

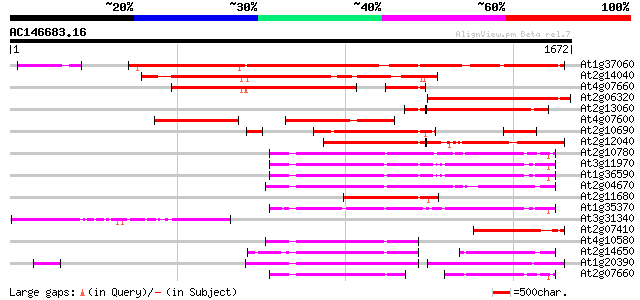
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.16 + phase: 0
(1672 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g37060 Athila retroelment ORF 1, putative 1465 0.0
At2g14040 putative retroelement pol polyprotein 849 0.0
At4g07660 putative athila transposon protein 627 e-179
At2g06320 putative retroelement pol polyprotein 594 e-169
At2g13060 pseudogene 489 e-138
At4g07600 473 e-133
At2g10690 putative retroelement pol polyprotein 466 e-131
At2g12040 T10J7.2 429 e-120
At2g10780 pseudogene 383 e-106
At3g11970 hypothetical protein 378 e-104
At1g36590 hypothetical protein 373 e-103
At2g04670 putative retroelement pol polyprotein 372 e-102
At2g11680 putative retroelement pol polyprotein 364 e-100
At1g35370 hypothetical protein 356 5e-98
At3g31340 Athila ORF 1, putative 321 3e-87
At2g07410 putative retroelement pol polyprotein 302 1e-81
At4g10580 putative reverse-transcriptase -like protein 300 5e-81
At2g14650 putative retroelement pol polyprotein 277 4e-74
At1g20390 hypothetical protein 269 9e-72
At2g07660 putative retroelement pol polyprotein 255 1e-67
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 1465 bits (3793), Expect = 0.0
Identities = 759/1351 (56%), Positives = 963/1351 (71%), Gaps = 112/1351 (8%)
Query: 353 NHPNKMNAVTLRSGKPL---EDPIRRTET-------------DNSEKEICEP----PSRE 392
N +A+TLRSGK L E P + TE +++EK I EP P+R
Sbjct: 427 NSKEYAHAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGNSAEKAIEEPILHQPTRP 486
Query: 393 TRAER----EKP---RVEEN--LPPPFKPKIPFPQRFAKSKLDEQFKKFIE-MMNKIYID 442
EKP + +EN +PPP+KP +PFP RF K + +++K +E + + +
Sbjct: 487 LAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMI-QKYKALLEKQLKNLEVT 545
Query: 443 VPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL-PPKLKDPGSFSI 501
+P + L +P K++K++++++ K E V L+ ECSAIIQ K+ P KL DPGSF++
Sbjct: 546 MPLVDCLALIPDSNKYVKDMITERIK-EVQGMVVLSHECSAIIQQKIIPKKLGDPGSFTL 604
Query: 502 PCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDV 561
PC +G K +CDLGASVSLMPL + ++LG + K ++L LADRSV+ G++ED+
Sbjct: 605 PCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILADRSVRISHGLLEDL 664
Query: 562 PVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKET-VEFE 620
PV +G V +P DFVV+EM+E+ + P++LGRPFLA A AIIDVKKGK+ N+G++ + F+
Sbjct: 665 PVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKIDLNLGRDLKMTFD 724
Query: 621 LAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGT-KLEGEAKAYEEL 679
+ MK P+I+ + I+ +D + D L+ L ++ L E E +
Sbjct: 725 ITNTMKKPTIEGNIFWIEEMDMLADKMLEELGETDHLQSALTKDSKEGDLHLETLGGEYI 784
Query: 680 LDR-------------TLPMKGLSVEELVKEEPTLLPKEAPKVELKTLPSNLRYEFLGPN 726
LD +L S LV ++ + + +APKV+LK LP LRY FLG N
Sbjct: 785 LDHITRPTAHSVYSTTSLDHNSSSEANLVSDDWSEI--KAPKVDLKPLPKGLRYVFLGLN 842
Query: 727 STYPVIVNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIE 786
STYPVIVN L + L+ L KY KAIGY++DDIKGI+P+LC HRI LE + SIE
Sbjct: 843 STYPVIVNDELTADQVNLLITELMKYRKAIGYSLDDIKGISPTLCTHRIHLENESYSSIE 902
Query: 787 HQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIAT 846
QRRLNPN+KEVVKKE+LKLLDAGVIYPISDS WVSPV VPKKGG+TV+KN K+E I T
Sbjct: 903 PQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPT 962
Query: 847 RTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPND 906
RT+TG RMCI+YRKLN A+RK+HFPLPFID MLERLA H ++C+LD YSGFFQIPIHPND
Sbjct: 963 RTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPND 1022
Query: 907 QEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFD 966
Q KTTFTCP+GTFAY+RMPFGLCNAPATFQRCM SIFSD +E+++EVFMDDFSV+GS+F
Sbjct: 1023 QGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFS 1082
Query: 967 DCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPP 1026
CL NL +VL+RCE+ NLVLNWEKCHFMVREGIVLG + + GIEVD+AKI+++ ++ PP
Sbjct: 1083 SCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPP 1142
Query: 1027 TSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITA 1086
+VK+IRSFLGHAGFYR FIKDFS + +PLT LL K+ +F FDD CL AF +KEALITA
Sbjct: 1143 KTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEALITA 1202
Query: 1087 PIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELL 1146
PI+Q P+W+ PFEI+ T+D AQV YATTEKELL
Sbjct: 1203 PIVQAPNWDFPFEII----------------------------TMDDAQVRYATTEKELL 1234
Query: 1147 AVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGV 1206
AVV+A +KFR YLVGSK+ +YTDH+A++++ KKD KPRL+RWILLLQEFD+EI DKKG+
Sbjct: 1235 AVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGI 1294
Query: 1207 ENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTD---APWYADFVNFLAAGVLPPEL 1263
EN VADHLSR+R +DE+ +DDS P++QL + Q + PWYAD VN+L +G PP L
Sbjct: 1295 ENGVADHLSRMR--IEDEVLIDDSMPEEQLMAIQQLNEKKLPWYADHVNYLVSGEEPPNL 1352
Query: 1264 NYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEVSSILTHCHSSSYGGHASTQK 1323
+ +KKKFF D+ H+YWDEPYL+ D I+R C+ E+E+ IL HCH +YGGH +T K
Sbjct: 1353 SSYEKKKFFKDINHFYWDEPYLYTLCKDKIYRTCVSEDEIEGILLHCHGFAYGGHFATFK 1412
Query: 1324 TSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFM 1383
T KIL +GFWWPS+FKD FISKCD E FDVWGIDFM
Sbjct: 1413 TMSKILQAGFWWPSMFKDAQEFISKCD--------------------SFENFDVWGIDFM 1452
Query: 1384 GPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGS 1443
GPFPSS+GN+YILVA+DYVSKWVEAIAS TNDA+VV+K+FK +IFPRFGVPR+VISDGG
Sbjct: 1453 GPFPSSYGNKYILVAIDYVSKWVEAIASHTNDARVVLKLFKTIIFPRFGVPRIVISDGGK 1512
Query: 1444 HFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDD 1503
HFI++ FE LL+K GV+HK TSGQVE+SNR+IKAILEKTV ++R DWS KL+D
Sbjct: 1513 HFINKGFENLLKKHGVKHK--------TSGQVEISNREIKAILEKTVGSTRKDWSAKLND 1564
Query: 1504 ALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNE 1563
LWAYRTA+KTPIG TPF L+YGKSCHLPVELE+KA WA++ LN D A +KR +QLN+
Sbjct: 1565 TLWAYRTAFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLND 1624
Query: 1564 LEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLLFNSRLKLFPGKLRSRWSGPF 1623
L ++R++AYE+++IYKERTK++HDKKI+ R FK GD VLLFNSRL+LFPGKL+SRWSGPF
Sbjct: 1625 LNKIRLEAYESSKIYKERTKSFHDKKIVSRDFKVGDQVLLFNSRLRLFPGKLKSRWSGPF 1684
Query: 1624 QVRTVYPYGAIEIFSEETGSFTVNGQRLKIY 1654
