRVDSFDFFRQ 746
Query: 733 VESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNIS--------KSFNPVFGSSKK----- 779
+ K + S S + + +G+ N+ SF+ V K+
Sbjct: 747 TKHADLKTIGETS-GGSCTSSIHSNMDIKGSFNLKGIRRLSRLTSFDSVQEEGKQRTKHC 805
Query: 780 ------------FFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTL-GGLTTPKIVVDD 826
+ +P SR S P+S + SP S T +T +++ +
Sbjct: 806 ASKSDTSSLIRHSVTCGLPFSRRGSVELFPLSIKSSPVESVATTSDFTTDITDHELLQEV 865
Query: 827 AKKTNDSLSQEVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEV 886
KK+N LS E+ L++QVE LT K++ E EL +TSK+L+ A+ +A ++ K K+++E+
Sbjct: 866 PKKSNQCLSHEISVLKAQVEELTLKSKKLETELGKTSKKLEVAVLMARDDAEKIKSSEEI 925
Query: 887 IKSLTAQLKDMAER 900
++SLT QL + ++
Sbjct: 926 VRSLTLQLMNTTKK 939
>At4g14370 disease resistance N like protein
Length = 1996
Score = 750 bits (1936), Expect = 0.0
Identities = 447/1027 (43%), Positives = 609/1027 (58%), Gaps = 132/1027 (12%)
Query: 12 ERDIEQTFEW--KEEKHLKLSHVSR--IISGQRTP-IFQRYPRPEKEYQSFSLIYN--DR 64
+RDI+Q K + LK S R S + +P +F+RY RPEK+Y SFSLIY+ DR
Sbjct: 997 DRDIDQALVSLKKGTQLLKYSRKGRPKFRSFRLSPAVFRRYLRPEKDYLSFSLIYHNGDR 1056
Query: 65 SLDLICKDKDEAEVWFSGLKALISRSHHRKWRTESRSDGIPSEANSPRTYTRRSSPLHSP 124
SLDLICKDK E EVWF+GLK+LI ++ +++ ++E IP +S T R S
Sbjct: 1057 SLDLICKDKAETEVWFAGLKSLIRQNRNKQAKSE-----IPEIHDSDCFSTGRPSTASID 1111
Query: 125 FGSNESSQKDSGDHLRLHSPYESPPKNGLDKALDVVLYAVPQKSFFPLDSASASVHSISS 184
F N + + + L + + SP K G S
Sbjct: 1112 FAPNNTRRGRTSIDLGIQN---SPTKFG-------------------------------S 1137
Query: 185 GGSDSMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGN 244
G+M D FR+S+SS S S+ SG DD ++LGDV++WGE DG+ G
Sbjct: 1138 SDVGYERGNMLRPSTDGFRISVSSTPSCSTGTSGPDDIESLGDVYVWGEVWSDGISPDG- 1196
Query: 245 HRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHG 304
V + VKID IACG RH ALVT+QGE+F+W EE+GGRLGHG
Sbjct: 1197 --VVNSTTVKID-----------------IACGVRHIALVTRQGEVFTWEEEAGGRLGHG 1237
Query: 305 VDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHW 364
+ DV PKL++ L+ TNI+ VACGEYHTCAV+ SGDL+TWG+G +N GLLGHG+ +SHW
Sbjct: 1238 IQVDVCRPKLVEFLALTNIDFVACGEYHTCAVSTSGDLFTWGDGIHNVGLLGHGSDLSHW 1297
Query: 365 VPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVES 424
+PKRV+GP+EG+ V ++CG WH+A+ T+ G+LFTFGDG FG LGHGDR+SVS P+EV+
Sbjct: 1298 IPKRVSGPVEGLQVLSVACGTWHSALATANGKLFTFGDGAFGVLGHGDRESVSYPKEVKM 1357
Query: 425 LKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPT 484
L GL+T++ +CGVWHT A+VEVM N + ++ SS KLFTWGDGDK RLGHG+KE L+PT
Sbjct: 1358 LSGLKTLKVACGVWHTVAIVEVM--NQTGTSTSSRKLFTWGDGDKNRLGHGNKETYLLPT 1415
Query: 485 CV-ALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKS 543
CV +L+++NF Q+ACGH+ TVALTTSGHV+ MG +GQLG+ +DGKLP V+ +L+
Sbjct: 1416 CVSSLIDYNFNQIACGHTFTVALTTSGHVFTMGGTSHGQLGSSNSDGKLPCLVQDRLVGE 1475
Query: 544 FVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACG 603
FVEEI+CG +HVAVLT R+EV+TWGKG+NGRLGHGD DDR PTLV+AL+++HVKSI+C
Sbjct: 1476 FVEEISCGDHHVAVLTSRSEVFTWGKGSNGRLGHGDKDDRKTPTLVEALRERHVKSISC- 1534
Query: 604 TNFTAAICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMAP 663
G DQS+CSGCR F F RKRHNCYNCGLV CH+CSSKK+LKA++AP
Sbjct: 1535 --------------GADQSVCSGCRQAFGFTRKRHNCYNCGLVHCHACSSKKALKAALAP 1580
Query: 664 NPNKPYRVCDGCFNKLRKTLETDSSSHSSVSRRGSIN-QGSLELIDKDDKLDTRSRNQLA 722
P KP+RVCD C+ KL+ +S +S+V+ R S SL+ + D+ D RS +++
Sbjct: 1581 TPGKPHRVCDACYTKLK---AGESGYNSNVANRNSTTPTRSLDGTGRPDR-DIRS-SRIL 1635
Query: 723 RFSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGAL------NISKSFNPVFGS 776
E K E RSS+ E + R S VP R I +F PV S
Sbjct: 1636 LSPKTEPVKYSEVRSSRS----ESSIVRASQVPALQQLRDVAFPSSLSAIQNAFKPVASS 1691
Query: 777 SKKFFSASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQ 836
S + SRI SPPRS+ G + + ++D KK+N +++
Sbjct: 1692 STSTLPSGTRSSRI-----------SSPPRSS-------GFS--RGMIDTLKKSNGVINK 1731
Query: 837 EVIKLRSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKD 896
E+ KL+SQ+++L K Q E++R K ++A +A + ++K KAA EV+KS+ L++
Sbjct: 1732 EMTKLQSQIKNLKEKCDNQGTEIQRLKKTAREASDLAVKHSSKHKAATEVMKSVAEHLRE 1791
Query: 897 MAERLPVGTAK-----SVKSPSIASFGSNELSFAAIDRLNIQATSPEADLTGSNTQLLSN 951
+ E+LP ++ S+ S + A ++E S + + ++ +NTQ +
Sbjct: 1792 LKEKLPPEVSRCEAFESMNSQAEAYLNASEASESCLPTTSL-GMGQRDPTPSTNTQ---D 1847
Query: 952 GSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGGVID 1011
+ S G N +SQ S S S E +EQ EPGVY+T G
Sbjct: 1848 QNIEEKQSSNGGNMRSQEPSGTTEAS---SSSKGGGKELIEQFEPGVYVTYVLHKNGGKI 1904
Query: 1012 LKRVRFS 1018
+RVRFS
Sbjct: 1905 FRRVRFS 1911
>At1g65920
Length = 1006
Score = 634 bits (1634), Expect = 0.0
Identities = 398/1058 (37%), Positives = 581/1058 (54%), Gaps = 136/1058 (12%)
Query: 19 FEWKEEKHLKLSHVSRIISGQRTPIFQRYPRPEKEYQSFSLIY--NDRSLDLICKDKDEA 76
+ +EE+ L+LS V I+ GQ TP FQ+ + +++ QSFSLIY + +LDLICKDK +A
Sbjct: 55 YSGEEERQLRLSSVITIVRGQITPNFQKQAQSDRKEQSFSLIYANGEHTLDLICKDKAQA 114
Query: 77 EVWFSGLKALISRSHHRKWRTESRSD-GIPSEANSPRTYTRRSSPLHSPFGSNESSQKDS 135
+ WF GL+A+I++ H+ + RS G S NSP + RR N +++
Sbjct: 115 DSWFKGLRAVITKHHNIRNSVNHRSSRGAQSCINSPAGFMRRKQ--------NLGLLEET 166
Query: 136 GDHLRLHSPYESPP--------KNGLDKALDVVLYAV---PQKSFFPLDSASASVHSISS 184
D ++ S SP NGL + D + P S++ D + S
Sbjct: 167 PDVTQIRSLCGSPSTLLEERCLSNGLSCSSDSFAESDALGPVSSYYETDYDFRNSDCDRS 226
Query: 185 GGSDSMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGDGVVGGGN 244
GS+ + R + S +S +Q + L D+ IWG TG ++ G
Sbjct: 227 TGSELCR-------FSSQRFAASPPLSIITQPVTRSN--VLKDIMIWGAITG--LIDGSK 275
Query: 245 HRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHG 304
++ D+L PK LESA + DVQ+I+ G +HAALVT+QGE+F WG + G+LG
Sbjct: 276 NQN--------DALSPKLLESATMFDVQSISLGAKHAALVTRQGEVFCWGNGNSGKLGLK 327
Query: 305 VDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHW 364
V+ D+ HPK +++L + + VAC ++ TCAVT SG+LY WG G S +
Sbjct: 328 VNIDIDHPKRVESLEDVAVRSVACSDHQTCAVTESGELYLWGIDGGTIEQSG-----SQF 382
Query: 365 VPKRVNGPLEG-IHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVE 423
+ ++++ L G + V ++CG WHTA+VTS+GQLFT+G GTFG LGHG +SV+ P+EVE
Sbjct: 383 LTRKISDVLGGSLTVLSVACGAWHTAIVTSSGQLFTYGSGTFGVLGHGSLESVTKPKEVE 442
Query: 424 SLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVP 483
SL+ ++ + SCG WHTAA+VE + S GKLFTWGDGDKGRLGH D + KLVP
Sbjct: 443 SLRRMKVISVSCGPWHTAAIVETANDRKFYNAKSCGKLFTWGDGDKGRLGHADSKRKLVP 502
Query: 484 TCVA-LVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLK 542
TCV L++H+F +V+CG +LTVAL+ SG VY MGS ++GQLG P+A K V G L +
Sbjct: 503 TCVTELIDHDFIKVSCGWTLTVALSISGTVYTMGSSIHGQLGCPRAKDKSVNVVLGNLTR 562
Query: 543 SFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIAC 602
FV++IA G++HVAVLT VYTWGKG NG+LG GD DRN+P LV+ L D+ V+SIAC
Sbjct: 563 QFVKDIASGSHHVAVLTSFGNVYTWGKGMNGQLGLGDVRDRNSPVLVEPLGDRLVESIAC 622
Query: 603 GTNFTAAICLHKWVSGVDQSMCSGCRLPFNFKRKRHNCYNCGLVFCHSCSSKKSLKASMA 662
G N TAAICLHK +S DQ+ CS C+ F F R++HNCYNCGL+FC++CSSKK++ AS+A
Sbjct: 623 GLNLTAAICLHKEISLNDQTACSSCKSAFGFTRRKHNCYNCGLLFCNACSSKKAVNASLA 682
Query: 663 PNPNKPYRVCDGCFNKLRKTLETDSSSHSSVSRRGSINQGSLELIDKDDKLDTRS-RNQL 721
PN +K RVCD CF+ L S+ ++ K+D + R Q+
