

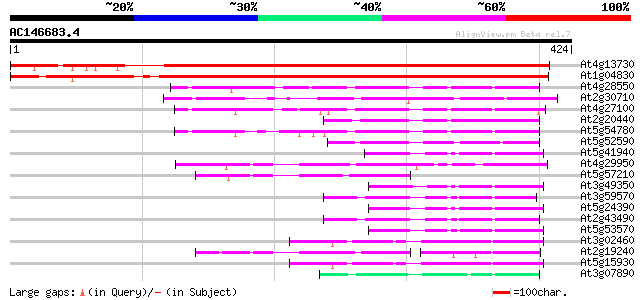
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.4 + phase: 0 /pseudo
(424 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g13730 unknown protein 505 e-143
At1g04830 unknown protein 477 e-135
At4g28550 putative protein 100 3e-21
At2g30710 unknown protein 99 6e-21
At4g27100 unknown protein 82 7e-16
At2g20440 unknown protein 78 1e-14
At5g54780 putative protein 77 2e-14
At5g52590 putative protein 74 1e-13
At5g41940 GTPase activator protein of Rab-like small GTPases-lik... 71 1e-12
At4g29950 unknown protein 70 2e-12
At5g57210 microtubule-associated protein-like 69 4e-12
At3g49350 GTPase activating -like protein 67 2e-11
At3g59570 putative protein 66 4e-11
At5g24390 GTPase activator-like protein of Rab-like small GTPases 65 9e-11
At2g43490 hypothetical protein 64 1e-10
At5g53570 GTPase activator protein of Rab-like small GTPases-lik... 64 2e-10
At3g02460 putative plant adhesion molecule 61 1e-09
At2g19240 hypothetical protein 59 7e-09
At5g15930 plant adhesion molecule 1 (PAM1) 58 9e-09
At3g07890 putative GTPase activator protein 44 2e-04
>At4g13730 unknown protein
Length = 449
Score = 505 bits (1301), Expect = e-143
Identities = 263/430 (61%), Positives = 318/430 (73%), Gaps = 54/430 (12%)
Query: 1 MAKKRVPDWLNSPMWSS--PTEQNRFSATSYPS-EPPSPPPVTVVEDPP--SIHNAITTS 55
MAKK +P+WLNS +WSS P + + S PPSPP +VE PP S +AI+T+
Sbjct: 1 MAKKGIPEWLNSSLWSSSPPIDDRHLRNPAATSITPPSPP---IVERPPFSSSPSAISTA 57
Query: 56 P--------PSSSSSS--ASTSSTDDMSISRLAQS-------QSQLLAEVPFTLKSKCL* 98
P PS S + + S D I A S ++Q++AE
Sbjct: 58 PAPPLPVRPPSKSEINDLQNGSGNDGAGIETPAVSSVEDVSRKAQVVAE----------- 106
Query: 99 LLFSIQLY*YC*LFVY**LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRS 158
LS+KVID++ELRKIASQG+PD G+RS +WKLLL YL PDRS
Sbjct: 107 ------------------LSKKVIDLKELRKIASQGLPDDAGIRSIVWKLLLDYLSPDRS 148
Query: 159 LWSSELAKKRSQYKRFKQDILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEH 218
LWSSELAKKRSQYK+FK+++L+NPSE+TR+M S D++D K E+ G LSRS+ITH +H
Sbjct: 149 LWSSELAKKRSQYKQFKEELLMNPSEVTRKMDKSKGGDSNDPKIESPGALSRSEITHEDH 208
Query: 219 PLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAK 278
PLSLG TS+WN FF+DT+++EQI+RDV RTHPDMHFF GDS +AKSNQ+ALKNIL IFAK
Sbjct: 209 PLSLGTTSLWNNFFKDTEVLEQIERDVMRTHPDMHFFSGDSAVAKSNQDALKNILTIFAK 268
Query: 279 LNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNS 338
LNPGIRYVQGMNE+LAP+FY+FKNDPD+ NAA++E+D FFCFVEL+SGFRDNFCQQLDNS
Sbjct: 269 LNPGIRYVQGMNEILAPIFYIFKNDPDKGNAAYAESDAFFCFVELMSGFRDNFCQQLDNS 328
Query: 339 IVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWD 398
+VGIR TITRLS LLK HDEELWRHLEVTTK+NPQFYAFRWITLLLTQEF+F +SL IWD
Sbjct: 329 VVGIRYTITRLSLLLKHHDEELWRHLEVTTKINPQFYAFRWITLLLTQEFNFVESLHIWD 388