V V PYGAI + + + G FTVNGQRLK Y
Sbjct: 1685 SVTAVRPYGAITL-AGKNGDFTVNGQRLKKY 1714
Score = 141 bits (355), Expect = 4e-33
Identities = 76/194 (39%), Positives = 109/194 (56%), Gaps = 22/194 (11%)
Query: 22 IVYPTVQGSNFKIKPALLILVQQNQFSGSPSEDPNLHISTFWRLSVTCKDD---QETVRL 78
IV P VQ +NF+IK L+ +VQ N+F G P EDP H+ F RL K + ++ +L
Sbjct: 29 IVPPPVQNNNFEIKSGLIAMVQSNKFHGLPMEDPLDHLDEFDRLCSLTKINRVSEDGFKL 88
Query: 79 YLFPFSLKDKASNWFNSLKPGSITSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQKNEES 138
LFPFSL DKA W SL GSITSW+ ++ FL +FF S+TA+LR + FTQ N E+
Sbjct: 89 RLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNET 148
Query: 139 LFDVWEHFKEILRRCPHHGLEKWLIIHTFYNGLTSSTKLTVDTAAGGALMNKDFTTAYAL 198
++ WE FK +CPHH ++ +DTA+ G +NKD + +
Sbjct: 149 FYEAWERFKGYQTQCPHH-------------------EMLLDTASNGNFLNKDVEDGWEV 189
Query: 199 IENMALNLFQWTEE 212
+EN+A + + E+
Sbjct: 190 VENLAQSDGNYNED 203
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 849 bits (2193), Expect = 0.0
Identities = 459/919 (49%), Positives = 589/919 (63%), Gaps = 128/919 (13%)
Query: 394 RAEREKPRVEENLPPPFKPKIPFPQRFAKSKLDEQFKKFIEMMNKIYIDVPFTEVLTQMP 453
R + + + ++ + PP+ PK+PFP R K + ++++ F E+M ++ P
Sbjct: 4 RKQTLEEKAKKTVLPPYVPKLPFPGRQRKIQREKEYSLFDEIMRQLQRYSP--------- 54
Query: 454 TYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVIGSEVVKKA 513
ECSAI+QN +P K +DPGSF +P IG +
Sbjct: 55 --------------------------ECSAILQNVIPVKREDPGSFVLPSRIGEYTFDRC 88
Query: 514 MCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPAD 573
+CDLGA VSLMP S+ +RLG T+M+L L DRS+ +P G+ EDV V+VG YIP D
Sbjct: 89 LCDLGAGVSLMPFSVAKRLGDTNFTPTKMSLVLGDRSISFPVGVAEDVQVRVGNFYIPTD 148
Query: 574 FVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDS 633
FV++E++E+ + ++LGRPFL A+IDV+K K+ +G EF + ++M P+ +
Sbjct: 149 FVIIELDEEPRHRLILGRPFLNIVAALIDVRKSKINLRIGDIVQEFNMERIMSKPTTECQ 208
Query: 634 YCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGT-KLEGEAKA-YEELLDRTLPMKG--- 688
+DI+D V E D L+ L GEA + +LD + PM
Sbjct: 209 TFWVDIMDELVNELLAELNTEDPLQTVLTKEESEFGYLGEATTRFARILDSSSPMTKVVA 268
Query: 689 ---LSVEELVKEEPTLLPKE--------APKVELKTLPSNLRYEFLGPNSTYPVIVNASL 737
L E+ K PK+ APK+ELK LP+ LR FLGPNSTY VI+N L
Sbjct: 269 FAELGDNEVEKALVVSSPKDCDDWSELNAPKMELKPLPARLRVAFLGPNSTYLVIINPEL 328
Query: 738 DEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKE 797
+ VE+ LL L+KY KA+GY++DDI GI+P+LCMH I LE + S+EHQRRLN N+++
Sbjct: 329 NNVESALLLCELRKYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNSNLRD 388
Query: 798 VVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCID 857
VVKKE++KLLDAG+IYPISDS WVSPV VVPKKGG+ VIKN+KNE I TRTVTG RMCID
Sbjct: 389 VVKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCID 448
Query: 858 YRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFG 917
YRKLN ATRKD+FPL FIDQMLERL+ ++C+LDGY GFFQI IHP+DQEKTTFTCP+G
Sbjct: 449 YRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYG 508
Query: 918 TFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLE 977
TFAYRRMPFGLCNAPATFQ CM IFSD +E MEVF+DDF VL+
Sbjct: 509 TFAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFMEVFIDDF---------------LVLQ 553
Query: 978 RCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLG 1037
RCE +LVLNWEK HFMVR+GIVLGH + ++G+EVDRAKIEI++ FLG
Sbjct: 554 RCEDKHLVLNWEKSHFMVRDGIVLGHKISEKGVEVDRAKIEIMR-------------FLG 600
Query: 1038 HAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLP 1097
HAGFYRRFIKDFS I +P T LL K+ FDD+CLQAF +KE+L++A I+QPP+W LP
Sbjct: 601 HAGFYRRFIKDFSKIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIVQPPEWELP 660
Query: 1098 FEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQ 1157
FE++CDASDY VGAVLGQR DKK+HAIYYAS+TLDGAQV
Sbjct: 661 FEVICDASDYVVGAVLGQRKDKKLHAIYYASRTLDGAQV--------------------- 699
Query: 1158 YLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRL 1217
IV+T H+A++YLL+KKDAKPRL+RWILLLQ+FDLEIKDKKG+EN VAD+LSRL
Sbjct: 700 -------IVHTVHAALRYLLSKKDAKPRLLRWILLLQDFDLEIKDKKGIENEVADNLSRL 752
Query: 1218 RETN----KDELP------LDDSFPDD---------QLFLL--AQTDAPWYADFVNFLAA 1256
R +D LP ++D + D+ +L L +++ PWYADF N+L+
Sbjct: 753 RVQEEVLMRDILPGENLASIEDCYMDEVGRLRVSTLELMTLHTGESNLPWYADFANYLSC 812
Query: 1257 GVLPPELNYQQKKKFFNDL 1275
V PP+ KKK ++
Sbjct: 813 EVPPPDFTGYFKKKLLKEV 831
>At4g07660 putative athila transposon protein
Length = 724
Score = 627 bits (1616), Expect = e-179
Identities = 328/596 (55%), Positives = 413/596 (69%), Gaps = 46/596 (7%)
Query: 483 AIIQNKL-PPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTR 541
AI Q K+ P KL DPGSF++PC +G K+ +CDLGA VSLMPLS+ +RLG + KS
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLGFTQYKSCN 62
Query: 542 MTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAII 601
++L LADRSV+ P + E++P+++G V IP DFVV+EM+E+ + P++LGRPFLATAGA+
Sbjct: 63 ISLILADRSVRIPHSLFENLPIRIGAVDIPTDFVVLEMDEEPKDPLILGRPFLATAGAMN 122
Query: 602 DVKKGKLAFNVGKET-VEFELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVC 660
DVKKGK+ N+GK + F++ MK P+IK I+ ID E D L
Sbjct: 123 DVKKGKIDLNLGKYCRMTFDVKDAMKKPTIKGQLFWIEEIDQLADELLEERAEEDHLYSA 182
Query: 661 LVNNAGTK-LEGEAKAYEELLDRTLPMK---------GLSVEELVKEEP----------- 699
L L E Y++LLD M+ G E +V E
Sbjct: 183 LTKRGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPETEVMVMSEEGSTQVQPAHSR 242
Query: 700 ----------------TLLPK-------EAPKVELKTLPSNLRYEFLGPNSTYPVIVNAS 736
++P +APKV+LK+LP LRY F GPNSTYPVI+N
Sbjct: 243 TYSTNNSTSTISNSGELIIPTSDDWSELKAPKVDLKSLPKGLRYVFFGPNSTYPVIINVE 302
Query: 737 LDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMK 796
L++ E LL L+KY +AIGY++ DIK I+PSLC HRI LE + SIE QRRLN N+K
Sbjct: 303 LNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLNLNLK 362
Query: 797 EVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCI 856
EVVKKE+LKLLDAGVIYPISDS WV PV VPKKGG+TV+KN+K+E I TRT+TG R+CI
Sbjct: 363 EVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCI 422
Query: 857 DYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPF 916
DYRKLN A+RKDHFPLPF +QMLE LA H + C+LDGYSGFFQIPIHPNDQEKTTFTCP+
Sbjct: 423 DYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPY 482