Sbjct: 683 PNKSKLSRVCDSCFDHL----------------------WSITEFSRNVKMDNHTPRMQM 720
Query: 722 ARFSSMESFKQVESRSSKKNKKLEFNSSRVSPVPNGGSQRGALNISKSFNPVFGSSKKFF 781
E + +S + +N SS P G G F+ + SS
Sbjct: 721 VTRRVSEDLTEKQSENEMQNLPQANRSS--DGQPRWGQVSGPSLF--RFDKISTSSSLNL 776
Query: 782 SASVPGSRIVSRATSPISRRPSPPRSTTPTPTLGGLTTPKIVVDDAKKTNDSLSQEVIKL 841
S S +RR S + +T + ++N L++E+ +L
Sbjct: 777 SVS--------------ARRTSSTKIST-----------------SSESNKILTEEIERL 805
Query: 842 RSQVESLTRKAQLQEIELERTSKQLKDAIAIAGEETAKCKAAKEVIKSLTAQLKDMAER- 900
++ +++L R+ +L ++E ++L +A EE K KAAKE+IK+L ++L+ E+
Sbjct: 806 KAVIKNLQRQCELGNEKMEECQQELDKTWEVAKEEAEKSKAAKEIIKALASKLQANKEKP 865
Query: 901 -LPVGTAKSVKSPSIASFGSNELSFAAI-----------DRLNIQATSPEADLTGSNTQL 948
P+ T + ++ + +S + + +++ ++ SN +L
Sbjct: 866 SNPLKTGIACNPSQVSPIFDDSMSIPYLTPITTARSQHETKQHVEKCVTKSSNRDSNIKL 925
Query: 949 LSNGSSTVSNRSTGQNKQSQSDSTNRNGSRTKDSESRSETEWVEQDEPGVYITLTSLPGG 1008
L + S ++ QN+ T+DS + E VEQ EPGVYIT T+LP G
Sbjct: 926 LVDASPAITRTGYLQNE-------------TQDSSA----EQVEQYEPGVYITFTALPCG 968
Query: 1009 VIDLKRVRFSRKRFSEKQAENWWAENRVRVYEQYNVRM 1046
LKRVRFSRKRFSEK+A+ WW E +V VY +Y+ +
Sbjct: 969 QKTLKRVRFSRKRFSEKEAQRWWEEKQVLVYNKYDAEI 1006
>At5g63860 UVB-resistance protein UVR8 (gb|AAD43920.1)
Length = 440
Score = 205 bits (522), Expect = 1e-52
Identities = 132/383 (34%), Positives = 195/383 (50%), Gaps = 30/383 (7%)
Query: 214 SQGSGHDDGDALGDVFI-WGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQ 272
S G+ H GD+ WG G DG +G G+ D P L + +
Sbjct: 21 SAGASHSVALLSGDIVCSWGRGE-DGQLGHGDAE---------DRPSPTQLSALDGHQIV 70
Query: 273 NIACGGRHAALVTKQG-EIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEY 331
++ CG H ++ G E++SWG GRLGHG SD+ P I AL I+ +ACG+
Sbjct: 71 SVTCGADHTVAYSQSGMEVYSWGWGDFGRLGHGNSSDLFTPLPIKALHGIRIKQIACGDS 130
Query: 332 HTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVV 391
H AVT+ G++ +WG G LG G+ VP+++ EGI + ++ G HTA V
Sbjct: 131 HCLAVTMEGEVQSWGRN--QNGQLGLGDTEDSLVPQKIQA-FEGIRIKMVAAGAEHTAAV 187
Query: 392 TSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNS 451
T G L+ +G G +G LG GDR +P V S G + +CG HT +V
Sbjct: 188 TEDGDLYGWGWGRYGNLGLGDRTDRLVPERVTSTGGEKMSMVACGWRHTISV-------- 239
Query: 452 SSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNFC-QVACGHSLTVALTTSG 510
SG L+T+G G+LGHGD E L+P + + ++F Q++ G T+ALT+ G
Sbjct: 240 ----SYSGALYTYGWSKYGQLGHGDLEDHLIPHKLEALSNSFISQISGGWRHTMALTSDG 295
Query: 511 HVYAMGSPVYGQLG-NPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGK 569
+Y G +GQ+G D P +V + V+ ++CG H +T RN V+ WG+
Sbjct: 296 KLYGWGWNKFGQVGVGNNLDQCSPVQVRFPDDQKVVQ-VSCGWRHTLAVTERNNVFAWGR 354
Query: 570 GANGRLGHGDTDDRNNPTLVDAL 592
G NG+LG G++ DRN P +++AL
Sbjct: 355 GTNGQLGIGESVDRNFPKIIEAL 377
Score = 184 bits (467), Expect = 2e-46
Identities = 120/369 (32%), Positives = 181/369 (48%), Gaps = 23/369 (6%)
Query: 274 IACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHT 333
I+ G H+ + + SWG G+LGHG D P + AL I V CG HT
Sbjct: 20 ISAGASHSVALLSGDIVCSWGRGEDGQLGHGDAEDRPSPTQLSALDGHQIVSVTCGADHT 79
Query: 334 CAVTLSG-DLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVT 392
A + SG ++Y+WG G ++G LGHGN + P + L GI + I+CG H VT
Sbjct: 80 VAYSQSGMEVYSWGWG--DFGRLGHGNSSDLFTPLPIKA-LHGIRIKQIACGDSHCLAVT 136
Query: 393 SAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSS 452
G++ ++G G LG GD + +P+++++ +G+R + G HTAAV E
Sbjct: 137 MEGEVQSWGRNQNGQLGLGDTEDSLVPQKIQAFEGIRIKMVAAGAEHTAAVTE------- 189
Query: 453 SSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVE-HNFCQVACGHSLTVALTTSGH 511
G L+ WG G G LG GD+ +LVP V VACG T++++ SG
Sbjct: 190 -----DGDLYGWGWGRYGNLGLGDRTDRLVPERVTSTGGEKMSMVACGWRHTISVSYSGA 244
Query: 512 VYAMGSPVYGQLGNPQADGKL-PTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKG 570
+Y G YGQLG+ + L P ++E L SF+ +I+ G H LT ++Y WG
Sbjct: 245 LYTYGWSKYGQLGHGDLEDHLIPHKLEA-LSNSFISQISGGWRHTMALTSDGKLYGWGWN 303
Query: 571 ANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAIC----LHKWVSGVDQSMCSG 626
G++G G+ D+ +P V D+ V ++CG T A+ + W G + + G
Sbjct: 304 KFGQVGVGNNLDQCSPVQVRFPDDQKVVQVSCGWRHTLAVTERNNVFAWGRGTNGQLGIG 363
Query: 627 CRLPFNFKR 635
+ NF +
Sbjct: 364 ESVDRNFPK 372
Score = 116 bits (291), Expect = 6e-26
Identities = 78/238 (32%), Positives = 115/238 (47%), Gaps = 20/238 (8%)
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGV 437
V IS G H+ + S + ++G G G LGHGD + P ++ +L G + + +CG
Sbjct: 17 VLIISAGASHSVALLSGDIVCSWGRGEDGQLGHGDAEDRPSPTQLSALDGHQIVSVTCGA 76
Query: 438 WHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQV 496
HT A + S ++++WG GD GRLGHG+ P + AL Q+
Sbjct: 77 DHTVAYSQ-----------SGMEVYSWGWGDFGRLGHGNSSDLFTPLPIKALHGIRIKQI 125
Query: 497 ACGHSLTVALTTSGHVYAMGSPVYGQLG-NPQADGKLPTRV---EGKLLKSFVEEIACGA 552
ACG S +A+T G V + G GQLG D +P ++ EG +K +A GA
Sbjct: 126 ACGDSHCLAVTMEGEVQSWGRNQNGQLGLGDTEDSLVPQKIQAFEGIRIKM----VAAGA 181
Query: 553 YHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
H A +T ++Y WG G G LG GD DR P V + + + +ACG T ++
Sbjct: 182 EHTAAVTEDGDLYGWGWGRYGNLGLGDRTDRLVPERVTSTGGEKMSMVACGWRHTISV 239
Score = 54.3 bits (129), Expect = 4e-07
Identities = 31/87 (35%), Positives = 47/87 (53%), Gaps = 2/87 (2%)
Query: 235 TGDG-VVGGGNHRVGS-GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFS 292
T DG + G G ++ G G+G +D P + V ++CG RH VT++ +F+
Sbjct: 292 TSDGKLYGWGWNKFGQVGVGNNLDQCSPVQVRFPDDQKVVQVSCGWRHTLAVTERNNVFA 351
Query: 293 WGEESGGRLGHGVDSDVLHPKLIDALS 319
WG + G+LG G D PK+I+ALS
Sbjct: 352 WGRGTNGQLGIGESVDRNFPKIIEALS 378
>At5g08710 unknown protein
Length = 434
Score = 171 bits (433), Expect = 2e-42
Identities = 137/455 (30%), Positives = 209/455 (45%), Gaps = 66/455 (14%)
Query: 186 GSDSMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDGDALGDVFIWGEGTGD-GVVGGGN 244
GS+ G +K + V S VSS S G F G+GD G +G GN
Sbjct: 18 GSNCTSGLLKDSKLGPIGVCCSRWVSSES-----------GKRFAAMWGSGDYGRLGLGN 66
Query: 245 HRVGSGLGVKIDSLFPKALESAVV-LDVQNIACGGRHAALVTKQGEIFSWGEESGGRLG- 302
+DS + A+ SA+ + +ACGG H +T+ +F+ G G+LG
Sbjct: 67 ----------LDSQWTPAVCSALSDHSITAVACGGAHTLFLTETRRVFATGLNDCGQLGV 116
Query: 303 HGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVS 362
V S + P + L +I ++ G YH+ A+T+ G+LY WG + G LG G + +
Sbjct: 117 SDVKSHAMDPLEVSGLDK-DILHISAGYYHSAAITVDGELYMWGKNSS--GQLGLGKKAA 173
Query: 363 HWV--PKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVS--- 417
V P +V L GI + ++ G H+ VT G++ ++G G G LGHG + S+
Sbjct: 174 RVVRVPTKVEA-LHGITIQSVALGSEHSVAVTDGGEVLSWGGGGSGRLGHGHQSSLFGIL 232
Query: 418 ------LPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGR 471
PR ++ L+G++ + G+ H+A E +G F +G+ +
Sbjct: 233 RSNSEFTPRLIKELEGIKVTNVAAGLLHSACTDE------------NGSAFMFGEKSINK 280
Query: 472 LGHGDKEAKLVPTCVALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQAD-G 530
+G G P+ ++ V + +VACG T +T G +Y GS G LG
Sbjct: 281 MGFGGVRNATTPSIISEVPYAE-EVACGGYHTCVVTRGGELYTWGSNENGCLGTDSTYVS 339
Query: 531 KLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGAN------------GRLGHG 578