Query: 399 TLVSDPDGPQ 408
TL+SDP+GPQ
Sbjct: 389 TLLSDPEGPQ 398
>At1g04830 unknown protein
Length = 448
Score = 477 bits (1228), Expect = e-135
Identities = 240/410 (58%), Positives = 309/410 (74%), Gaps = 20/410 (4%)
Query: 1 MAKKRVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPP---SIHNAITTSPP 57
M +K+VP+WLNS MWS+P + SY PVT +++ S+ + ++PP
Sbjct: 1 MVRKKVPEWLNSTMWSTPPPPS-----SYDDGLLRHSPVTKMKEEAESISVAPRLNSAPP 55
Query: 58 SSSSSSASTSSTDDMSISRLAQSQSQLLAEVPFTLKSKCL*LLFSIQLY*YC*LFVY**L 117
SS++S + S + + ++ + V + + FS Q + V L
Sbjct: 56 PSSNTSVPSPSHRPRNGNSISGGSGEYGHSVGPSAED------FSRQAH------VSAEL 103
Query: 118 SRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQD 177
S+KVI+M+ELR +A Q +PDSPG+RST+WKLLLGYLPP+RSLWS+EL +KRSQYK +K +
Sbjct: 104 SKKVINMKELRSLALQSLPDSPGIRSTVWKLLLGYLPPERSLWSTELKQKRSQYKHYKDE 163
Query: 178 ILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDI 237
+L +PSEIT +M S +D D+K E+R ML+RS+IT +HPLSLGK SIWN +FQDT+
Sbjct: 164 LLTSPSEITWKMVRSKGFDNYDLKSESRCMLARSRITDEDHPLSLGKASIWNTYFQDTET 223
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
IEQIDRDVKRTHPD+ FF G+S A+SNQE++KNIL++FAKLN GIRYVQGMNE+LAP+F
Sbjct: 224 IEQIDRDVKRTHPDIPFFSGESSFARSNQESMKNILLVFAKLNQGIRYVQGMNEILAPIF 283
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
YVF+NDPDE++++ +EAD FFCFVELLSGFRD +CQQLDNS+VGIRS ITRLSQL+++HD
Sbjct: 284 YVFRNDPDEDSSSHAEADAFFCFVELLSGFRDFYCQQLDNSVVGIRSAITRLSQLVRKHD 343
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGP 407
EELWRHLE+TTKVNPQFYAFRWITLLLTQEF F DSL IWD L+SDP+GP
Sbjct: 344 EELWRHLEITTKVNPQFYAFRWITLLLTQEFSFFDSLHIWDALLSDPEGP 393
>At4g28550 putative protein
Length = 424
Score = 99.8 bits (247), Expect = 3e-21
Identities = 80/282 (28%), Positives = 137/282 (48%), Gaps = 30/282 (10%)
Query: 122 IDM-RELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAK--KRSQYKRFKQDI 178
+DM R LR+I GI P ++ +W+ LLG PD + + +R QY +K++
Sbjct: 57 LDMERVLRRIQRGGI--HPSIKGEVWEFLLGAYDPDSTFEERNKLRNHRREQYYAWKEEC 114
Query: 179 LINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDII 238
+ F + + A+D + L S + + E + T + Q ++
Sbjct: 115 KNMVPLVGSGKFVTMAVVAEDGQ-----PLEESSVDNQEWVVKTAITD--KRVLQWMLVL 167
Query: 239 EQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFY 298
QI DV RT + F+ ++SNQ L +IL I+ LNP I YVQGMN++ +P+
Sbjct: 168 SQIGLDVVRTDRYLCFY-----ESESNQARLWDILSIYTWLNPDIGYVQGMNDICSPMII 222
Query: 299 VFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDE 358
+ ++ EAD F+CF + R+NF + + +G+++ + LSQ++K D
Sbjct: 223 LLED----------EADAFWCFERAMRRLRENF--RTTATSMGVQTQLGMLSQVIKTVDP 270
Query: 359 ELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
L +HLE +A R + +L +EF F D+L +W+ +
Sbjct: 271 RLHQHLE-DLDGGEYLFAIRMLMVLFRREFSFLDALYLWELM 311