Query: 917 GTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVL 976
GTFAY+RMPFGLCNAP TFQRCM SIFSD +EK++EVFMDDFSV+G +F CL NL +VL
Sbjct: 483 GTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRVL 542
Query: 977 ERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEI 1032
+ E+ NLVLNWEKC+FMV+EGIVLGH + ++GIEVD+ KI+++ ++ PP +VK+I
Sbjct: 543 TKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
Score = 146 bits (368), Expect = 1e-34
Identities = 73/121 (60%), Positives = 95/121 (78%), Gaps = 2/121 (1%)
Query: 1119 KKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLN 1178
K + IYYAS+TLD AQ YATTEKELL VV+A KFR YLV SK+ VYTDH+A++++
Sbjct: 593 KTVKDIYYASRTLDEAQGRYATTEKELLVVVFAFKKFRSYLVESKVTVYTDHAALRHMYA 652
Query: 1179 KKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFL 1238
KKD KPRL+R ILLLQEFD+EI +KKG+EN ADHLSR+R ++ + +DDS P++QL +
Sbjct: 653 KKDTKPRLLRGILLLQEFDMEIVEKKGIENGAADHLSRMR--IEEPILIDDSMPEEQLMV 710
Query: 1239 L 1239
+
Sbjct: 711 V 711
>At2g06320 putative retroelement pol polyprotein
Length = 466
Score = 594 bits (1532), Expect = e-169
Identities = 278/427 (65%), Positives = 342/427 (79%), Gaps = 1/427 (0%)
Query: 1244 APWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEV 1303
+PWYAD VN+LA G+ PP L ++K FF D+ HYYWDEPYL+ D I+R + E+EV
Sbjct: 37 SPWYADHVNYLACGIEPPNLTSYERKNFFRDIHHYYWDEPYLYTLCKDKIYRSYVSEDEV 96
Query: 1304 SSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNE 1363
IL HCH S+YGGH +T KT KIL +GFWWP++FKD F+SKCD CQR G+I++RNE
Sbjct: 97 EGILLHCHDSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNE 156
Query: 1364 MPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMF 1423
MP N ILEV+IFDVWGIDFMGPFPSS+GN+YILV VDYVSKWVEAIASPTNDA+VV+K+F
Sbjct: 157 MPQNPILEVDIFDVWGIDFMGPFPSSYGNKYILVVVDYVSKWVEAIASPTNDAKVVLKLF 216
Query: 1424 KKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIK 1483
K +IFPRFGV VVISDGG HFI++ FE LL+K GV+HK+ATPYHPQTSGQVE+SNR+IK
Sbjct: 217 KTIIFPRFGVSWVVISDGGKHFINKVFENLLKKHGVKHKVATPYHPQTSGQVEISNREIK 276
Query: 1484 AILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAYWAI 1543
ILEKTV +R DWS KLDDALWAY+TA+KTPIG TPF L+ KSCHL VELE+KA WA+
Sbjct: 277 TILEKTVGITRKDWSTKLDDALWAYKTAFKTPIGTTPFNLLCVKSCHLHVELEYKAMWAV 336
Query: 1544 RNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLL 1603
+ LN D A +KR +QL+EL+E+R++AYE+++IYKERTK +HDKKII + F+ GD VLL
Sbjct: 337 KLLNFDIKTAEEKRLIQLSELDEIRLEAYESSKIYKERTKLFHDKKIITKDFQVGDQVLL 396
Query: 1604 FNSRLKLFPGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSFTVNGQRLKIYNTGEVNEVV 1663
FNSRLK+FPGKL+SRWSGPF + V PYGA+ + + ++G FTVNGQRLK Y ++ V
Sbjct: 397 FNSRLKIFPGKLKSRWSGPFCITEVRPYGAVTL-AGKSGVFTVNGQRLKKYLANQILPEV 455
Query: 1664 ADFTLSD 1670
L +
Sbjct: 456 TSIHLQE 462
>At2g13060 pseudogene
Length = 930
Score = 489 bits (1259), Expect = e-138
Identities = 228/364 (62%), Positives = 282/364 (76%), Gaps = 6/364 (1%)
Query: 1241 QTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPE 1300
+ D PWYAD VN+LAA V P KK+F +++ Y WDEPYL++ DGI+RRCI
Sbjct: 133 EKDYPWYADIVNYLAADVEPDNFTDYNKKRFLREIRRYQWDEPYLYKHSYDGIYRRCIAA 192
Query: 1301 NEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITK 1360
EV SIL+HCHSSSYGGH +T KT K+L + FWWP++F+D FIS+CD CQR G I+K
Sbjct: 193 TEVPSILSHCHSSSYGGHFATFKTVSKVLQADFWWPTMFRDTQKFISQCDPCQRRGKISK 252
Query: 1361 RNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVI 1420
RNEMP N LEVE+FD WGIDFMGPFP S N YILV VDYVSKWVEAIAS ND+ VV+
Sbjct: 253 RNEMPPNFRLEVEVFDRWGIDFMGPFPPSNKNLYILVDVDYVSKWVEAIASLKNDSAVVM 312
Query: 1421 KMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNR 1480
K+FK +IFPRFGVPR+VISDG HFI++ EKLL + GV+H++ATPYHPQTSGQVEVSNR
Sbjct: 313 KLFKSIIFPRFGVPRIVISDGDKHFINKILEKLLLQYGVQHRVATPYHPQTSGQVEVSNR 372
Query: 1481 QIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAY 1540
QIK ILEKTV ++ +WS KL DALWAY+TA+KTP+G TPF L+YGK+CHLPVELEHKA
Sbjct: 373 QIKEILEKTVGKAKKEWSYKLYDALWAYKTAFKTPLGTTPFHLLYGKACHLPVELEHKAA 432
Query: 1541 WAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDL 1600
WA++ +N D AG+ +EL+E+R+ AY+N+++YKERTK +HDKKI+ R F+ D
Sbjct: 433 WAVKMMNFDIKSAGE------SELDEIRIHAYDNSKLYKERTKAYHDKKILTRTFEPNDQ 486
Query: 1601 VLLF 1604
+L F
Sbjct: 487 ILRF 490
Score = 83.6 bits (205), Expect = 9e-16
Identities = 37/63 (58%), Positives = 53/63 (83%), Gaps = 2/63 (3%)
Query: 1177 LNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQL 1236
+ KKDAKPRL+ WILLLQEFD+E++DKKGVEN VADHLSR+R D++P++D P++ +
Sbjct: 1 MQKKDAKPRLLIWILLLQEFDIEVRDKKGVENGVADHLSRIR--IDDDVPINDFLPEENI 58
Query: 1237 FLL 1239
+++
Sbjct: 59 YMI 61
>At4g07600
Length = 630
Score = 473 bits (1218), Expect = e-133
Identities = 221/325 (68%), Positives = 262/325 (80%), Gaps = 18/325 (5%)
Query: 821 VSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLE 880
VSPVQ +PKKGG+TVIKN+K+E I RT+TG RMCIDYR LN A+R DHFPLPF DQMLE
Sbjct: 324 VSPVQCIPKKGGITVIKNEKDELIPIRTITGLRMCIDYRNLNAASRNDHFPLPFTDQMLE 383
Query: 881 RLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMM 940
RLA H ++C+LDGYSGFFQIPIHPND EKTTFTCP+GTFAY RMPFGLCNAPATFQRCM
Sbjct: 384 RLANHPYYCFLDGYSGFFQIPIHPNDHEKTTFTCPYGTFAYERMPFGLCNAPATFQRCMT 443
Query: 941 SIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIV 1000
SIFSD +E+++EVFMDDFSV+G +F CL NL +VL RCE+ NLVLNWEKCHFMV+EGI+
Sbjct: 444 SIFSDIIEEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNLVLNWEKCHFMVKEGIM 503
Query: 1001 LGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLL 1060
LGH + ++GIEVD+ FLGHAGFYRRFIKDFS I +PLT LL
Sbjct: 504 LGHKISEKGIEVDKG------------------CFLGHAGFYRRFIKDFSKIARPLTRLL 545
Query: 1061 LKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKK 1120
K+ +F FD+ C ++F +KEAL++AP+++ +W+ PFEIMCDA DYAVGAVLGQ+ DKK
Sbjct: 546 CKETEFEFDEDCPKSFHTIKEALVSAPVVRATNWDYPFEIMCDALDYAVGAVLGQKIDKK 605
Query: 1121 MHAIYYASKTLDGAQVNYATTEKEL 1145
+H IYYAS+TLD AQ YATTEKEL
Sbjct: 606 LHVIYYASRTLDEAQGRYATTEKEL 630
Score = 178 bits (451), Expect = 3e-44
Identities = 99/253 (39%), Positives = 156/253 (61%), Gaps = 4/253 (1%)
Query: 432 FIEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL-P 490
F + + ++ + +P + L + KFLK+++ ++ + E V L+ ECSAIIQ K+ P