P RVEG L+S V +++CG H A ++ N V+TWG G + G+LGHG
Sbjct: 340 HSPVRVEGPFLESTVSQVSCGWKHTAAIS-DNNVFTWGWGGSHGTFSVDGHSSGGQLGHG 398
Query: 579 DTDDRNNPTLVDALKDKHVKSIACGTNFTAAICLH 613
D P +VD K+ I+CG N TAA+ H
Sbjct: 399 SDVDYARPAMVDLGKNVRAVHISCGFNHTAAVLEH 433
>At1g27060 hypothetical protein
Length = 376
Score = 162 bits (409), Expect = 1e-39
Identities = 130/409 (31%), Positives = 197/409 (47%), Gaps = 56/409 (13%)
Query: 213 SSQGSGHDDGDALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQ 272
+ G D + V+ WG GT DG +G + D L P+ L + +
Sbjct: 2 AEHGETQADEEEEEQVWSWGAGT-DGQLGTTKLQ---------DELLPQLLSLTSLPSIS 51
Query: 273 NIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYH 332
+ACGG H +T G++F+WG S G+LGHG ++ PKL+ ++ I A G H
Sbjct: 52 MLACGGAHVIALTSGGKVFTWGRGSSGQLGHGDILNITLPKLVSFFDDSVITQAAAGWSH 111
Query: 333 TCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVT 392
+ V+ SG ++T GNG ++G LGHG+ +S P +V+ V ++CG
Sbjct: 112 SGFVSDSGCIFTCGNG--SFGQLGHGDTLSLSTPAKVS-HFNNDSVKMVACG-------- 160
Query: 393 SAGQLFTFGDGTFGALG-HGDR-KSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGN 450
Q+ FG G G LG DR KSV+LP V LK + +R S H+AA+
Sbjct: 161 --NQVCGFGSGKRGQLGFSSDRIKSVNLPCVVSGLKDVEVVRISANGDHSAAI------- 211
Query: 451 SSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNFCQVACGHSLTVALTTSG 510
+ G+ F+WG G G G + +P+ + +F +VA G + + LT G
Sbjct: 212 -----SADGQFFSWGRGFCG--GPDVHAPQSLPSPL-----SFREVAVGWNHALLLTVDG 259
Query: 511 HVYAMGS-----PVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVY 565
V+ +GS P QL ++ + +K V +IA GA H A +T EV
Sbjct: 260 EVFKLGSTLNKQPEKQQLQIDSSEALFEKVPDFDGVK--VMQIAAGAEHSAAVTENGEVK 317
Query: 566 TWGKGANGRLGHGDTDDRNNPTLVDA----LKDKHVKSIACGTNFTAAI 610
TWG G +G+LG G+T+D+ +P LV L+ K +K + CG+ FT A+
Sbjct: 318 TWGWGEHGQLGLGNTNDQTSPELVSLGSIDLRTKEIK-VYCGSGFTYAV 365
Score = 78.2 bits (191), Expect = 2e-14
Identities = 69/237 (29%), Positives = 105/237 (44%), Gaps = 31/237 (13%)
Query: 396 QLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRA-SCGVWHTAAVVEVMVGNSSSS 454
Q++++G GT G LG + LP ++ SL L ++ +CG H A+
Sbjct: 16 QVWSWGAGTDGQLGTTKLQDELLP-QLLSLTSLPSISMLACGGAHVIALT---------- 64
Query: 455 NCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHN-FCQVACGHSLTVALTTSGHVY 513
S GK+FTWG G G+LGHGD +P V+ + + Q A G S + ++ SG ++
Sbjct: 65 --SGGKVFTWGRGSSGQLGHGDILNITLPKLVSFFDDSVITQAAAGWSHSGFVSDSGCIF 122
Query: 514 AMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANG 573
G+ +GQLG+ V+ +ACG N+V +G G G
Sbjct: 123 TCGNGSFGQLGHGDTLSLSTPAKVSHFNNDSVKMVACG----------NQVCGFGSGKRG 172
Query: 574 RLGHGDTDDR----NNPTLVDALKDKHVKSIACGTNFTAAICLHKWVSGVDQSMCSG 626
+LG + DR N P +V LKD V I+ + +AAI + C G
Sbjct: 173 QLGF--SSDRIKSVNLPCVVSGLKDVEVVRISANGDHSAAISADGQFFSWGRGFCGG 227
>At3g15430 unknown protein
Length = 488
Score = 148 bits (374), Expect = 1e-35
Identities = 107/338 (31%), Positives = 157/338 (45%), Gaps = 31/338 (9%)
Query: 259 FPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGH-GVDSDVLHPKLIDA 317
FP + A V QN H+A V + G++ + G+ S GH + PKL++A
Sbjct: 159 FPFPAQVAQVSATQN------HSAFVLQSGQVLTCGDNSSHCCGHLDTSRPIFRPKLVEA 212
Query: 318 LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIH 377
L T + VA G + T ++ G YT G+ + G LGHG+ + VPK V
Sbjct: 213 LKGTPCKQVAAGLHFTVFLSREGHAYTCGSNTH--GQLGHGDTLDRPVPKVVEFLKTIGP 270
Query: 378 VSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLK--GLRTMRASC 435
V I+ GP + VT G +++FG G+ LGHG+++ PR +++ K G+ +R S
Sbjct: 271 VVQIAAGPSYVLAVTQDGSVYSFGSGSNFCLGHGEQQDELQPRVIQAFKRKGIHILRVSA 330
Query: 436 GVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNFC- 494
G H A+ S+G+++TWG G G LGHGD+ K+ P LV N C
Sbjct: 331 GDEHAVALD------------SNGRVYTWGKGYCGALGHGDENDKITPQ--VLVNLNNCL 376
Query: 495 --QVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQ---ADGKLPTRVEGKLLKSFVEEIA 549
QV T L G +Y G +G LG P +D L RV L V +++
Sbjct: 377 AVQVCARKRKTFVLVEGGLLYGFGWMGFGSLGFPDRGVSDKVLRPRVLECLKPHRVSQVS 436
Query: 550 CGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPT 587
G YH V+T R ++ +G +LGH PT
Sbjct: 437 TGLYHTIVVTQRGRIFGFGDNERAQLGHDSLRGCLEPT 474
Score = 110 bits (274), Expect = 6e-24
Identities = 82/259 (31%), Positives = 117/259 (44%), Gaps = 21/259 (8%)
Query: 236 GDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLD-VQNIACGGRHAALVTKQGEIFSWG 294
G G N G G +D PK +E + V IA G + VT+ G ++S+G
Sbjct: 235 GHAYTCGSNTHGQLGHGDTLDRPVPKVVEFLKTIGPVVQIAAGPSYVLAVTQDGSVYSFG 294
Query: 295 EESGGRLGHGVDSDVLHPKLIDALSNTNIEL--VACGEYHTCAVTLSGDLYTWGNGAYNY 352
S LGHG D L P++I A I + V+ G+ H A+ +G +YTWG G
Sbjct: 295 SGSNFCLGHGEQQDELQPRVIQAFKRKGIHILRVSAGDEHAVALDSNGRVYTWGKG--YC 352
Query: 353 GLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGD 412
G LGHG++ P +V L + T V+ G L+ FG FG+LG D
Sbjct: 353 GALGHGDENDKITP-QVLVNLNNCLAVQVCARKRKTFVLVEGGLLYGFGWMGFGSLGFPD 411
Query: 413 R---KSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDK 469
R V PR +E LK R + S G++HT V + G++F +GD ++
Sbjct: 412 RGVSDKVLRPRVLECLKPHRVSQVSTGLYHTIVVTQ------------RGRIFGFGDNER 459
Query: 470 GRLGHGDKEAKLVPTCVAL 488
+LGH L PT + L
Sbjct: 460 AQLGHDSLRGCLEPTEIFL 478
Score = 108 bits (270), Expect = 2e-23
Identities = 81/272 (29%), Positives = 124/272 (44%), Gaps = 26/272 (9%)
Query: 326 VACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLE---GIHVSYIS 382
+A G+YHT + S + G G+L HG++ + V P+E V+ +S
Sbjct: 116 IATGKYHTLLINNSK---VYSCGVSLSGVLAHGSETTQCVAFT---PIEFPFPAQVAQVS 169
Query: 383 CGPWHTAVVTSAGQLFTFGDGTFGALGHGDR-KSVSLPREVESLKGLRTMRASCGVWHTA 441
H+A V +GQ+ T GD + GH D + + P+ VE+LKG + + G+ T
Sbjct: 170 ATQNHSAFVLQSGQVLTCGDNSSHCCGHLDTSRPIFRPKLVEALKGTPCKQVAAGLHFTV 229
Query: 442 AVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEH--NFCQVACG 499
+ G +T G G+LGHGD + VP V ++ Q+A G
Sbjct: 230 FLSR------------EGHAYTCGSNTHGQLGHGDTLDRPVPKVVEFLKTIGPVVQIAAG 277
Query: 500 HSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVE--EIACGAYHVAV 557
S +A+T G VY+ GS LG+ + +L RV + + ++ G H
Sbjct: 278 PSYVLAVTQDGSVYSFGSGSNFCLGHGEQQDELQPRVIQAFKRKGIHILRVSAGDEHAVA 337
Query: 558 LTLRNEVYTWGKGANGRLGHGDTDDRNNPTLV 589
L VYTWGKG G LGHGD +D+ P ++
Sbjct: 338 LDSNGRVYTWGKGYCGALGHGDENDKITPQVL 369
Score = 86.3 bits (212), Expect = 9e-17
Identities = 68/236 (28%), Positives = 108/236 (44%), Gaps = 19/236 (8%)
Query: 381 ISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLP-REVESLKGLRTMRASCGVWH 439
I+ G +HT ++ ++ ++++ G G L HG + + +E + + S H
Sbjct: 116 IATGKYHTLLINNS-KVYSCGVSLSGVLAHGSETTQCVAFTPIEFPFPAQVAQVSATQNH 174
Query: 440 TAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLV-PTCVALVEHNFC-QVA 497
+A V++ SG++ T GD GH D + P V ++ C QVA
Sbjct: 175 SAFVLQ------------SGQVLTCGDNSSHCCGHLDTSRPIFRPKLVEALKGTPCKQVA 222
Query: 498 CGHSLTVALTTSGHVYAMGSPVYGQLGNPQA-DGKLPTRVEGKLLKSFVEEIACGAYHVA 556
G TV L+ GH Y GS +GQLG+ D +P VE V +IA G +V
Sbjct: 223 AGLHFTVFLSREGHAYTCGSNTHGQLGHGDTLDRPVPKVVEFLKTIGPVVQIAAGPSYVL 282
Query: 557 VLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDK--HVKSIACGTNFTAAI 610