>At2g30710 unknown protein
Length = 440
Score = 98.6 bits (244), Expect = 6e-21
Identities = 83/312 (26%), Positives = 136/312 (42%), Gaps = 70/312 (22%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
LS + + +LR++A G+P +R +W+LLLGY PP+ + L +KR +Y
Sbjct: 126 LSETTVILEKLRELAWNGVPHY--MRPDVWRLLLGYAPPNSDRREAVLRRKRLEYLESVG 183
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
YD D + RS + +
Sbjct: 184 QF----------------YDLPDSE--------RSD--------------------DEIN 199
Query: 237 IIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPL 296
++ QI D RT PD+ FF + Q++L+ IL +A +P YVQG+N+++ P
Sbjct: 200 MLRQIAVDCPRTVPDVSFFQQEQV-----QKSLERILYTWAIRHPASGYVQGINDLVTPF 254
Query: 297 FYVF-------------KNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIR 343
+F +D E + EAD ++C +LL G +D++ GI+
Sbjct: 255 LVIFLSEYLDGGVDSWSMDDLSAEKVSDVEADCYWCLTKLLDGMQDHYTFAQP----GIQ 310
Query: 344 STITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
+ +L +L++ DE + RH+E QF AFRW LL +E F R+WDT +++
Sbjct: 311 RLVFKLKELVRRIDEPVSRHMEEHGLEFLQF-AFRWYNCLLIREIPFNLINRLWDTYLAE 369
Query: 404 PDG-PQVQIYSF 414
D P +Y +
Sbjct: 370 GDALPDFLVYIY 381
>At4g27100 unknown protein
Length = 436
Score = 81.6 bits (200), Expect = 7e-16
Identities = 80/300 (26%), Positives = 134/300 (44%), Gaps = 47/300 (15%)
Query: 125 RELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRS--QYKRFKQDILINP 182
+ LR+I GI P +R +W+ LLG P + E ++R QY +K++
Sbjct: 54 KTLRRIRRGGI--HPSIRGEVWEFLLGCYDPMSTFEEREQIRQRRRLQYASWKEECKQMF 111
Query: 183 SEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQ--------D 234
I F +A ++ G + + E ++LG S + FF+ D
Sbjct: 112 PVIGSGRFTTAPVITEN------GQPNYDPLVLQE--INLGTNSNGSVFFKELTSRGPLD 163
Query: 235 TDIIE------QIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQG 288
II+ QI DV RT + F+ K N L +IL ++A ++ + Y QG
Sbjct: 164 KKIIQWLLTLHQIGLDVNRTDRALVFY-----EKKENLSKLWDILSVYAWIDNDVGYCQG 218
Query: 289 MNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITR 348
M+++ +P+ + ++ EAD F+CF L+ R NF + VG+ + +T
Sbjct: 219 MSDLCSPMIILLED----------EADAFWCFERLMRRLRGNF--RSTGRSVGVEAQLTH 266
Query: 349 LSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWD---TLVSDPD 405
LS + + D +L +HL+ +A R + + +EF F DSL +W+ L DPD
Sbjct: 267 LSSITQVVDPKLHQHLD-KLGGGDYLFAIRMLMVQFRREFSFCDSLYLWEMMWALEYDPD 325
>At2g20440 unknown protein
Length = 371
Score = 77.8 bits (190), Expect = 1e-14
Identities = 50/164 (30%), Positives = 85/164 (51%), Gaps = 18/164 (10%)
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
+ QI DV RT + F+ D NQ L ++L I+ LN I YVQGMN++ +P+
Sbjct: 51 LHQIGLDVARTDRYLCFYENDR-----NQSKLWDVLAIYTWLNLDIGYVQGMNDICSPMI 105
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