Sbjct: 2 FAKNIKEVELRIPLVDALALILDTHKFLKDLIVERIQ-EVQGMVVLSHECSAIIQKKIVP 60
Query: 491 PKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRS 550
KL DPGSF++PC +G+ + +CDLGA VS MPLS+ +RLG + KS ++L LADRS
Sbjct: 61 KKLSDPGSFTLPCFLGTVAFNRCLCDLGALVSPMPLSIAKRLGFTQYKSCNISLILADRS 120
Query: 551 VKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAF 610
V+ G+++++P+++G I DFV++EM+E+ + P++L RPFLATAGA+IDVKKGK+
Sbjct: 121 VRISHGLLKNLPIRIGAAEISTDFVILEMDEEPKDPLILRRPFLATAGAMIDVKKGKIDL 180
Query: 611 NVGKE-TVEFELAKLMKGPSIKDSYCMIDIIDHCVKECSLASTAHDGLEVCLVNNAGTK- 668
N+GK+ + F++ MK P+I+ I+ +D E D L L
Sbjct: 181 NLGKDFRMTFDIKDAMKKPNIEGQLFWIEEMDQLADELLEELAQEDHLNSALTKTGQDGF 240
Query: 669 LEGEAKAYEELLD 681
L + Y++LLD
Sbjct: 241 LHLKVLGYQKLLD 253
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 466 bits (1198), Expect = e-131
Identities = 231/382 (60%), Positives = 288/382 (74%), Gaps = 27/382 (7%)
Query: 907 QEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFD 966
++KTTFTCP GTFAY+RM FGLCNAP TFQR M SIFSDF+E+IMEVFMDDFS F
Sbjct: 163 KKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDDFS----GFS 218
Query: 967 DCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPP 1026
CL NL +VLERCE+ NLVLNWEKCHFMV EGIVLGH + +GIEVD+AKI+++ ++ PP
Sbjct: 219 SCLLNLCRVLERCEETNLVLNWEKCHFMVHEGIVLGHKISGKGIEVDKAKIDVMIQLQPP 278
Query: 1027 TSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQAFCRLKEALITA 1086
+VK+IRSFLGHA FYRRFIKDFS I +PLT LL K+ +F FD+ CL+AF +KE L++A
Sbjct: 279 KTVKDIRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCLKAFHLIKETLVSA 338
Query: 1087 PIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGAQVNYATTEKELL 1146
PI Q P+W+ PFEIMCDA DYAVGAVL Q+ D K+H IYYAS+T+D AQ YAT EKELL
Sbjct: 339 PIFQAPNWDHPFEIMCDAFDYAVGAVLDQKIDDKLHVIYYASRTMDEAQTRYATIEKELL 398
Query: 1147 AVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGV 1206
AVV+A +K R YLVG K+ VY DH+A++++ KK+ K RL+RWILLLQEFD+EI DKKGV
Sbjct: 399 AVVFAFEKIRSYLVGFKVKVYIDHAALRHIYAKKETKSRLLRWILLLQEFDMEIIDKKGV 458
Query: 1207 ENVVADHLSRLRETNKDELPLDDSFPDDQLFL-------------------LAQTDAPWY 1247
EN VADHLSR+R +D +P+DD+ P++QL + + PWY
Sbjct: 459 ENGVADHLSRMR--IEDSVPIDDTMPEEQLMFYDLVNKSFDTKDMPEEADAVEEEKLPWY 516
Query: 1248 ADFVNFLAAGVLPPELNYQQKK 1269
AD VN+L +G + E++ +Q K
Sbjct: 517 ADLVNYLTSGQV--EVSNRQMK 536
Score = 140 bits (354), Expect = 5e-33
Identities = 66/98 (67%), Positives = 80/98 (81%)
Query: 1471 TSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCH 1530
TSGQVEVSNRQ+K IL K V S+ DWS KLD+ LWAYRTAYKTPIG TPF+++YGKSCH
Sbjct: 524 TSGQVEVSNRQMKDILTKVVGVSKRDWSTKLDETLWAYRTAYKTPIGRTPFQMLYGKSCH 583
Query: 1531 LPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELR 1568
LPVE+E+KA WA + LNL+ A +KR + L+ELEE+R
Sbjct: 584 LPVEVEYKAIWATKLLNLEIKGAQEKRPIDLHELEEIR 621
Score = 60.8 bits (146), Expect = 6e-09
Identities = 27/48 (56%), Positives = 35/48 (72%)
Query: 705 EAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKY 752
+APK++LK LP LRY FLGPN TYPVI+N L + + +LL L+KY
Sbjct: 99 KAPKLDLKPLPKGLRYAFLGPNDTYPVIINDGLSDEQVNQLLNELRKY 146
>At2g12040 T10J7.2
Length = 976
Score = 429 bits (1103), Expect = e-120
Identities = 202/304 (66%), Positives = 254/304 (83%), Gaps = 2/304 (0%)
Query: 936 QRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMV 995
+R MMSIF+D +E IMEVFMDDFSV+GS F+DCL NL KVL RCE+ +LVLNWEKCHF V
Sbjct: 79 ERGMMSIFTDMIEDIMEVFMDDFSVYGSLFEDCLENLYKVLARCEEKHLVLNWEKCHFRV 138
Query: 996 REGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKP 1055
++GIVLGH + + GIE DRAKIE++ + ++K +RSFLGHAGFYRRFIKDFS I +P
Sbjct: 139 QDGIVLGHRISEYGIEADRAKIEVMTSLQALDNLKAVRSFLGHAGFYRRFIKDFSKIARP 198
Query: 1056 LTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQ 1115
LTSLL K+ F F C AF ++K+ALI+API+QPPDW+LPFE+MCDASD+AVGAVLGQ
Sbjct: 199 LTSLLCKEVKFEFTQECHDAFQQIKQALISAPIVQPPDWDLPFEVMCDASDFAVGAVLGQ 258
Query: 1116 RNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKY 1175
R DKK+HAIYYAS+TLD A+ NYATTEKE LAVV+A +KFR YLVG K+IV+TDH+A+KY
Sbjct: 259 RKDKKLHAIYYASRTLDDAKRNYATTEKEFLAVVFAFEKFRSYLVGLKVIVHTDHAALKY 318
Query: 1176 LLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQ 1235
L+ KDAKPRL+RWILLLQEFD+E++DKKGVE VADHLSR+R D++P++D P++
Sbjct: 319 LMQNKDAKPRLLRWILLLQEFDIEVRDKKGVEKCVADHLSRIR--IDDDVPINDFLPEEN 376
Query: 1236 LFLL 1239
++++
Sbjct: 377 IYMI 380
Score = 346 bits (887), Expect = 7e-95
Identities = 194/420 (46%), Positives = 255/420 (60%), Gaps = 54/420 (12%)
Query: 1241 QTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPE 1300
+ D PWYAD V +L A V KK+F +++ YYWD+PYL
Sbjct: 455 EKDYPWYADIVIYLGADVELDNFIDYNKKRFLREIRRYYWDKPYL--------------- 499
Query: 1301 NEVSSILT------HCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQR 1354
+S +L HC + S Q +S ++L + + S ++H+ D CQR
Sbjct: 500 -PISIVLMEFIGEMHCCNRD-----SIQSSSSRLLVA--YNVSGCSEIHI---ASDPCQR 548
Query: 1355 TGSITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTN 1414
G I+ NEMP ILEV++FD WGI+FMGP P S N YILV VDYVSKWVEAIASP N
Sbjct: 549 RGKISNCNEMPQKFILEVKVFDCWGINFMGPIPPSNKNLYILVVVDYVSKWVEAIASPKN 608
Query: 1415 DAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQ 1474
D+ VV+K+FK +IFP FGVPR+VISDGG HFI++ KLL + GV+H++ATPYHPQTSGQ
Sbjct: 609 DSAVVMKLFKCIIFPHFGVPRIVISDGGKHFINKILTKLLLQYGVQHRVATPYHPQTSGQ 668
Query: 1475 VEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVE 1534
VEVSNRQIK ILEKTV A+KTP+G TPF L+YGK+CHLPVE
Sbjct: 669 VEVSNRQIKEILEKTV--------------------AFKTPLGTTPFHLLYGKACHLPVE 708
Query: 1535 LEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRH 1594
LEHKA W ++ LN D AG++R +QLNEL+E+++ AY+N+++YKERTK +HDKKI+ R
Sbjct: 709 LEHKAAWTVKMLNFDIKSAGERRLIQLNELDEIQIHAYDNSKLYKERTKAYHDKKILTRT 768
Query: 1595 FKSGDLVLLFNSRLKLFPGKLRSRWSGPFQVRTVYPYGAIEIFSEE--TGSFTVNGQRLK 1652
F+ D K F G+ + + ++ + I EE T FT++ LK
Sbjct: 769 FEPKDQKKKKTRETKPFKGRGKKSERVGSLLIPIFEHFRIRFEGEEVNTTRFTMDESYLK 828
Score = 38.9 bits (89), Expect = 0.025
Identities = 18/33 (54%), Positives = 25/33 (75%)
Query: 772 MHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVL 804
MHRI LE++ K S+EHQRRLN K+ ++K +L
Sbjct: 1 MHRIHLEDESKSSVEHQRRLNLIQKKRLRKRLL 33