+T VY++G G+N LGHG+ D P ++ A K K H+ ++ G A+
Sbjct: 283 AVTQDGSVYSFGSGSNFCLGHGEQQDELQPRVIQAFKRKGIHILRVSAGDEHAVAL 338
>At5g60870 UVB-resistance protein UVR8 - like
Length = 445
Score = 148 bits (373), Expect = 2e-35
Identities = 110/350 (31%), Positives = 167/350 (47%), Gaps = 45/350 (12%)
Query: 289 EIFSWGEESGGRLGHGVDSDVLHPK--------------------LIDALSNTNIELVAC 328
++FSWG + G+LG G++ ++P IDA S++ ++C
Sbjct: 49 QLFSWGRGASGQLGGGIEEIRMYPSPVANLLFRSDQSFSLAQTPGRIDADSSSFRIGISC 108
Query: 329 GEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHT 388
G +H+ +T+ GDL+ WG G + G LG G + S +VP +N E + I+ G H+
Sbjct: 109 GLFHS-GLTIDGDLWIWGKG--DGGRLGFGQENSVFVP-NLNPLFEEHSIRCIALGGLHS 164
Query: 389 AVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMV 448
+T G +FT+G G FGALGH +PR V+ + + HTAA+ E
Sbjct: 165 VALTHQGDVFTWGYGGFGALGHKVYTRELVPRRVDDSWDCKISAIATSGTHTAAITE--- 221
Query: 449 GNSSSSNCSSGKLFTWG--DGDKGRLGHG------DKEAKLVPTCVALVEHNFCQVACGH 500
SG+L+ WG +GD GRLG G + VP+ V + V+CG
Sbjct: 222 ---------SGELYMWGREEGD-GRLGLGPGRGPNEGGGLSVPSKVKALTVPVASVSCGG 271
Query: 501 SLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTL 560
T+ALT G ++ G+ +LG G L + +IACG YH LT
Sbjct: 272 FFTMALTKEGQLWNWGANSNYELGRGDNLGGWEPMPVPSLEGVRITQIACGGYHSLALTE 331
Query: 561 RNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
+V +WG G +G+LG ++ PT ++AL DK + IA G + +AAI
Sbjct: 332 EGKVLSWGHGGHGQLGSSSLRNQKVPTEIEALADKKIVFIASGGSSSAAI 381
Score = 144 bits (364), Expect = 2e-34
Identities = 112/350 (32%), Positives = 165/350 (47%), Gaps = 37/350 (10%)
Query: 226 GDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVT 285
GD++IWG+G G G +G G V + +L P E ++ + IA GG H+ +T
Sbjct: 119 GDLWIWGKGDG-GRLGFGQEN-----SVFVPNLNPLFEEHSI----RCIALGGLHSVALT 168
Query: 286 KQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTW 345
QG++F+WG G LGH V + L P+ +D + I +A HT A+T SG+LY W
Sbjct: 169 HQGDVFTWGYGGFGALGHKVYTRELVPRRVDDSWDCKISAIATSGTHTAAITESGELYMW 228
Query: 346 GNGAYNYGL-LGHGNQVSHW----VPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTF 400
G + L LG G + VP +V + V+ +SCG + T +T GQL+ +
Sbjct: 229 GREEGDGRLGLGPGRGPNEGGGLSVPSKVKALT--VPVASVSCGGFFTMALTKEGQLWNW 286
Query: 401 GDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGK 460
G + LG GD P V SL+G+R + +CG +H+ A+ E GK
Sbjct: 287 GANSNYELGRGDNLGGWEPMPVPSLEGVRITQIACGGYHSLALTE------------EGK 334
Query: 461 LFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACGHSLTVALTTSGHVYAMGSPV 519
+ +WG G G+LG + VPT + AL + +A G S + A+T G ++ G+
Sbjct: 335 VLSWGHGGHGQLGSSSLRNQKVPTEIEALADKKIVFIASGGSSSAAITDGGELWMWGNAK 394
Query: 520 YGQLGNPQADGKLPTRVEGKLLKSFVE-------EIACGAYHVAVLTLRN 562
QLG P T VE L E I+ GA H L R+
Sbjct: 395 DFQLGVPGLPEIQTTPVEVNFLTEEDECQPHKVISISIGASHALCLVSRS 444
Score = 144 bits (362), Expect = 3e-34
Identities = 123/411 (29%), Positives = 192/411 (45%), Gaps = 46/411 (11%)
Query: 228 VFIWGEGTGDGVVGGGNHRVG------SGLGVKIDSLFPKALESAVVLDVQN------IA 275
+F WG G G +GGG + + L + D F A ++ +D + I+
Sbjct: 50 LFSWGRGAS-GQLGGGIEEIRMYPSPVANLLFRSDQSFSLA-QTPGRIDADSSSFRIGIS 107
Query: 276 CGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCA 335
CG H+ L T G+++ WG+ GGRLG G ++ V P L +I +A G H+ A
Sbjct: 108 CGLFHSGL-TIDGDLWIWGKGDGGRLGFGQENSVFVPNLNPLFEEHSIRCIALGGLHSVA 166
Query: 336 VTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAG 395
+T GD++TWG G +G LGH VP+RV+ + +S I+ HTA +T +G
Sbjct: 167 LTHQGDVFTWGYG--GFGALGHKVYTRELVPRRVDDSWD-CKISAIATSGTHTAAITESG 223
Query: 396 QLFTF----GDGTFG---ALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMV 448
+L+ + GDG G G + +S+P +V++L + SCG + T A+ +
Sbjct: 224 ELYMWGREEGDGRLGLGPGRGPNEGGGLSVPSKVKALT-VPVASVSCGGFFTMALTK--- 279
Query: 449 GNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEH-NFCQVACGHSLTVALT 507
G+L+ WG LG GD P V +E Q+ACG ++ALT
Sbjct: 280 ---------EGQLWNWGANSNYELGRGDNLGGWEPMPVPSLEGVRITQIACGGYHSLALT 330
Query: 508 TSGHVYAMGSPVYGQLGNPQ-ADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYT 566
G V + G +GQLG+ + K+PT +E K V IA G A +T E++
Sbjct: 331 EEGKVLSWGHGGHGQLGSSSLRNQKVPTEIEALADKKIV-FIASGGSSSAAITDGGELWM 389
Query: 567 WGKGANGRLG-HGDTDDRNNPTLVDALKDKHV----KSIACGTNFTAAICL 612
WG + +LG G + + P V+ L ++ K I+ + A+CL
Sbjct: 390 WGNAKDFQLGVPGLPEIQTTPVEVNFLTEEDECQPHKVISISIGASHALCL 440
>At5g16040 UVB-resistance protein-like
Length = 396
Score = 135 bits (341), Expect = 9e-32
Identities = 106/365 (29%), Positives = 169/365 (46%), Gaps = 48/365 (13%)
Query: 228 VFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQ 287
V WG G DG +G G L +D+L P +V+++ G R++ +
Sbjct: 7 VIAWGSGE-DGQLGLGTDEEKE-LASVVDALEP--------FNVRSVVGGSRNSLAICDD 56
Query: 288 GEIFSWGEESGGRLGHGVDSDVLH-PKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWG 346
G++F+WG G LGH ++ P L+ +L+N I A G +H AV G Y WG
Sbjct: 57 GKLFTWGWNQRGTLGHPPETKTESTPSLVKSLANVKIVQAAIGGWHCLAVDDQGRAYAWG 116
Query: 347 NGAYNYGLLGH-------GNQVSH--WVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQL 397
YG G G V +PKR + + V ++ G H+ V+T G +
Sbjct: 117 GN--EYGQCGEEPSKDETGRPVRRDIVIPKRC---AQQLTVRQVAAGGTHSVVLTREGYV 171
Query: 398 FTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCS 457
+T+G GD K +S+P V+ L+ +R + + G +H A+ E
Sbjct: 172 WTWGQ----PWPPGDIKQISVPVRVQGLENVRLI--AVGAFHNLALKE------------ 213
Query: 458 SGKLFTWGDGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACGHSLTVALTTSGHVYAMG 516
G L+ WG+ + G+LG GD + + P V L + +A G + ALT G VY G
Sbjct: 214 DGTLWAWGNNEYGQLGTGDTQPRSYPIPVQGLDDLTLVDIAAGGWHSTALTNEGEVYGWG 273
Query: 517 SPVYGQLG---NPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANG 573
+G+LG N ++ LP +V L + +++CG H LT ++++G+G +G
Sbjct: 274 RGEHGRLGFGDNDKSSKMLPQKVN-LLAGEDIIQVSCGGTHSVALTRDGRIFSFGRGDHG 332
Query: 574 RLGHG 578
RLG+G
Sbjct: 333 RLGYG 337
Score = 121 bits (304), Expect = 2e-27
Identities = 81/257 (31%), Positives = 128/257 (49%), Gaps = 26/257 (10%)
Query: 223 DALGDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAA 282
D G + WG G G G + +G V+ D + PK A L V+ +A GG H+
Sbjct: 107 DDQGRAYAWG-GNEYGQCGEEPSKDETGRPVRRDIVIPK--RCAQQLTVRQVAAGGTHSV 163
Query: 283 LVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDL 342
++T++G +++WG+ G + P + L N + L+A G +H A+ G L
Sbjct: 164 VLTREGYVWTWGQP----WPPGDIKQISVPVRVQGLEN--VRLIAVGAFHNLALKEDGTL 217
Query: 343 YTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGD 402
+ WGN YG LG G+ P V G L+ + + I+ G WH+ +T+ G+++ +G
Sbjct: 218 WAWGNN--EYGQLGTGDTQPRSYPIPVQG-LDDLTLVDIAAGGWHSTALTNEGEVYGWGR 274
Query: 403 GTFGALGHGD--RKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGK 460
G G LG GD + S LP++V L G ++ SCG H+ A+ G+
Sbjct: 275 GEHGRLGFGDNDKSSKMLPQKVNLLAGEDIIQVSCGGTHSVALTR------------DGR 322
Query: 461 LFTWGDGDKGRLGHGDK 477
+F++G GD GRLG+G K
Sbjct: 323 IFSFGRGDHGRLGYGRK 339
Score = 114 bits (285), Expect = 3e-25
Identities = 90/333 (27%), Positives = 140/333 (42%), Gaps = 34/333 (10%)
Query: 290 IFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGA 349