+F + E D F+CF + R+NF + + +G+++ + LSQ++K D
Sbjct: 106 ILFDD----------EGDAFWCFERAMRRLRENF--RATATSMGVQTQLGVLSQVIKTVD 153
Query: 358 EELWRHLEVTTKVNPQF-YAFRWITLLLTQEFDFADSLRIWDTL 400
L +HL ++ +A R + +L +EF F D+L +W+ +
Sbjct: 154 PRLHQHLGKKDLDGGEYLFAIRMLMVLFRREFSFLDALYLWELM 197
>At5g54780 putative protein
Length = 435
Score = 76.6 bits (187), Expect = 2e-14
Identities = 76/299 (25%), Positives = 132/299 (43%), Gaps = 60/299 (20%)
Query: 125 RELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRS--QYKRFKQDILINP 182
+ L +I GI P +R +W+ LLG P + E ++R QY +K++
Sbjct: 54 KTLSRIQRGGI--HPSIRGEVWEFLLGCYDPKSTFEEREQIRQRRRLQYASWKEEC---- 107
Query: 183 SEITRRMFNSASYDADDVKCETRGMLSRSQITH-GE---HPLSLGKTSIW---NQFFQDT 235
++MF + G ++ IT+ GE P+ L +T++ + FF+D
Sbjct: 108 ----KQMFPVIG---------SGGFITAPVITNKGEPIYDPIVLQETNLGANGSDFFKDL 154
Query: 236 D--------------IIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNP 281
+ QI DV RT + F+ K N L +IL ++A ++
Sbjct: 155 ASRGPLDQKVIQWLLTLHQIGLDVNRTDRTLVFY-----EKKENLSKLWDILALYAWIDN 209
Query: 282 GIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVG 341
+ Y QGM+++ +P+ + ++ EAD F+CF L+ R NF + VG
Sbjct: 210 DVGYCQGMSDLCSPMIMLLED----------EADAFWCFERLMRRLRGNF--RDTGRSVG 257
Query: 342 IRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
+ + +T L+ + + D +L HLE +A R I + +EF F DSL +W+ +
Sbjct: 258 VEAQLTHLASITQIIDPKLHHHLE-KLGGGDYLFAIRMIMVQFRREFSFCDSLYLWEMM 315
>At5g52590 putative protein
Length = 338
Score = 74.3 bits (181), Expect = 1e-13
Identities = 42/160 (26%), Positives = 88/160 (54%), Gaps = 18/160 (11%)
Query: 241 IDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVF 300
ID+DV RT ++ GD L + ++++IL+ ++ N + Y QGM++ L+P+ +V
Sbjct: 104 IDKDVVRTDRAFEYYEGDDNL---HVNSMRDILLTYSFYNFDLGYCQGMSDYLSPILFVM 160
Query: 301 KNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEEL 360
++ E+++F+CFV L+ NF + + G+ + + LS+L++ D L
Sbjct: 161 ED----------ESESFWCFVALMERLGPNFNRDQN----GMHTQLFALSKLVELLDSPL 206
Query: 361 WRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
+ + +N F+ FRWI + +EF++ ++++W+ +
Sbjct: 207 HNYFKENDCLN-YFFCFRWILIQFKREFEYEKTMQLWEVM 245
>At5g41940 GTPase activator protein of Rab-like small GTPases-like
protein
Length = 506
Score = 71.2 bits (173), Expect = 1e-12
Identities = 45/135 (33%), Positives = 74/135 (54%), Gaps = 15/135 (11%)
Query: 269 LKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFR 328
L IL +A +P I Y QGM+++L+PL V ++D F+CFV +S R
Sbjct: 304 LVGILEAYAVYDPEIGYCQGMSDLLSPLIAVMEDD----------VLAFWCFVGFMSKAR 353
Query: 329 DNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEF 388
NF +LD VGIR ++ +S+++K D L+RHLE + F+ +R + +L +E
Sbjct: 354 HNF--RLDE--VGIRRQLSMVSKIIKFKDIHLYRHLE-NLEAEDCFFVYRMVVVLFRREL 408