>At2g10780 pseudogene
Length = 1611
Score = 383 bits (983), Expect = e-106
Identities = 267/880 (30%), Positives = 441/880 (49%), Gaps = 81/880 (9%)
Query: 775 ILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLT 834
I LE P + R+ P +KK++ +LLD G I P S S W +PV V KK G
Sbjct: 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRP-SSSPWGAPVLFVKKKDG-- 707
Query: 835 VIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGY 894
+R+CIDYR LNK T K+ +PLP ID+++++L F +D
Sbjct: 708 ----------------SFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLA 751
Query: 895 SGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVF 954
SG+ QIPI P D KT F + F + MPFGL NAPA F + M +F DF+++ + +F
Sbjct: 752 SGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIF 811
Query: 955 MDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDR 1014
++D V+ +++ +L VLER + L KC F R LGH++ D+G+ VD
Sbjct: 812 INDILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDP 871
Query: 1015 AKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQ 1074
KI IK+ P + EIRSFLG AG+YRRF+ F+S+ +PLT L KD F + D C +
Sbjct: 872 EKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEK 931
Query: 1075 AFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGA 1134
+F LK L AP++ P+ P+ + DAS +G VL Q+ I YAS+ L
Sbjct: 932 SFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKH 987
Query: 1135 QVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQ 1194
+ NY T + E+ AVV+ + +R YL G+K+ +YTDH ++KY+ + + R RW+ L+
Sbjct: 988 EKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVA 1047
Query: 1195 EFDLEIKDKKGVENVVADHLSRLR---ETNKDELPLDDSFPDDQLFLLAQTDAP---WYA 1248
+++L+I G N VAD LSR R E + ++ L + + L++ P A
Sbjct: 1048 DYNLDIAYHPGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLGLGAA 1107
Query: 1249 DFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIF----RRCIPENEV- 1303
D + L+ L E + ++K + + ++ ++G R C+P +
Sbjct: 1108 DQADLLSRIRLAQERD--------EEIKGWAQNNKTEYQTSNNGTIVVNGRVCVPNDRAL 1159
Query: 1304 -SSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRN 1362
IL H S + H + K ++ L + W + KDV +++KC CQ + +
Sbjct: 1160 KEEILREAHQSKFSIHPGSNK-MYRDLKRYYHWVGMKKDVARWVAKCPTCQ---LVKAEH 1215
Query: 1363 EMPLNNILEVEI----FDVWGIDFMGPFPSSFGNQY--ILVAVDYVSKWVEAIA-SPTND 1415
++P + + I +D +DF+ P+ +++ + V VD ++K +A S +
Sbjct: 1216 QVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDG 1275
Query: 1416 AQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQV 1475
A+++ + + I G+P ++SD + F S+ ++ + LG R ++T YHPQT Q
Sbjct: 1276 AEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQS 1335
Query: 1476 EVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVEL 1535
E + + ++ +L V +W L +AY +++ IGM+P++ +YG++C P+
Sbjct: 1336 ERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPL-- 1393
Query: 1536 EHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHF 1595
+ + L D+ ++ L+ +A ++R K++ +K+ + F
Sbjct: 1394 ---CWTPVGERRLFGPTIVDETTERMKFLKIKLKEA-------QDRQKSYANKRRKELEF 1443
Query: 1596 KSGDLVLL----------FNSRLKLFPGKLRSRWSGPFQV 1625
+ GDLV L F SR KL P R+ GP++V
Sbjct: 1444 QVGDLVYLKAMTYKGAGRFTSRKKLSP-----RYVGPYKV 1478
Score = 39.7 bits (91), Expect = 0.015
Identities = 20/84 (23%), Positives = 44/84 (51%), Gaps = 3/84 (3%)
Query: 544 LQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDV 603
++ A YP G++ + V V V +PAD +++ +++ + ++LG +L A ID
Sbjct: 507 VRAAGGQAMYPTGLVRGISVVVNGVNMPADLIIVPLKKHD---VILGMDWLGKYKAHIDC 563
Query: 604 KKGKLAFNVGKETVEFELAKLMKG 627
+G++ F + ++F+ + G
Sbjct: 564 HRGRIQFERDEGMLKFQGIRTTSG 587
>At3g11970 hypothetical protein
Length = 1499
Score = 378 bits (970), Expect = e-104
Identities = 280/865 (32%), Positives = 421/865 (48%), Gaps = 69/865 (7%)
Query: 773 HRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGG 832
H+I L E P + R + + K + K V LL G + S S + SPV +V KK G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQA-SSSPYASPVVLVKKKDG 649
Query: 833 LTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLD 892
WR+C+DYR+LN T KD FP+P I+ +++ L F +D
Sbjct: 650 T------------------WRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKID 691
Query: 893 GYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIME 952
+G+ Q+ + P+D +KT F G F Y MPFGL NAPATFQ M IF F+ K +
Sbjct: 692 LRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVL 751
Query: 953 VFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEV 1012
VF DD V+ S+ ++ +L++V E L KC F V + LGH + +GIE
Sbjct: 752 VFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIET 811
Query: 1013 DRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSC 1072
D AKI+ +K+ PT++K++R FLG AG+YRRF++ F I PL +L DA F +
Sbjct: 812 DPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDA-FEWTAVA 870
Query: 1073 LQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLD 1132
QAF LK AL AP++ P ++ F + DA +GAVL Q H + Y S+ L
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEG----HPLAYISRQLK 926
Query: 1133 GAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILL 1192
G Q++ + EKELLAV++A+ K+R YL+ S I+ TD ++KYLL ++ P +W+
Sbjct: 927 GKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPK 986
Query: 1193 LQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFVN 1252
L EFD EI+ ++G ENVVAD LSR+ + L + + + L Q + +
Sbjct: 987 LLEFDYEIQYRQGKENVVADALSRVE--GSEVLHMAMTVVECDLLKDIQAGYANDSQLQD 1044
Query: 1253 FLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENE--VSSILTHC 1310
+ A P+ KK+F+ W + L R+ + +P N+ ++IL
Sbjct: 1045 IITALQRDPD-----SKKYFS------WSQNILRRKS-----KIVVPANDNIKNTILLWL 1088
Query: 1311 HSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNI- 1369
H S GGH+ T ++ F+W + KD+ +I C CQ+ S + L +
Sbjct: 1089 HGSGVGGHSGRDVTHQRV-KGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLP 1147
Query: 1370 LEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIA-SPTNDAQVVIKMFKKVIF 1428
+ I+ +DF+ P S G I+V VD +SK IA S A V + +F
Sbjct: 1148 IPDTIWSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVF 1207
Query: 1429 PRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEK 1488
G P ++SD F S + + GV K+ + YHPQ+ GQ EV NR ++ L