+ +WG G+LG G D + ++DAL N+ V G ++ A+ G L+TWG
Sbjct: 7 VIAWGSGEDGQLGLGTDEEKELASVVDALEPFNVRSVVGGSRNSLAICDDGKLFTWGWN- 65
Query: 350 YNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALG 409
G LGH + + L + + + G WH V G+ + +G +G G
Sbjct: 66 -QRGTLGHPPETKTESTPSLVKSLANVKIVQAAIGGWHCLAVDDQGRAYAWGGNEYGQCG 124
Query: 410 HGD---------RKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGK 460
R+ + +P+ +R + A G S G
Sbjct: 125 EEPSKDETGRPVRRDIVIPKRCAQQLTVRQVAAG--------------GTHSVVLTREGY 170
Query: 461 LFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVY 520
++TWG GD + VP V +E N +A G +AL G ++A G+ Y
Sbjct: 171 VWTWGQP----WPPGDIKQISVPVRVQGLE-NVRLIAVGAFHNLALKEDGTLWAWGNNEY 225
Query: 521 GQLGNPQADGK-LPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGD 579
GQLG + P V+G L + +IA G +H LT EVY WG+G +GRLG GD
Sbjct: 226 GQLGTGDTQPRSYPIPVQG-LDDLTLVDIAAGGWHSTALTNEGEVYGWGRGEHGRLGFGD 284
Query: 580 TDDRNN--PTLVDALKDKHVKSIACGTNFTAAI 610
D + P V+ L + + ++CG + A+
Sbjct: 285 NDKSSKMLPQKVNLLAGEDIIQVSCGGTHSVAL 317
Score = 91.3 bits (225), Expect = 3e-18
Identities = 55/192 (28%), Positives = 92/192 (47%), Gaps = 17/192 (8%)
Query: 270 DVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACG 329
+V+ IA G H + + G +++WG G+LG G +P + L + + +A G
Sbjct: 197 NVRLIAVGAFHNLALKEDGTLWAWGNNEYGQLGTGDTQPRSYPIPVQGLDDLTLVDIAAG 256
Query: 330 EYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTA 389
+H+ A+T G++Y WG G + G ++ S +P++VN L G + +SCG H+
Sbjct: 257 GWHSTALTNEGEVYGWGRGEHGRLGFGDNDKSSKMLPQKVN-LLAGEDIIQVSCGGTHSV 315
Query: 390 VVTSAGQLFTFGDGTFGALGHGDRKSVSLPREV----------------ESLKGLRTMRA 433
+T G++F+FG G G LG+G + + P E+ E
Sbjct: 316 ALTRDGRIFSFGRGDHGRLGYGRKVTTGQPLELPIHIPPPEGRFNHTDEEDDGKWIAKHV 375
Query: 434 SCGVWHTAAVVE 445
+CG HT A+VE
Sbjct: 376 ACGGRHTLAIVE 387
Score = 71.2 bits (173), Expect = 3e-12
Identities = 41/127 (32%), Positives = 71/127 (55%), Gaps = 5/127 (3%)
Query: 243 GNHRVGS-GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRL 301
GN+ G G G +P ++ L + +IA GG H+ +T +GE++ WG GRL
Sbjct: 221 GNNEYGQLGTGDTQPRSYPIPVQGLDDLTLVDIAAGGWHSTALTNEGEVYGWGRGEHGRL 280
Query: 302 GHGVD--SDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGN 359
G G + S + P+ ++ L+ +I V+CG H+ A+T G ++++G G ++G LG+G
Sbjct: 281 GFGDNDKSSKMLPQKVNLLAGEDIIQVSCGGTHSVALTRDGRIFSFGRG--DHGRLGYGR 338
Query: 360 QVSHWVP 366
+V+ P
Sbjct: 339 KVTTGQP 345
Score = 53.1 bits (126), Expect = 8e-07
Identities = 36/127 (28%), Positives = 58/127 (45%), Gaps = 24/127 (18%)
Query: 226 GDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVT 285
G+V+ WG G + R+G G K + P+ + D+ ++CGG H+ +T
Sbjct: 267 GEVYGWGRGE--------HGRLGFGDNDKSSKMLPQKVNLLAGEDIIQVSCGGTHSVALT 318
Query: 286 KQGEIFSWGEESGGRLGHG--------VDSDVLHPKLIDALSNTNIE--------LVACG 329
+ G IFS+G GRLG+G ++ + P ++T+ E VACG
Sbjct: 319 RDGRIFSFGRGDHGRLGYGRKVTTGQPLELPIHIPPPEGRFNHTDEEDDGKWIAKHVACG 378
Query: 330 EYHTCAV 336
HT A+
Sbjct: 379 GRHTLAI 385
Score = 52.4 bits (124), Expect = 1e-06
Identities = 43/172 (25%), Positives = 71/172 (41%), Gaps = 15/172 (8%)
Query: 457 SSGKLFTWGDGDKGRLGHG-DKEAKLVPTCVALVEHNFCQVACGHSLTVALTTSGHVYAM 515
S+ + WG G+ G+LG G D+E +L AL N V G ++A+ G ++
Sbjct: 3 SATSVIAWGSGEDGQLGLGTDEEKELASVVDALEPFNVRSVVGGSRNSLAICDDGKLFTW 62
Query: 516 GSPVYGQLGNPQADGKLPTRVEGKLLKSF----VEEIACGAYHVAVLTLRNEVYTWGKGA 571
G G LG+P + T L+KS + + A G +H + + Y WG
Sbjct: 63 GWNQRGTLGHPP---ETKTESTPSLVKSLANVKIVQAAIGGWHCLAVDDQGRAYAWGGNE 119
Query: 572 NGRLGHGDT-DDRNNPTLVDALKDKH------VKSIACGTNFTAAICLHKWV 616
G+ G + D+ P D + K V+ +A G + + +V
Sbjct: 120 YGQCGEEPSKDETGRPVRRDIVIPKRCAQQLTVRQVAAGGTHSVVLTREGYV 171
Score = 36.2 bits (82), Expect = 0.10
Identities = 27/99 (27%), Positives = 44/99 (44%), Gaps = 1/99 (1%)
Query: 506 LTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVY 565
+ ++ V A GS GQLG + K V L V + G+ + + +++
Sbjct: 1 MASATSVIAWGSGEDGQLGLGTDEEKELASVVDALEPFNVRSVVGGSRNSLAICDDGKLF 60
Query: 566 TWGKGANGRLGH-GDTDDRNNPTLVDALKDKHVKSIACG 603
TWG G LGH +T + P+LV +L + + A G
Sbjct: 61 TWGWNQRGTLGHPPETKTESTPSLVKSLANVKIVQAAIG 99
>At3g02510 unknown protein
Length = 393
Score = 134 bits (338), Expect = 2e-31
Identities = 108/405 (26%), Positives = 177/405 (43%), Gaps = 51/405 (12%)
Query: 235 TGDGVVGGGNHRVGS-GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSW 293
T ++ G+ G G+G + + +E+ V+++ G R++ + G +F+W
Sbjct: 3 TATSIIAWGSGEDGQLGIGTNEEKEWACVVEALEPYSVRSVVSGSRNSLAICDDGTMFTW 62
Query: 294 GEESGGRLGHGVDSDVLH-PKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNY 352
G G LGH ++ + P + AL+N I A G +H AV G Y WG Y
Sbjct: 63 GWNQRGTLGHQPETKTENIPSRVKALANVKITQAAIGGWHCLAVDDQGRAYAWGGNEYGQ 122
Query: 353 -------GLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTF 405
+G + +PKR L V ++ G H+ V+T G ++T+G
Sbjct: 123 CGEEPLKDEMGRPVRRDIVIPKRCAPKLT---VRQVAAGGTHSVVLTREGHVWTWGQ--- 176
Query: 406 GALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWG 465
GD K +S+P V+ L+ +R + + G +H A+ E G+L WG
Sbjct: 177 -PWPPGDIKQISVPVRVQGLENVRLI--AVGAFHNLALEE------------DGRLLAWG 221
Query: 466 DGDKGRLGHGDKEAKLVPTCV-ALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLG 524
+ + G+LG GD + P V L + +A G + ALT G VY G +G+LG
Sbjct: 222 NNEYGQLGTGDTQPTSHPVPVQGLDDLTLVDIAAGGWHSTALTDKGEVYGWGRGEHGRLG 281
Query: 525 ---NPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHG--- 578
N ++ +P +V L + + +++CG H LT ++++G+G +GRLG+G
Sbjct: 282 LGDNDKSSKMVPQKVN-LLAEEDIIQVSCGGTHSVALTRDGRIFSFGRGDHGRLGYGRKV 340
Query: 579 -------------DTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
+ N T + KSIACG T AI
Sbjct: 341 TTGQPLELPIKIPPPEGSFNHTDEEEEGKWSAKSIACGGRHTLAI 385
Score = 96.3 bits (238), Expect = 8e-20
Identities = 58/192 (30%), Positives = 96/192 (49%), Gaps = 17/192 (8%)
Query: 270 DVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACG 329
+V+ IA G H + + G + +WG G+LG G HP + L + + +A G
Sbjct: 197 NVRLIAVGAFHNLALEEDGRLLAWGNNEYGQLGTGDTQPTSHPVPVQGLDDLTLVDIAAG 256
Query: 330 EYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTA 389
+H+ A+T G++Y WG G + LG ++ S VP++VN L + +SCG H+
Sbjct: 257 GWHSTALTDKGEVYGWGRGEHGRLGLGDNDKSSKMVPQKVN-LLAEEDIIQVSCGGTHSV 315
Query: 390 VVTSAGQLFTFGDGTFGALGHGDRKSVSLPREV---------------ESLKGLRTMRA- 433
+T G++F+FG G G LG+G + + P E+ E +G + ++
Sbjct: 316 ALTRDGRIFSFGRGDHGRLGYGRKVTTGQPLELPIKIPPPEGSFNHTDEEEEGKWSAKSI 375
Query: 434 SCGVWHTAAVVE 445
+CG HT A+VE
Sbjct: 376 ACGGRHTLAIVE 387
Score = 70.5 bits (171), Expect = 5e-12
Identities = 63/236 (26%), Positives = 110/236 (45%), Gaps = 23/236 (9%)
Query: 178 SVHSISSGGSDSM----HGHMKTMGM-----DAFRVSLSSAVSSSSQGSGHDDGDALGDV 228
+V +++GG+ S+ GH+ T G D ++S+ V QG + A+G
Sbjct: 151 TVRQVAAGGTHSVVLTREGHVWTWGQPWPPGDIKQISVPVRV----QGLENVRLIAVGAF 206
Query: 229 FIWGEGTGDGVVGGGNHRVGS-GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQ 287
++ GN+ G G G + P ++ L + +IA GG H+ +T +
Sbjct: 207 HNLALEEDGRLLAWGNNEYGQLGTGDTQPTSHPVPVQGLDDLTLVDIAAGGWHSTALTDK 266