Query: 389 DFADSLRIWDTLVSD 403
F +L +W+ + +D
Sbjct: 409 TFEQTLCLWEVMWAD 423
>At4g29950 unknown protein
Length = 828
Score = 70.1 bits (170), Expect = 2e-12
Identities = 68/300 (22%), Positives = 130/300 (42%), Gaps = 53/300 (17%)
Query: 126 ELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSS---ELAKKRSQYKRFKQDILINP 182
E+ + + LR W++ LG LP S A+ R +Y ++ +LI+P
Sbjct: 5 EIESLGDESDRRFANLRGLRWRVNLGVLPFQSSSIDDLRKATAESRRRYAALRRRLLIDP 64
Query: 183 SEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQID 242
+++ + NS D+ PLS S W +FF++ ++ + +D
Sbjct: 65 -HLSKDVRNSPDLSIDN-------------------PLSQNPDSTWGRFFRNAELEKTLD 104
Query: 243 RDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKN 302
+D+ R +P+ + A Q L+ IL+++ +P Y QGM+E+LAPL YV
Sbjct: 105 QDLSRLYPEHWSYFQ----APGCQGMLRRILLLWCLKHPEYGYRQGMHELLAPLLYVLHV 160
Query: 303 DPDE----------------ENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTI 346
D D + +F E D + F F ++F D+ I GI+
Sbjct: 161 DVDRLSEVRKSYEDHFIDRFDGLSFEERDITYNFE--FKKFLEDF---TDDEIGGIQGNS 215
Query: 347 TRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDG 406
++ L +EL ++ +++ + A + ++L+++F D+ ++D L++ G
Sbjct: 216 KKIKSL-----DELDPEIQSIVRLSDAYGAEGELGIVLSEKFMEHDAYCMFDALMNGVHG 270
Score = 38.9 bits (89), Expect = 0.005
Identities = 21/56 (37%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Query: 347 TRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVS 402
T LL D L HL V V PQ++ RW+ +L +EF D L +WD + S
Sbjct: 298 TAFYHLLSFVDSSLHSHL-VELGVEPQYFGLRWLRVLFGREFLLQDLLIVWDEIFS 352
>At5g57210 microtubule-associated protein-like
Length = 737
Score = 69.3 bits (168), Expect = 4e-12
Identities = 49/167 (29%), Positives = 80/167 (47%), Gaps = 28/167 (16%)
Query: 141 LRSTIWKLLLGYLPPDRSLWSSEL----AKKRSQYKRFKQDILINPSEITRRMFNSASYD 196
LR W++ LG LP S EL A R +Y ++ +LI+P + ++ NS
Sbjct: 33 LRGVRWRINLGILPSSPSSTIDELRRVTADSRRRYAALRRRLLIDP-HLPKKGTNSPDLT 91
Query: 197 ADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFFC 256
D+ PLS S W +FF++ ++ + +D+D+ R +P+
Sbjct: 92 IDN-------------------PLSQNPDSTWGRFFRNAELEKTLDQDLSRLYPEH---- 128
Query: 257 GDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKND 303
G + Q L+ IL+++ +P I Y QGM+E+LAPL YV + D
Sbjct: 129 GSYFQSSGCQGMLRRILLLWCLKHPEIGYRQGMHELLAPLLYVLQVD 175
Score = 41.2 bits (95), Expect = 0.001
Identities = 25/70 (35%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query: 336 DNSIVGIRSTITR---LSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFAD 392
++SI G+ I L LL D L HL V V PQ++A RW+ +L +EF ++
Sbjct: 305 NDSITGLPPVIEASGALYHLLSLVDASLHSHL-VELGVEPQYFALRWLRVLFGREFPLSN 363
Query: 393 SLRIWDTLVS 402
L +WD + S
Sbjct: 364 LLIVWDEIFS 373
>At3g49350 GTPase activating -like protein
Length = 554