Sbjct: 1208 KLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRC 1267
Query: 1489 TVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGK--SCHLPVELEHKAYWAIRNL 1546
WS L A + Y T Y + MTPF++VYG+ HLP L
Sbjct: 1268 MCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPY------------L 1315
Query: 1547 NLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLL--- 1603
+ +A R LQ E E++ + + + R K + D+ +R F+ GD V +
Sbjct: 1316 PGESKVAVVARSLQ--EREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQ 1373
Query: 1604 ---FNSRLKLFPGKLRSRWSGPFQV 1625
S + KL ++ GP+++
Sbjct: 1374 PYRQQSVVMRANQKLSPKYFGPYKI 1398
>At1g36590 hypothetical protein
Length = 1499
Score = 373 bits (958), Expect = e-103
Identities = 279/865 (32%), Positives = 421/865 (48%), Gaps = 69/865 (7%)
Query: 773 HRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGG 832
H+I L E P + R + + K + K V LL G + S S + SPV +V KK G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQA-SSSPYASPVVLVKKKDG 649
Query: 833 LTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLD 892
WR+C+DYR+LN T KD FP+P I+ +++ L F +D
Sbjct: 650 T------------------WRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKID 691
Query: 893 GYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIME 952
+G+ Q+ + P+D +KT F G F Y MPFGL NAPATFQ M IF F+ K +
Sbjct: 692 LRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVL 751
Query: 953 VFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEV 1012
VF DD V+ S+ ++ +L++V E L KC F V + LGH + +GIE
Sbjct: 752 VFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIET 811
Query: 1013 DRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSC 1072
D AKI+ +K+ PT++K++R FLG AG+YRRF++ F I PL +L DA F +
Sbjct: 812 DPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDA-FEWTAVA 870
Query: 1073 LQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLD 1132
QAF LK AL AP++ P ++ F + DA +GAVL Q H + Y S+ L
Sbjct: 871 QQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEG----HPLAYISRQLK 926
Query: 1133 GAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILL 1192
G Q++ + EKELLAV++A+ K+R YL+ S I+ TD ++KYLL ++ P +W+
Sbjct: 927 GKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPK 986
Query: 1193 LQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFVN 1252
L EFD EI+ ++G ENVVAD LSR+ + L + + + L Q + +
Sbjct: 987 LLEFDYEIQYRQGKENVVADALSRVE--GSEVLHMAMTVVECDLLKDIQAGYANDSQLQD 1044
Query: 1253 FLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENE--VSSILTHC 1310
+ A P+ KK+F+ W + L R+ + +P N+ ++IL
Sbjct: 1045 IITALQRDPD-----SKKYFS------WSQNILRRKS-----KIVVPANDNIKNTILLWL 1088
Query: 1311 HSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNI- 1369
H S GGH+ T ++ F+ + KD+ +I C CQ+ S + L +
Sbjct: 1089 HGSGVGGHSGRDVTHQRV-KGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLP 1147
Query: 1370 LEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIA-SPTNDAQVVIKMFKKVIF 1428
+ I+ +DF+ P S G I+V VD +SK IA S A V + + +F
Sbjct: 1148 IPDTIWSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVF 1207
Query: 1429 PRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEK 1488
G P ++SD F S + + GV K+ + YHPQ+ GQ EV NR ++ L
Sbjct: 1208 KLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRC 1267
Query: 1489 TVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGK--SCHLPVELEHKAYWAIRNL 1546
WS L A + Y T Y + MTPF++VYG+ HLP L
Sbjct: 1268 MCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPY------------L 1315
Query: 1547 NLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLL--- 1603
+ +A R LQ E E++ + + + R K + D+ +R F+ GD V +
Sbjct: 1316 PGESKVAVVARSLQ--EREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQ 1373
Query: 1604 ---FNSRLKLFPGKLRSRWSGPFQV 1625
S + KL ++ GP+++
Sbjct: 1374 PYRQQSVVMRANQKLSPKYFGPYKI 1398
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 372 bits (955), Expect = e-102
Identities = 268/874 (30%), Positives = 421/874 (47%), Gaps = 79/874 (9%)
Query: 763 IKGINPSLC-MHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWV 821
++G+ PS I LE P + R+ P +KK++ LL G I P S S W
Sbjct: 496 LQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRP-STSPWG 554
Query: 822 SPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLER 881
+PV V KK G +R+CIDYR LN T K+ +PLP ID++L++
Sbjct: 555 APVLFVKKKDG------------------SFRLCIDYRGLNWVTVKNKYPLPRIDELLDQ 596
Query: 882 LAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMS 941
L + F +D SG+ QIPI D KT F +G F + MPF L NAPA F R M S
Sbjct: 597 LRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNS 656
Query: 942 IFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVL 1001
+F +F+++ + +F+DD V+ + ++ +L +V+E+ + L KC F RE L
Sbjct: 657 VFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFL 716
Query: 1002 GHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLL 1061
GH+V G+ VD KIE I+ PT+ EIRSFL G+YRRF+K F+S+ +P+T L
Sbjct: 717 GHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTG 776
Query: 1062 KDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKM 1121
KD F + C + F LKE L + P++ P+ P+ + DAS +G VL QR
Sbjct: 777 KDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQRG---- 832
Query: 1122 HAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKD 1181
I YAS+ L + NY T + E+ V++A+ +R YL G K+ V+TDH ++KY+ N+ +
Sbjct: 833 KVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKYIFNQPE 892
Query: 1182 AKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPD-DQLFLLA 1240
R +RW+ L+ ++DLEI G NVVAD LSR R ++ + L L
Sbjct: 893 LNLRQMRWMELVADYDLEIAYHPGKANVVADALSRKRVGAAPGQSVEALVSEIGALRLCV 952
Query: 1241 QTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRR-CIP 1299
P + V+ A +L Q+K + E G+ ++ R C+P
Sbjct: 953 VAREPLGLEAVD--RADLLTRARLAQEKDEGLIAASKAEGSEYQFAANGTIFVYGRVCVP 1010
Query: 1300 ENEV--SSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGS 1357
++E IL+ H+S + H K ++ L + W + +DV ++++CD CQ
Sbjct: 1011 KDEELRREILSEAHASMFSIHPGATK-MYRDLKRYYQWVGMKRDVANWVAECDVCQ---L 1066
Query: 1358 ITKRNEMPLNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIA-SPTNDA 1416