Query: 288 GEIFSWGEESGGRLGHGVD--SDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTW 345
GE++ WG GRLG G + S + P+ ++ L+ +I V+CG H+ A+T G ++++
Sbjct: 267 GEVYGWGRGEHGRLGLGDNDKSSKMVPQKVNLLAEEDIIQVSCGGTHSVALTRDGRIFSF 326
Query: 346 GNGAYNYGLLGHGNQVSHWVPKRVN---GPLEGI--HVSYISCGPWHTAVVTSAGQ 396
G G ++G LG+G +V+ P + P EG H G W + G+
Sbjct: 327 GRG--DHGRLGYGRKVTTGQPLELPIKIPPPEGSFNHTDEEEEGKWSAKSIACGGR 380
Score = 54.3 bits (129), Expect = 4e-07
Identities = 35/127 (27%), Positives = 59/127 (45%), Gaps = 24/127 (18%)
Query: 226 GDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVT 285
G+V+ WG G + R+G G K + P+ + D+ ++CGG H+ +T
Sbjct: 267 GEVYGWGRGE--------HGRLGLGDNDKSSKMVPQKVNLLAEEDIIQVSCGGTHSVALT 318
Query: 286 KQGEIFSWGEESGGRLGHG--------VDSDVLHPKLIDALSNTNIE--------LVACG 329
+ G IFS+G GRLG+G ++ + P + ++T+ E +ACG
Sbjct: 319 RDGRIFSFGRGDHGRLGYGRKVTTGQPLELPIKIPPPEGSFNHTDEEEEGKWSAKSIACG 378
Query: 330 EYHTCAV 336
HT A+
Sbjct: 379 GRHTLAI 385
Score = 38.9 bits (89), Expect = 0.016
Identities = 16/48 (33%), Positives = 29/48 (60%)
Query: 564 VYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAIC 611
+ WG G +G+LG G +++ +V+AL+ V+S+ G+ + AIC
Sbjct: 7 IIAWGSGEDGQLGIGTNEEKEWACVVEALEPYSVRSVVSGSRNSLAIC 54
>At3g02300 unknown protein
Length = 471
Score = 126 bits (316), Expect = 7e-29
Identities = 108/373 (28%), Positives = 168/373 (44%), Gaps = 62/373 (16%)
Query: 273 NIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGEYH 332
+I+CG H A V G +F+WG G+LG G + HPK + L + ++ V+CG +
Sbjct: 65 DISCGREHTAAVASDGSLFAWGANEYGQLGDGTEVGRKHPKKVKQLQSEFVKFVSCGAFC 124
Query: 333 TCAV--------TLS-GDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEG-IHVSYIS 382
T A+ TLS L+ WG NQ S+ +P+ +G + +S
Sbjct: 125 TAAIAEPRENDGTLSTSRLWVWGQ-----------NQGSN-LPRLFSGAFPATTAIRQVS 172
Query: 383 CGPWHTAVVTSAGQLFTFGDGTFGALGHG-DRKSVSLPREV--------ESLKGLRTMRA 433
CG H ++ G L +G G LG G + + PR + E+ + ++ M+
Sbjct: 173 CGTAHVVALSEEGLLQAWGYNEQGQLGRGVTCEGLQAPRVINAYAKFLDEAPELVKIMQL 232
Query: 434 SCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGH-----GDKEAKLVP-TCVA 487
SCG +HTAA+ + +G+++TWG G G+LGH GDKE L+P V
Sbjct: 233 SCGEYHTAALSD------------AGEVYTWGLGSMGQLGHVSLQSGDKE--LIPRRVVG 278
Query: 488 LVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLG-NPQA---------DGKLPTRVE 537
L + +VACG T AL+ G +YA G GQLG PQ+ L V
Sbjct: 279 LDGVSMKEVACGGVHTCALSLEGALYAWGGGQAGQLGLGPQSGFFFSVSNGSEMLLRNVP 338
Query: 538 GKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHV 597
++ + V +ACG H V + WG + G+ + + P+ VD + V
Sbjct: 339 VLVIPTDVRLVACGHSHTLVYMREGRICGWGYNSYGQAANEKSSYAWYPSPVDWCVGQ-V 397
Query: 598 KSIACGTNFTAAI 610
+ +A G +A +
Sbjct: 398 RKLAAGGGHSAVL 410
Score = 105 bits (261), Expect = 2e-22
Identities = 89/325 (27%), Positives = 143/325 (43%), Gaps = 73/325 (22%)
Query: 248 GSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDS 307
G G + LF A + ++ ++CG H ++++G + +WG G+LG GV
Sbjct: 147 GQNQGSNLPRLFSGAFPATTA--IRQVSCGTAHVVALSEEGLLQAWGYNEQGQLGRGVTC 204
Query: 308 DVLH-PKLIDALSN--------TNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHG 358
+ L P++I+A + I ++CGEYHT A++ +G++YTWG G + G LGH
Sbjct: 205 EGLQAPRVINAYAKFLDEAPELVKIMQLSCGEYHTAALSDAGEVYTWGLG--SMGQLGHV 262
Query: 359 NQVS---HWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKS 415
+ S +P+RV G L+G+ + ++CG HT ++ G L+ +G G G LG G +
Sbjct: 263 SLQSGDKELIPRRVVG-LDGVSMKEVACGGVHTCALSLEGALYAWGGGQAGQLGLGPQ-- 319
Query: 416 VSLPREVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHG 475
SG F+ +G + L
Sbjct: 320 ------------------------------------------SGFFFSVSNGSEMLL--R 335
Query: 476 DKEAKLVPTCVALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQAD-GKLPT 534
+ ++PT V L VACGHS T+ G + G YGQ N ++ P+
Sbjct: 336 NVPVLVIPTDVRL-------VACGHSHTLVYMREGRICGWGYNSYGQAANEKSSYAWYPS 388
Query: 535 RVEGKLLKSFVEEIACGAYHVAVLT 559
V+ + V ++A G H AVLT
Sbjct: 389 PVDWCV--GQVRKLAAGGGHSAVLT 411
Score = 81.6 bits (200), Expect = 2e-15
Identities = 54/195 (27%), Positives = 94/195 (47%), Gaps = 17/195 (8%)
Query: 264 ESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGH----GVDSDVLHPKLIDALS 319
E+ ++ + ++CG H A ++ GE+++WG S G+LGH D +++ P+ + L
Sbjct: 222 EAPELVKIMQLSCGEYHTAALSDAGEVYTWGLGSMGQLGHVSLQSGDKELI-PRRVVGLD 280
Query: 320 NTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQV---------SHWVPKRVN 370
+++ VACG HTCA++L G LY WG G G LG G Q S + + V
Sbjct: 281 GVSMKEVACGGVHTCALSLEGALYAWGGG--QAGQLGLGPQSGFFFSVSNGSEMLLRNVP 338
Query: 371 GPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRT 430
+ V ++CG HT V G++ +G ++G + P V+ G +
Sbjct: 339 VLVIPTDVRLVACGHSHTLVYMREGRICGWGYNSYGQAANEKSSYAWYPSPVDWCVG-QV 397
Query: 431 MRASCGVWHTAAVVE 445
+ + G H+A + +
Sbjct: 398 RKLAAGGGHSAVLTD 412
Score = 64.7 bits (156), Expect = 3e-10
Identities = 50/183 (27%), Positives = 83/183 (45%), Gaps = 23/183 (12%)
Query: 226 GDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVT 285
G+V+ WG G+ G +G + + G L P+ + + ++ +ACGG H ++
Sbjct: 246 GEVYTWGLGSM-GQLGHVSLQSGD------KELIPRRVVGLDGVSMKEVACGGVHTCALS 298
Query: 286 KQGEIFSWGEESGGRLGHGVD----------SDVLHPKLIDALSNTNIELVACGEYHTCA 335
+G +++WG G+LG G S++L + + T++ LVACG HT
Sbjct: 299 LEGALYAWGGGQAGQLGLGPQSGFFFSVSNGSEMLLRNVPVLVIPTDVRLVACGHSHTLV 358
Query: 336 VTLSGDLYTWGNGAYN-YGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSA 394
G + WG YN YG + W P V+ + V ++ G H+AV+T A
Sbjct: 359 YMREGRICGWG---YNSYGQAANEKSSYAWYPSPVDWCVG--QVRKLAAGGGHSAVLTDA 413
Query: 395 GQL 397
L
Sbjct: 414 FSL 416
Score = 56.2 bits (134), Expect = 9e-08
Identities = 41/162 (25%), Positives = 70/162 (42%), Gaps = 14/162 (8%)
Query: 461 LFTWGDGDKGRLGHGDKEAKL-VPT--------CVALVEHNFCQVACGHSLTVALTTSGH 511
++ WG G+ G ++E L +P C A + ++CG T A+ + G
Sbjct: 22 IYVWGYNQSGQTGRNEQEKLLRIPKQLPPELFGCPAGANSRWLDISCGREHTAAVASDGS 81
Query: 512 VYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGA 571
++A G+ YGQLG+ G+ + +L FV+ ++CGA+ A + E G +
Sbjct: 82 LFAWGANEYGQLGDGTEVGRKHPKKVKQLQSEFVKFVSCGAFCTAAIAEPRE--NDGTLS 139
Query: 572 NGRLG-HGDTDDRNNPTLVDAL--KDKHVKSIACGTNFTAAI 610
RL G N P L ++ ++CGT A+
Sbjct: 140 TSRLWVWGQNQGSNLPRLFSGAFPATTAIRQVSCGTAHVVAL 181
>At3g55580 regulator of chromosome condensation-like protein
Length = 488
Score = 123 bits (309), Expect = 5e-28
Identities = 88/262 (33%), Positives = 126/262 (47%), Gaps = 34/262 (12%)
Query: 204 VSLSSAVSSSSQGSGHDDGDALGDVF-IWGEGTGDGVVGGGNHRVGSGLGVKI------- 255
VSL+ V S +G AL D+ +WG G G G ++G G V++
Sbjct: 228 VSLAPGVRIVSVAAGGRHTLALSDIGQVWGWGYG------GEGQLGLGSRVRLVSSPHPI 281
Query: 256 ----DSLFPKALESAVVLD------------VQNIACGGRHAALVTKQGEIFSWGEESGG 299
S + KA S V + V+ IACGGRH+A++T G + ++G G
Sbjct: 282 PCIEPSSYGKATSSGVNMSSVVQCGRVLGSYVKKIACGGRHSAVITDTGALLTFGWGLYG 341
Query: 300 RLGHGVDSDVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGN 359
+ G G D L P + +L IE VA G +HT + GD+Y +G +G LG G
Sbjct: 342 QCGQGSTDDELSPTCVSSLLGIRIEEVAAGLWHTTCASSDGDVYAFGGN--QFGQLGTGC 399
Query: 360 QVSHWVPKRVNGP-LEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSL 418