Score = 67.0 bits (162), Expect = 2e-11
Identities = 39/132 (29%), Positives = 71/132 (53%), Gaps = 15/132 (11%)
Query: 272 ILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNF 331
+L +A +P I Y QGM+++L+P+ V +D + F+CFV + R NF
Sbjct: 350 VLEAYALYDPDIGYCQGMSDLLSPILSVIPDDHE----------VFWCFVGFMKKARHNF 399
Query: 332 CQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFA 391
+LD VGIR + +S+++K D +L+RHLE + F+ +R + ++ +E
Sbjct: 400 --RLDE--VGIRRQLNIVSKIIKSKDSQLYRHLE-KLQAEDCFFVYRMVVVMFRRELTLD 454
Query: 392 DSLRIWDTLVSD 403
+L +W+ + +D
Sbjct: 455 QTLCLWEVMWAD 466
>At3g59570 putative protein
Length = 549
Score = 65.9 bits (159), Expect = 4e-11
Identities = 43/161 (26%), Positives = 83/161 (50%), Gaps = 19/161 (11%)
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
+ +I DV RT + F+ L + + +IL ++A ++P Y QGM+++++P
Sbjct: 389 LHRIVVDVVRTDSHLEFYEDPGNLGR-----MSDILAVYAWVDPATGYCQGMSDLVSPFV 443
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
++F+++ AD F+CF L+ R NF Q++ G+ + L ++L+ D
Sbjct: 444 FLFEDN----------ADAFWCFEMLIRRTRANF--QMEGP-TGVMDQLQSLWRILQLTD 490
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWD 398
+E++ HL +AFR + +L +E F +LR+W+
Sbjct: 491 KEMFSHLS-RIGAESLHFAFRMLLVLFRRELSFNKALRMWE 530
>At5g24390 GTPase activator-like protein of Rab-like small GTPases
Length = 528
Score = 64.7 bits (156), Expect = 9e-11
Identities = 37/132 (28%), Positives = 70/132 (53%), Gaps = 15/132 (11%)
Query: 272 ILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNF 331
+L +A +P I Y QGM+++L+P+ V +D + F+CFV + R NF
Sbjct: 322 VLEAYALHDPEIGYCQGMSDLLSPILSVIPDD----------YEAFWCFVGFMKKARQNF 371
Query: 332 CQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFA 391
++D VGI + +S+++K D +L++HLE K F+ +R + ++ +E
Sbjct: 372 --RVDE--VGITRQLNIVSKIIKSKDSQLYKHLE-KVKAEDCFFVYRMVLVMFRRELTLE 426
Query: 392 DSLRIWDTLVSD 403
+L +W+ + +D
Sbjct: 427 QTLHLWEVIWAD 438
>At2g43490 hypothetical protein
Length = 756
Score = 64.3 bits (155), Expect = 1e-10
Identities = 42/163 (25%), Positives = 83/163 (50%), Gaps = 19/163 (11%)
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
+ +I DV RT + F+ L + + +IL ++A ++P Y QGM+++++P
Sbjct: 359 LHRIVVDVVRTDSHLEFYEDPGNLGR-----MSDILAVYAWVDPATGYCQGMSDLVSPFV 413
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
+F+++ AD F+CF L+ R NF Q++ G+ + L +L+ D
Sbjct: 414 VLFEDN----------ADAFWCFEMLIRRTRANF--QMEGP-TGVMDQLQSLWHILQITD 460
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
++++ HL +AFR + +L +E F ++LR+W+ +
Sbjct: 461 KDIFSHLS-RIGAESLHFAFRMLLVLFRRELSFNEALRMWEMM 502
>At5g53570 GTPase activator protein of Rab-like small GTPases-like
protein
Length = 550
Score = 63.9 bits (154), Expect = 2e-10
Identities = 37/132 (28%), Positives = 71/132 (53%), Gaps = 15/132 (11%)