+ +++P I V +D ++K +A T+ A
Sbjct: 1067 VKAEHQVP---------------------------DAIWVIMDRLTKSAHFLAIRKTDGA 1099
Query: 1417 QVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVE 1476
V+ K + I GVP ++SD S F + K+G + +++T YHPQT GQ E
Sbjct: 1100 AVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSE 1159
Query: 1477 VSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELE 1536
+ + ++ +L V W++ L +AY +Y+ IGM PF+ +YG+ C P+
Sbjct: 1160 RTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPLRWT 1219
Query: 1537 HKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFK 1596
+I + R L+LN E + R +++ DK+ + F+
Sbjct: 1220 QVEERSIYGADYVQETTERIRVLKLNMKEA------------QARQRSYADKRRRELEFE 1267
Query: 1597 SGDLVLLFNSRLK-----LFPGKLRSRWSGPFQV 1625
GD V L + L+ + KL R+ GPF++
Sbjct: 1268 VGDRVYLKMAMLRGPNRSILETKLSPRYMGPFRI 1301
>At2g11680 putative retroelement pol polyprotein
Length = 301
Score = 364 bits (934), Expect = e-100
Identities = 176/303 (58%), Positives = 229/303 (75%), Gaps = 20/303 (6%)
Query: 994 MVREGIVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSIT 1053
MVREGIVLGH + +G++V++AK++++ ++ PP + K+IRSFLGHAGFYRRFIKDFS +
Sbjct: 1 MVREGIVLGHKISKKGVQVNKAKVDVMMQLQPPKTGKDIRSFLGHAGFYRRFIKDFSKLA 60
Query: 1054 KPLTSLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVL 1113
+PLT LL K+A+F F++ CL AF KEAL++API+Q P+ + PFEIMCD SDYAVGAVL
Sbjct: 61 RPLTRLLCKEAEFIFEEECLTAFKSTKEALVSAPIVQAPNRDYPFEIMCDDSDYAVGAVL 120
Query: 1114 GQRNDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAI 1173
GQ+ D+K+H IYYAS T+D QV YATTEKELLAVV+A +KFR YLVGSK+ VYTDH+A+
Sbjct: 121 GQKIDRKLHVIYYASGTMDDTQVRYATTEKELLAVVFAFEKFRSYLVGSKVTVYTDHAAL 180
Query: 1174 KYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPD 1233
+++ KKD KPRL+RWILLLQEFD+EI DKKG+EN VADHLSR+R +DE+P+DDS P+
Sbjct: 181 RHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGVADHLSRMR--IEDEVPIDDSMPE 238
Query: 1234 DQLFLLAQTDAP------------------WYADFVNFLAAGVLPPELNYQQKKKFFNDL 1275
+QL + Q + WYAD V +L G PP L+ +KKKFF D+
Sbjct: 239 EQLMAIQQLNESAQINKSLDQVCAVEDKLMWYADHVKYLFGGEEPPNLSSYEKKKFFKDI 298
Query: 1276 KHY 1278
H+
Sbjct: 299 NHF 301
>At1g35370 hypothetical protein
Length = 1447
Score = 356 bits (914), Expect = 5e-98
Identities = 273/867 (31%), Positives = 420/867 (47%), Gaps = 81/867 (9%)
Query: 773 HRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGG 832
H+I L E P + R + K+ + K V ++ +G I +S S + SPV +V KK G
Sbjct: 570 HKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQ-VSSSPFASPVVLVKKKDG 628
Query: 833 LTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLD 892
WR+C+DY +LN T KD F +P I+ +++ L F +D
Sbjct: 629 T------------------WRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKID 670
Query: 893 GYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIME 952
+G+ Q+ + P+D +KT F G F Y M FGL NAPATFQ M S+F DF+ K +
Sbjct: 671 LRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVL 730
Query: 953 VFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEV 1012
VF DD ++ S+ ++ +L V E L K H LGH + R IE
Sbjct: 731 VFFDDILIYSSSIEEHKEHLRLVFEVMRLHKLFAKGSKEH--------LGHFISAREIET 782
Query: 1013 DRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSC 1072
D AKI+ +K+ PT+VK++R FLG AG+YRRF+++F I PL +L D F +
Sbjct: 783 DPAKIQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIAGPLHALTKTDG-FCWSLEA 841
Query: 1073 LQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLD 1132
AF LK L AP++ P ++ F + DA + AVL Q K H + Y S+ L
Sbjct: 842 QSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQ----KGHPLAYISRQLK 897
Query: 1133 GAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILL 1192
G Q++ + EKELLA ++A+ K+R YL+ S I+ TD ++KYLL ++ P +W+
Sbjct: 898 GKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPK 957
Query: 1193 LQEFDLEIKDKKGVENVVADHLSRLRETNKDELPLDDSFPDDQLFLLAQTDAPWYADFVN 1252
L EFD EI+ ++G EN+VAD LSR+ + + L D L + + +D
Sbjct: 958 LLEFDYEIQYRQGKENLVADALSRVEGSEVLHMALSIVECD----FLKEIQVAYESD--- 1010
Query: 1253 FLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEV---SSILTH 1309
GVL ++ Q+ + KHY W + L R+ + + N+V + +L
Sbjct: 1011 ----GVLKDIISALQQHP--DAKKHYSWSQDILRRKS------KIVVPNDVEITNKLLQW 1058
Query: 1310 CHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNI 1369
H S GG S + S + + S F+W + KD+ FI C CQ+ S L +
Sbjct: 1059 LHCSGMGGR-SGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPL 1117
Query: 1370 -LEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVE--AIASPTNDAQVVIKMFKKV 1426
+ +I+ +DF+ P+S G I+V VD +SK A+A P + A V + F
Sbjct: 1118 PIPDKIWCDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYS-ALTVAQAFLDN 1176
Query: 1427 IFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAIL 1486
++ G P ++SD F S +++ + GV ++++ YHPQ+ GQ EV NR ++ L
Sbjct: 1177 VYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYL 1236
Query: 1487 EKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKS--CHLPVELEHKAYWAIR 1544
W+ L A + Y T Y + MTPF+LVYG++ HLP
Sbjct: 1237 RCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPY----------- 1285
Query: 1545 NLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLL- 1603
L +A R LQ E E + + + + R K + D+ +R F GD V +
Sbjct: 1286 -LPGKSKVAVVARSLQ--ERENMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVK 1342
Query: 1604 -----FNSRLKLFPGKLRSRWSGPFQV 1625
S + KL ++ GP+++
Sbjct: 1343 LQPYRQQSVVLRVNQKLSPKYFGPYKI 1369
>At3g31340 Athila ORF 1, putative
Length = 781
Score = 321 bits (822), Expect = 3e-87
Identities = 221/680 (32%), Positives = 339/680 (49%), Gaps = 64/680 (9%)
Query: 5 RTLKEYSIPSSDEPCTIIVYPTVQGSNFKIKPALLILVQQNQFSGSPSEDPNLHISTF-- 62
+TL+++ P+ D + + ++F+IK LL LV+QNQF SE+ H+ F
Sbjct: 60 QTLRDHDAPNIDFFRDRNQHLPINRNDFEIKTGLLALVKQNQFFVHSSENHVYHLDNFED 119
Query: 63 -WRLSVTCKDDQETVRLYLFPFSLKDKASNWFNSLKPGSITSWDQLRREFLCRFFPPSKT 121
R++ + ++ LF FSL D A W SL P ++ SW+ + FL ++F S+T
Sbjct: 120 ICRITRMNGVPDDIIKCRLFIFSLADNAHRWLKSLDPINLRSWEDYKAAFLGQYFTQSRT 179
Query: 122 AQLRGKLYQFTQKNEESLFDVWEHFKEILRRCPHHGLEKWLIIHTFYNGLTSSTKLTVDT 181
A LR K+ F Q ES + WE FK+ R CPHHG + +I TFY G+ + K+ +DT
Sbjct: 180 AILRNKISSFQQGGTESFPEAWERFKDYYRECPHHGFSRATLISTFYQGVDKAYKMALDT 239