+ +PK + P LE ++V ISCG HTAV+T G++F +G +G LG GD +
Sbjct: 400 DQAETLPKLLEAPNLENVNVKTISCGARHTAVITDEGRVFCWGWNKYGQLGIGDVIDRNA 459
Query: 419 PREVESLKGLRTMRASCGVWHT 440
P EV +K +CG WHT
Sbjct: 460 PAEVR-IKDCFPKNIACGWWHT 480
Score = 110 bits (276), Expect = 3e-24
Identities = 124/443 (27%), Positives = 187/443 (41%), Gaps = 54/443 (12%)
Query: 199 MDAFRVSLSSAVSSSSQG-SGHDDGDAL-----GDVFIWG--EGTGDGVVGGGNHRVGSG 250
M V + AV SS + SG G A+ G + WG + G V G H
Sbjct: 50 MSPVEVKIPPAVESSWKDVSGGGCGFAMATAESGKLITWGSTDDLGQSYVTSGKHG---- 105
Query: 251 LGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEES---GGRLGHGVDS 307
+ FP E V Q G H VT+ ++++WG GR+ VD
Sbjct: 106 ---ETPEPFPLPPEVCV----QKAEAGWAHCVAVTENQQVYTWGWRECIPTGRVFGQVDG 158
Query: 308 DVLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPK 367
D + + S + + G+ + T S G + Q + +
Sbjct: 159 DSCERNI--SFSTEQVSSSSQGK-KSSGGTSSQVEGRGGGEPTKKRRISPSKQAAENSSQ 215
Query: 368 RVNGPLE----------GIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDR-KSV 416
N L G+ + ++ G HT ++ GQ++ +G G G LG G R + V
Sbjct: 216 SDNIDLSALPCLVSLAPGVRIVSVAAGGRHTLALSDIGQVWGWGYGGEGQLGLGSRVRLV 275
Query: 417 SLPREVESLKGLRTMRASCGVWHTAAVVE------------VMVGNSSSSNCSSGKLFTW 464
S P + ++ +A+ + ++VV+ G S+ +G L T+
Sbjct: 276 SSPHPIPCIEPSSYGKATSSGVNMSSVVQCGRVLGSYVKKIACGGRHSAVITDTGALLTF 335
Query: 465 GDGDKGRLGHGDKEAKLVPTCVA-LVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQL 523
G G G+ G G + +L PTCV+ L+ +VA G T ++ G VYA G +GQL
Sbjct: 336 GWGLYGQCGQGSTDDELSPTCVSSLLGIRIEEVAAGLWHTTCASSDGDVYAFGGNQFGQL 395
Query: 524 GN--PQADGKLPTRVEGKLLKSF-VEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDT 580
G QA+ LP +E L++ V+ I+CGA H AV+T V+ WG G+LG GD
Sbjct: 396 GTGCDQAE-TLPKLLEAPNLENVNVKTISCGARHTAVITDEGRVFCWGWNKYGQLGIGDV 454
Query: 581 DDRNNPTLVDALKDKHVKSIACG 603
DRN P V +KD K+IACG
Sbjct: 455 IDRNAPAEV-RIKDCFPKNIACG 476
Score = 104 bits (259), Expect = 3e-22
Identities = 91/334 (27%), Positives = 143/334 (42%), Gaps = 53/334 (15%)
Query: 210 VSSSSQGSGHDDGDALGDVFIWGEGTGDG-----VVGGGNHRVGSGLGVKID-SLFPKAL 263
VSSSSQG G + + G G G+ + S ID S P +
Sbjct: 172 VSSSSQGKKSSGGTSSQ---VEGRGGGEPTKKRRISPSKQAAENSSQSDNIDLSALPCLV 228
Query: 264 ESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHG-----VDS----DVLHPKL 314
A + + ++A GGRH ++ G+++ WG G+LG G V S + P
Sbjct: 229 SLAPGVRIVSVAAGGRHTLALSDIGQVWGWGYGGEGQLGLGSRVRLVSSPHPIPCIEPSS 288
Query: 315 IDALSNTNIEL----------------VACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHG 358
+++ + + +ACG H+ +T +G L T+G G YG G G
Sbjct: 289 YGKATSSGVNMSSVVQCGRVLGSYVKKIACGGRHSAVITDTGALLTFGWGL--YGQCGQG 346
Query: 359 NQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSL 418
+ P V+ L GI + ++ G WHT +S G ++ FG FG LG G ++ +L
Sbjct: 347 STDDELSPTCVSS-LLGIRIEEVAAGLWHTTCASSDGDVYAFGGNQFGQLGTGCDQAETL 405
Query: 419 PR--EVESLKGLRTMRASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGD 476
P+ E +L+ + SCG HTA + + G++F WG G+LG GD
Sbjct: 406 PKLLEAPNLENVNVKTISCGARHTAVITD------------EGRVFCWGWNKYGQLGIGD 453
Query: 477 KEAKLVPTCVALVEHNFCQVACG--HSLTVALTT 508
+ P V + + +ACG H+L + T
Sbjct: 454 VIDRNAPAEVRIKDCFPKNIACGWWHTLLLGQPT 487
Score = 80.9 bits (198), Expect = 4e-15
Identities = 107/446 (23%), Positives = 171/446 (37%), Gaps = 109/446 (24%)
Query: 228 VFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQ 287
V++WG + G R S L ++ P A+ES+ DV CG A +
Sbjct: 33 VYMWGY-----LPGASPQR--SPLMSPVEVKIPPAVESSWK-DVSGGGCG--FAMATAES 82
Query: 288 GEIFSWGEESGGRLGHGVDSDVLH---PKLIDALSNTNIELVACGEYHTCAVTLSGDLYT 344
G++ +WG S LG + H P+ ++ G H AVT + +YT
Sbjct: 83 GKLITWG--STDDLGQSYVTSGKHGETPEPFPLPPEVCVQKAEAGWAHCVAVTENQQVYT 140
Query: 345 WGNGAYNYGLLGHGNQVSHW---VPK-RVNGPLEGIHVSYISCG---PWHTAVVTSAGQL 397
WG W +P RV G ++G SC + T V+S+ Q
Sbjct: 141 WG-----------------WRECIPTGRVFGQVDGD-----SCERNISFSTEQVSSSSQG 178
Query: 398 FTFGDGTFGAL---GHGD---RKSVS-------------------LPREVESLKGLRTMR 432
GT + G G+ ++ +S LP V G+R +
Sbjct: 179 KKSSGGTSSQVEGRGGGEPTKKRRISPSKQAAENSSQSDNIDLSALPCLVSLAPGVRIVS 238
Query: 433 ASCGVWHTAAVVEVMVGNSSSSNCSSGKLFTWGDGDKGRLGHGDK--------------- 477
+ G HT A+ ++ G+++ WG G +G+LG G +
Sbjct: 239 VAAGGRHTLALSDI------------GQVWGWGYGGEGQLGLGSRVRLVSSPHPIPCIEP 286
Query: 478 -----------EAKLVPTCVALVEHNFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNP 526
V C ++ ++ACG + +T +G + G +YGQ G
Sbjct: 287 SSYGKATSSGVNMSSVVQCGRVLGSYVKKIACGGRHSAVITDTGALLTFGWGLYGQCGQG 346
Query: 527 QADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNP 586
D +L LL +EE+A G +H + +VY +G G+LG G P
Sbjct: 347 STDDELSPTCVSSLLGIRIEEVAAGLWHTTCASSDGDVYAFGGNQFGQLGTGCDQAETLP 406
Query: 587 TLVDA--LKDKHVKSIACGTNFTAAI 610
L++A L++ +VK+I+CG TA I
Sbjct: 407 KLLEAPNLENVNVKTISCGARHTAVI 432
>At3g26100 unknown protein (MPE11.29)
Length = 532
Score = 122 bits (306), Expect = 1e-27
Identities = 116/435 (26%), Positives = 180/435 (40%), Gaps = 58/435 (13%)
Query: 169 FFPLDSASASVHSISSGGSDSMHGHMKTMGMDAFRVSLSSAVSSSSQGSGHDDG-DALGD 227
F P++ GGS + G R S AV+ G GH + G+
Sbjct: 108 FKPMNEEERQEMKRRCGGSWKLVLRFLLAGEACCRREKSQAVA----GPGHSVAVTSKGE 163
Query: 228 VFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQ 287
V+ +G G +G G+ D + + S + + A G L++
Sbjct: 164 VYTFGYNNS-GQLGHGHTE---------DEARIQPVRSLQGVRIIQAAAGAARTMLISDD 213
Query: 288 GEIFSWGEESGGRLGHGVDSD--VLHPKLIDALSNTNIELVACGEYHTCAVTLSGDLYT- 344
G++++ G+ES G +G V P+L+ +L N + A G Y T ++ G +YT
Sbjct: 214 GKVYACGKESFGEAEYGGQGTKPVTTPQLVTSLKNIFVVQAAIGNYFTAVLSREGKVYTF 273
Query: 345 -WGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVT---SAGQLFTF 400
WGN G LGH + + P+ + GPLE + V I+ G + + + +++
Sbjct: 274 SWGND----GRLGHQTEAADVEPRPLLGPLENVPVVQIAAGYCYLLALACQPNGMSVYSV 329
Query: 401 GDGTFGALGHGDRKSVSLPREVESLK--GLRTMRASCGVWHTAAVVEVMVGNSSSSNCSS 458
G G G LGHG R PR +E + L+ + G WH A V +
Sbjct: 330 GCGLGGKLGHGSRTDEKYPRVIEQFQILNLQPRVVAAGAWHAAVVGQ------------D 377
Query: 459 GKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEH-NFCQVACGHSLTVALTTSGHVYAMGS 517
G++ TWG G G LGHG++E + VP V + H VA G T ++ G VY+ G
Sbjct: 378 GRVCTWGWGRYGCLGHGNEECESVPKVVEGLSHVKAVHVATGDYTTFVVSDDGDVYSFGC 437
Query: 518 ---------PVYGQLGNPQADGKLPTRV-------EGKLLKSFVEEIACGAYHVAVLTLR 561
P + + GN A+ PT V E + S I A H LT
Sbjct: 438 GESASLGHHPSFDEQGNRHANVLSPTVVTSLKQVNERMVQISLTNSIYWNA-HTFALTES 496
Query: 562 NEVYTWGKGANGRLG 576
+++ +G G G+LG
Sbjct: 497 GKLFAFGAGDQGQLG 511
Score = 113 bits (283), Expect = 5e-25
Identities = 87/285 (30%), Positives = 144/285 (50%), Gaps = 26/285 (9%)
Query: 329 GEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHT 388
G H+ AVT G++YT+G N G LGHG+ + V L+G+ + + G T
Sbjct: 151 GPGHSVAVTSKGEVYTFGYN--NSGQLGHGHTEDEARIQPVRS-LQGVRIIQAAAGAART 207
Query: 389 AVVTSAGQLFTFGDGTFGALGHGDR--KSVSLPREVESLKGLRTMRASCGVWHTAAVVEV 446
+++ G+++ G +FG +G + K V+ P+ V SLK + ++A+ G + TA +
Sbjct: 208 MLISDDGKVYACGKESFGEAEYGGQGTKPVTTPQLVTSLKNIFVVQAAIGNYFTAVLSR- 266
Query: 447 MVGNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNF--CQVACGHSLTV 504