Query: 272 ILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNF 331
IL +A +P I Y QGM+++L+P+ V D + F+CFV + R NF
Sbjct: 343 ILEAYAMYDPEIGYCQGMSDLLSPILAVISED----------HEAFWCFVGFMKKARHNF 392
Query: 332 CQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFA 391
+LD + GI+ ++ +S+++K D +L++HLE + + +R + ++ +E F
Sbjct: 393 --RLDEA--GIQRQLSIVSKIIKNKDSQLYKHLE-NLQAEDCSFVYRMVLVMFRRELSFE 447
Query: 392 DSLRIWDTLVSD 403
+L +W+ + +D
Sbjct: 448 QTLCLWEVMWAD 459
Score = 35.0 bits (79), Expect = 0.078
Identities = 39/170 (22%), Positives = 81/170 (46%), Gaps = 17/170 (10%)
Query: 15 WSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTDDMSI 74
++S + N S++S PS S P + PPS ++ ++ SSSSSS+ + +
Sbjct: 8 YTSTSSGNSSSSSSLPSSSSSSLPSSSSSSPPSSNSNSYSNSNSSSSSSSWIHLRSVLFV 67
Query: 75 SRLAQ----SQSQLLAEVPFTLKSKCL*LLFSIQLY*YC*LFVY**LSRKVID--MRELR 128
+ L+ + S + P++ + + +++ + + LF K+ D + L+
Sbjct: 68 ANLSSPSSVTSSDRRRKSPWSRRKR----KWALTPHQWRSLFT---PEGKLRDGGVGFLK 120
Query: 129 KIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAK--KRSQYKRFKQ 176
K+ S+G+ P +R+ +W LLG + + E K KR +Y++ ++
Sbjct: 121 KVRSRGV--DPSIRAEVWLFLLGVYDLNSTSEEREAVKTQKRKEYEKLQR 168
>At3g02460 putative plant adhesion molecule
Length = 353
Score = 61.2 bits (147), Expect = 1e-09
Identities = 52/195 (26%), Positives = 86/195 (43%), Gaps = 18/195 (9%)
Query: 212 QITHGEHPLSLGKTSIWNQFFQDTDIIEQID--RDVKRTHPDMHFFCGDSQLAKSNQEAL 269
Q+ G L L ++ Q ++D RD+ RT P FF + Q +L
Sbjct: 99 QLISGSRDLLLMNPGVYEQLVIYETSASELDIIRDISRTFPSHVFF---QKRHGPGQRSL 155
Query: 270 KNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRD 329
N+L ++ + + YVQGM +A L ++ SE D F+ V LL G
Sbjct: 156 YNVLKAYSVYDRDVGYVQGMG-FIAGLLLLY----------MSEEDAFWLLVALLKGAVH 204
Query: 330 NFCQQLDNS-IVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEF 388
+ L ++ + ++ + +L L+KE +L H +NP YA +W + + F
Sbjct: 205 APMEGLYHAGLPLVQQYLFQLESLVKELIPKLGEHF-TQEMINPSMYASQWFITVFSYSF 263
Query: 389 DFADSLRIWDTLVSD 403
F +LRIWD +S+
Sbjct: 264 PFPLALRIWDVFLSE 278
>At2g19240 hypothetical protein
Length = 840
Score = 58.5 bits (140), Expect = 7e-09
Identities = 43/163 (26%), Positives = 79/163 (48%), Gaps = 26/163 (15%)
Query: 141 LRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILINPSEITRRMFNSASYDADDV 200
LR W++ LG LP S S + ++ + R ++ +L++P + + +S ++ D+
Sbjct: 29 LRGVRWRVNLGVLPSLAS--SIDEFRRAAANSRRRRRLLMDP-HVLKHEDSSPNFIIDN- 84
Query: 201 KCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFFCGDSQ 260
PLS S W QFF++ ++ + +D+D+ R +P+ + +
Sbjct: 85 ------------------PLSQNPNSTWGQFFRNAELEKTLDQDLSRLYPEHWCYFQTPR 126
Query: 261 LAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKND 303
Q L+ IL+++ +P Y QGM+E+LAPL YV D
Sbjct: 127 Y----QGMLRRILLLWCLKHPEYGYRQGMHELLAPLLYVLHVD 165
Score = 42.0 bits (97), Expect = 6e-04
Identities = 29/101 (28%), Positives = 43/101 (41%), Gaps = 11/101 (10%)
Query: 311 FSEADTFFCFVELLSGFRDNFCQQ-------LDNSIVGIRSTITRLS---QLLKEHDEEL 360
F E D + F L+SG F S G+ + S ++L D L
Sbjct: 254 FMEHDAYCMFDALMSGIHGCFAMASFFSYSPASGSHTGLTPVLEACSAFYRILAVVDSSL 313
Query: 361 WRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLV 401
HL V V PQ++ RW+ +L +EF D L +WD ++
Sbjct: 314 HSHL-VELGVEPQYFGLRWLRVLFGREFLLQDLLLVWDEII 353
>At5g15930 plant adhesion molecule 1 (PAM1)
Length = 356
Score = 58.2 bits (139), Expect = 9e-09
Identities = 49/195 (25%), Positives = 85/195 (43%), Gaps = 18/195 (9%)
Query: 212 QITHGEHPLSLGKTSIWNQFFQDTDIIEQID--RDVKRTHPDMHFFCGDSQLAKSNQEAL 269
Q+ G L L ++ Q ++D RD+ RT P FF + Q +L
Sbjct: 96 QLISGSRDLLLMNPGVYVQLVIYETSASELDIIRDISRTFPSHVFF---QKRHGPGQRSL 152
Query: 270 KNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRD 329
N+L ++ + + YVQGM +A L ++ SE D F+ V LL G
Sbjct: 153 YNVLKAYSVYDRDVGYVQGMG-FIAGLLLLY----------MSEEDAFWLLVALLKGAVH 201
Query: 330 NFCQQLDNS-IVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEF 388
+ + L + + ++ + + QL++E +L H +NP YA +W + +
Sbjct: 202 SPIEGLYQAGLPLVQQYLLQFDQLVRELMPKLGEHF-TQEMINPSMYASQWFITVFSYSL 260
Query: 389 DFADSLRIWDTLVSD 403
F +LRIWD +++
Sbjct: 261 PFHSALRIWDVFLAE 275
>At3g07890 putative GTPase activator protein
Length = 400
Score = 43.5 bits (101), Expect = 2e-04
Identities = 40/166 (24%), Positives = 62/166 (37%), Gaps = 18/166 (10%)
Query: 235 TDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLA 294
T QID D+ RT P + AL+ +L+ ++ + + Y QG+N V A
Sbjct: 156 TPATRQIDHDLPRTFPGHPWLD-----TPEGHAALRRVLVGYSFRDSDVGYCQGLNYVAA 210
Query: 295 PLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLK 354
L V K +E D F+ LL C + S + + LL
Sbjct: 211 LLLLVMK----------TEEDAFWMLAVLLENVLVRDCYTTNLSGCHVEQRV--FKDLLA 258
Query: 355 EHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
+ + HLE + A W L ++ +LR+WD L
Sbjct: 259 QKCSRIATHLE-DMGFDVSLVATEWFLCLFSKSLPSETTLRVWDVL 303
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.139 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,672,262
Number of Sequences: 26719
Number of extensions: 407036
Number of successful extensions: 4969
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 81
Number of HSP's that attempted gapping in prelim test: 3880
Number of HSP's gapped (non-prelim): 765
length of query: 424
length of database: 11,318,596
effective HSP length: 102
effective length of query: 322
effective length of database: 8,593,258
effective search space: 2767029076
effective search space used: 2767029076
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146683.4