GK++T+ G+ GRLGH + A + P + N Q+A G+ +
Sbjct: 267 -----------EGKVYTFSWGNDGRLGHQTEAADVEPRPLLGPLENVPVVQIAAGYCYLL 315
Query: 505 ALTTSGH---VYAMGSPVYGQLGN-PQADGKLPTRVEG-KLLKSFVEEIACGAYHVAVLT 559
AL + VY++G + G+LG+ + D K P +E ++L +A GA+H AV+
Sbjct: 316 ALACQPNGMSVYSVGCGLGGKLGHGSRTDEKYPRVIEQFQILNLQPRVVAAGAWHAAVVG 375
Query: 560 LRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGT 604
V TWG G G LGHG+ + + P +V+ L HVK++ T
Sbjct: 376 QDGRVCTWGWGRYGCLGHGNEECESVPKVVEGL--SHVKAVHVAT 418
Score = 48.1 bits (113), Expect = 3e-05
Identities = 35/118 (29%), Positives = 53/118 (44%), Gaps = 2/118 (1%)
Query: 495 QVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYH 554
Q G +VA+T+ G VY G GQLG+ + + + L + + A GA
Sbjct: 147 QAVAGPGHSVAVTSKGEVYTFGYNNSGQLGHGHTEDEARIQPVRSLQGVRIIQAAAGAAR 206
Query: 555 VAVLTLRNEVYTWGKGANGRLGHG--DTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
+++ +VY GK + G +G T P LV +LK+ V A G FTA +
Sbjct: 207 TMLISDDGKVYACGKESFGEAEYGGQGTKPVTTPQLVTSLKNIFVVQAAIGNYFTAVL 264
>At3g53830 putative protein
Length = 487
Score = 119 bits (298), Expect = 9e-27
Identities = 61/171 (35%), Positives = 97/171 (56%), Gaps = 4/171 (2%)
Query: 271 VQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGE 330
++ I+CGGRH+A +T G + ++G G+ GHG +D L P + + + +E VA G
Sbjct: 312 IKAISCGGRHSAAITDAGGLITFGWGLYGQCGHGNTNDQLRPMAVSEVKSVRMESVAAGL 371
Query: 331 YHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGP-LEGIHVSYISCGPWHTA 389
+HT ++ G +Y +G +G LG G + +P+ ++G LEG H +SCG H+A
Sbjct: 372 WHTICISSDGKVYAFGGN--QFGQLGTGTDHAEILPRLLDGQNLEGKHAKAVSCGARHSA 429
Query: 390 VVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHT 440
V+ GQL +G +G LG GD ++P +V+ L G R + +CG WHT
Sbjct: 430 VLAEDGQLLCWGWNKYGQLGLGDTNDRNIPTQVQ-LDGCRLRKVACGWWHT 479
Score = 111 bits (277), Expect = 2e-24
Identities = 114/424 (26%), Positives = 173/424 (39%), Gaps = 65/424 (15%)
Query: 226 GDVFIWGEGTGDGVVGGGNHRVGSGLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVT 285
G + WG +G V SG + FP E+ VV + G H A+VT
Sbjct: 71 GKLITWGSTDDEG-----QSYVASGKHGETPEPFPLPTEAPVV----QASSGWAHCAVVT 121
Query: 286 KQGEIFSWG-------------EESGGRLGHGVDSDVLHPKLIDALSNTNIEL------- 325
+ GE F+WG ++SG + S + L N N ++
Sbjct: 122 ETGEAFTWGWKECIPSKDPVGKQQSGSSEQGDIASQGSNAASGTTLQNENQKVGEESVKR 181
Query: 326 --VACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVS----HWVPKRVNGPLEGIHVS 379
V+ + T T GD + + GL V+ H + V E HV
Sbjct: 182 RRVSTAKDETEGHTSGGDFFATTPSLVSVGLGVRITSVATGGRHTLALSVEDSFEDDHVF 241
Query: 380 YISCGPWHTAVVTSAGQLFTFGDGTFGALGHGDR-KSVSLPREVESLKGLRTMRASCGVW 438
V+ GQ++ +G G G LG G R K VS P + L+ + + + +
Sbjct: 242 ---------TEVSDLGQIWGWGYGGEGQLGLGSRIKMVSSPHLIPCLESIGSGKERSFIL 292
Query: 439 HTAAVVEVMV----------------GNSSSSNCSSGKLFTWGDGDKGRLGHGDKEAKLV 482
H G S++ +G L T+G G G+ GHG+ +L
Sbjct: 293 HQGGTTTTSAQASREPGQYIKAISCGGRHSAAITDAGGLITFGWGLYGQCGHGNTNDQLR 352
Query: 483 PTCVALVEH-NFCQVACGHSLTVALTTSGHVYAMGSPVYGQLGNPQADGK-LPTRVEGKL 540
P V+ V+ VA G T+ +++ G VYA G +GQLG + LP ++G+
Sbjct: 353 PMAVSEVKSVRMESVAAGLWHTICISSDGKVYAFGGNQFGQLGTGTDHAEILPRLLDGQN 412
Query: 541 LKS-FVEEIACGAYHVAVLTLRNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKS 599
L+ + ++CGA H AVL ++ WG G+LG GDT+DRN PT V L ++
Sbjct: 413 LEGKHAKAVSCGARHSAVLAEDGQLLCWGWNKYGQLGLGDTNDRNIPTQVQ-LDGCRLRK 471
Query: 600 IACG 603
+ACG
Sbjct: 472 VACG 475
Score = 105 bits (261), Expect = 2e-22
Identities = 81/296 (27%), Positives = 128/296 (42%), Gaps = 61/296 (20%)
Query: 260 PKALESAVVLDVQNIACGGRHAALVTKQ---------------GEIFSWGEESGGRLGHG 304
P + + + + ++A GGRH ++ + G+I+ WG G+LG G
Sbjct: 205 PSLVSVGLGVRITSVATGGRHTLALSVEDSFEDDHVFTEVSDLGQIWGWGYGGEGQLGLG 264
Query: 305 VDSDVLH-PKLIDALSNTN----------------------------IELVACGEYHTCA 335
++ P LI L + I+ ++CG H+ A
Sbjct: 265 SRIKMVSSPHLIPCLESIGSGKERSFILHQGGTTTTSAQASREPGQYIKAISCGGRHSAA 324
Query: 336 VTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGPWHTAVVTSAG 395
+T +G L T+G G Y G GHGN P V+ ++ + + ++ G WHT ++S G
Sbjct: 325 ITDAGGLITFGWGLY--GQCGHGNTNDQLRPMAVS-EVKSVRMESVAAGLWHTICISSDG 381
Query: 396 QLFTFGDGTFGALGHGDRKSVSLPR--EVESLKGLRTMRASCGVWHTAAVVEVMVGNSSS 453
+++ FG FG LG G + LPR + ++L+G SCG H+A + E
Sbjct: 382 KVYAFGGNQFGQLGTGTDHAEILPRLLDGQNLEGKHAKAVSCGARHSAVLAE-------- 433
Query: 454 SNCSSGKLFTWGDGDKGRLGHGDKEAKLVPTCVALVEHNFCQVACGHSLTVALTTS 509
G+L WG G+LG GD + +PT V L +VACG T+ L S
Sbjct: 434 ----DGQLLCWGWNKYGQLGLGDTNDRNIPTQVQLDGCRLRKVACGWWHTLLLGDS 485
Score = 74.7 bits (182), Expect = 3e-13
Identities = 42/144 (29%), Positives = 73/144 (50%), Gaps = 6/144 (4%)
Query: 250 GLGVKIDSLFPKALESAVVLDVQNIACGGRHAALVTKQGEIFSWGEESGGRLGHGVDSDV 309
G G D L P A+ + ++++A G H ++ G+++++G G+LG G D
Sbjct: 343 GHGNTNDQLRPMAVSEVKSVRMESVAAGLWHTICISSDGKVYAFGGNQFGQLGTGTDHAE 402
Query: 310 LHPKLIDA--LSNTNIELVACGEYHTCAVTLSGDLYTWGNGAYNYGLLGHGNQVSHWVPK 367
+ P+L+D L + + V+CG H+ + G L WG YG LG G+ +P
Sbjct: 403 ILPRLLDGQNLEGKHAKAVSCGARHSAVLAEDGQLLCWGWN--KYGQLGLGDTNDRNIPT 460
Query: 368 RVNGPLEGIHVSYISCGPWHTAVV 391
+V L+G + ++CG WHT ++
Sbjct: 461 QVQ--LDGCRLRKVACGWWHTLLL 482
Score = 58.9 bits (141), Expect = 1e-08
Identities = 79/350 (22%), Positives = 139/350 (39%), Gaps = 44/350 (12%)
Query: 276 CGGR--HAALVTKQGEIFSWG---EESGGRLGHGVDSDVLHPKLIDALSNTNIELVACGE 330
CGG A ++++G++ +WG +E + G + P+ + + + G
Sbjct: 57 CGGGCGFAMAISEKGKLITWGSTDDEGQSYVASGKHGET--PEPFPLPTEAPVVQASSGW 114
Query: 331 YHTCAVTLSGDLYTWG-----NGAYNYGLLGHGNQVSHWVPKRVNGPLEGIHVSYISCGP 385
H VT +G+ +TWG G G+ + + + G + +
Sbjct: 115 AHCAVVTETGEAFTWGWKECIPSKDPVGKQQSGSSEQGDIASQGSNAASGTTLQNENQKV 174
Query: 386 WHTAVVTSAGQLFTFGDGTFGALGHGDRKSVSLPREVESLKGLRTMRASCGVWHTAA--V 443
+V ++ T D T G GD + + P V G+R + G HT A V
Sbjct: 175 GEESV--KRRRVSTAKDETEGHTSGGDFFATT-PSLVSVGLGVRITSVATGGRHTLALSV 231
Query: 444 VEVMVGNSSSSNCSS-GKLFTWGDGDKGRLGHGDKEAKLV--PTCVALVEHNFCQVACGH 500
+ + + S G+++ WG G +G+LG G + K+V P + +E + G
Sbjct: 232 EDSFEDDHVFTEVSDLGQIWGWGYGGEGQLGLGSR-IKMVSSPHLIPCLE----SIGSGK 286
Query: 501 SLTVALTTSGHVYAMGSPVYGQLGNPQADGKLPTRVEGKLLKSFVEEIACGAYHVAVLTL 560
+ L G +R G+ +K+ I+CG H A +T
Sbjct: 287 ERSFILHQGGTTTTSAQA---------------SREPGQYIKA----ISCGGRHSAAITD 327
Query: 561 RNEVYTWGKGANGRLGHGDTDDRNNPTLVDALKDKHVKSIACGTNFTAAI 610
+ T+G G G+ GHG+T+D+ P V +K ++S+A G T I
Sbjct: 328 AGGLITFGWGLYGQCGHGNTNDQLRPMAVSEVKSVRMESVAAGLWHTICI 377
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,015,871
Number of Sequences: 26719
Number of extensions: 1165149
Number of successful extensions: 5188
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 81
Number of HSP's that attempted gapping in prelim test: 4040
Number of HSP's gapped (non-prelim): 468
length of query: 1061
length of database: 11,318,596
effective HSP length: 110
effective length of query: 951
effective length of database: 8,379,506
effective search space: 7968910206
effective search space used: 7968910206
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146692.2