Query: 182 AAGGALMNKDFTTAYALIENMALNLFQWTEEKATIDPSPSKKEAGMDETSSIDYLSTKVN 241
A+ G M K T A LI+N+A + +D S + G E + LS KV
Sbjct: 240 ASNGDFMTKTETEATKLIKNLAASNINHN-----VDYDRSNRGGG-GELKQLAELSVKVE 293
Query: 242 ALSQRLDRMSTPTFSPPCETCDLSSHSNTDCNLSSVEQLNFVQNGQRVVQKSHIELLMEN 301
L +R ++ F + + DC+ ++NFV NG R +Q + +N
Sbjct: 294 QLLRRDQKVIN--FCEDSSKGMVHQEYSGDCSEDLQAEMNFV-NGIRYIQLRQAHKV-QN 349
Query: 302 YFLKQSEQLQELKDQTRL------LNNSLATLTTKIDS-----------ISSHNKISETQ 344
+ ++D + L L + A + K+DS +SSH K E Q
Sbjct: 350 FSPTDFRIKGTIRDNSNLQQILEGLKKNAADINVKVDSMYNDLNVKFATLSSHVKTLENQ 409
Query: 345 LSQVVRKVNHPNKMNAVTLRSGKPLEDPIRRTETDNSEKEICEPPSRETRAEREKPRVEE 404
+SQVV M SGK I + + + E + E ++P EE
Sbjct: 410 VSQVVSA-----SMRPAGAHSGKEEPREIDVAKQVETNAVVVETLVEDKIVEDDEPLSEE 464
Query: 405 NLPPPFKPKIPFP--QRFAKSKLDEQFKKFIEMMNKIYIDVPFTEVLTQMPTYAKFLKEI 462
PPP+ PK+PFP +R +S+ E++ +E + + T+V+ E+
Sbjct: 465 --PPPYVPKLPFPGRERQIQSRKREEYALLVESQRQ-QKEAQLTDVV-----------EV 510
Query: 463 LSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVS 522
L+ K + CSAI +P KL DPGSF +PC IG ++ +CDLGA V+
Sbjct: 511 LAGKEMVST---------CSAIPPATIPEKLGDPGSFVLPCRIGKSAFERCLCDLGAGVN 561
Query: 523 LMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEED 582
LMPLS+ +RLGI K +R++L LADRSV++P G+ E+V V+VG+ YIP DFVV+E++++
Sbjct: 562 LMPLSMSKRLGITNFKPSRISLILADRSVRFPVGLAENVHVRVGDFYIPTDFVVLELDKE 621
Query: 583 NQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDSYCMIDIID- 641
P+ LGRPFL T GAIIDV++ + +G +EF++ K P+I+ +D D
Sbjct: 622 PHDPLTLGRPFLNTVGAIIDVRRSTINLQIGDFALEFDMKGTRKNPTIEGHAFSVDTNDE 681
Query: 642 ---HCVKECSLASTAHDGLE 658
C K+C S + L+
Sbjct: 682 PGAECAKDCIELSDVEEVLD 701
>At2g07410 putative retroelement pol polyprotein
Length = 411
Score = 302 bits (773), Expect = 1e-81
Identities = 157/276 (56%), Positives = 197/276 (70%), Gaps = 36/276 (13%)
Query: 1383 MGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGG 1442
MG FPSS+GN+YILVAVDYVSKWVEAI SPTNDA+VV+K+FK +IF RFGV VVISDG
Sbjct: 1 MGSFPSSYGNKYILVAVDYVSKWVEAIVSPTNDAKVVLKLFKTIIFTRFGVHMVVISDGR 60
Query: 1443 SHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLD 1502
HFI++ FE LL+K GV+HK+AT YHPQTSGQVE+SNR+IKAILEKTV +R D S KLD
Sbjct: 61 KHFINKAFENLLKKHGVKHKVATSYHPQTSGQVEISNREIKAILEKTVGVTRKDLSAKLD 120
Query: 1503 DALWAYRTAYKT----PIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRK 1558
DALWAY+TA+KT PIG TPF L+Y KS HLP+ELE KA WA++ +N D A ++R
Sbjct: 121 DALWAYKTAFKTAFKTPIGTTPFNLLYAKSGHLPIELEFKAMWAVKLMNFDIKTAEEERL 180
Query: 1559 LQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVLLFNSRLKLFPGKLRSR 1618
+QLN+L+E+R++AYE+++ YK + K L+SR
Sbjct: 181 IQLNDLDEIRLEAYESSKNYKGKRK-------------------------------LKSR 209
Query: 1619 WSGPFQVRTVYPYGAIEIFSEETGSFTVNGQRLKIY 1654
GPF V + PYGA+ + + ++G FTVNGQRLK Y
Sbjct: 210 LFGPFCVTEIRPYGAVTL-AVKSGDFTVNGQRLKKY 244
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 300 bits (768), Expect = 5e-81
Identities = 175/457 (38%), Positives = 254/457 (55%), Gaps = 24/457 (5%)
Query: 763 IKGINPSLC-MHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWV 821
++G+ PS I LE P + R+ P +KK++ LL G I P S S W
Sbjct: 470 LQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRP-STSPWG 528
Query: 822 SPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLER 881
+PV V KK G +R+CIDYR+LN+ T K+ +PLP ID++L++
Sbjct: 529 APVLFVKKKDG------------------SFRLCIDYRELNRVTVKNRYPLPRIDELLDQ 570
Query: 882 LAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMS 941
L + F +D SG+ QIPI D KT F +G F + MPFGL NAPA F R M S
Sbjct: 571 LRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNS 630
Query: 942 IFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVL 1001
+F +F+++ + +F+DD V+ + ++ +L +V+E+ + L KC F RE L
Sbjct: 631 VFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFL 690
Query: 1002 GHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLL 1061
GH+V G+ VD KIE I+ PT+ EIRSFLG AG+YRRF+K F+S+ +P+T L
Sbjct: 691 GHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTG 750
Query: 1062 KDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKM 1121
KD F + C + F LKE L + P++ P+ P+ + DAS +G VL Q
Sbjct: 751 KDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHG---- 806
Query: 1122 HAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKD 1181
I YAS+ L + NY T + E+ AV++A+ +R YL G K+ V+TDH ++KY+ + +
Sbjct: 807 KVIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPE 866
Query: 1182 AKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLR 1218
R RW+ L+ ++DLEI G NVV D LSR R
Sbjct: 867 LNLRQRRWMELVADYDLEIAYHPGKANVVVDALSRKR 903
Score = 31.2 bits (69), Expect = 5.3
Identities = 30/115 (26%), Positives = 47/115 (40%), Gaps = 10/115 (8%)
Query: 1326 FKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGIDFMG- 1384
++ L + W + DV ++++CD CQ + K L +L+ W DF+
Sbjct: 962 YRDLKRYYQWVGMKMDVANWVAECDVCQ----LVKAEHQVLGGMLQSLPIPEWKWDFITM 1017
Query: 1385 ----PFPSSFGNQYILVAVDYVSKWVEAIA-SPTNDAQVVIKMFKKVIFPRFGVP 1434
S I V VD ++K +A T+ A V+ K F I GVP
Sbjct: 1018 DLVVGLRVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVP 1072
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 277 bits (708), Expect = 4e-74
Identities = 177/512 (34%), Positives = 263/512 (50%), Gaps = 41/512 (8%)
Query: 708 KVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGIN 767
+V + +L Y+ + P S VI V+ EK++ + ++G+
Sbjct: 407 RVSFERPEGSLVYQGVRPTSGSLVI-----SAVQAEKMIEKGCEAYLEFEDVFQSLQGLP 461
Query: 768 PSLC-MHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQV 826
PS I LE P + R+ P +KK++ LL A PV
Sbjct: 462 PSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGA-------------PVLF 508
Query: 827 VPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHS 886
V KK G +R+CIDYR LN T K+ +PLP ID++L++L +
Sbjct: 509 VKKKDG------------------SFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGAT 550
Query: 887 HFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDF 946
F +D SG+ IPI D KT F +G F + MPFGL NAPA F R M S+F +
Sbjct: 551 CFSKIDLTSGYHLIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEV 610
Query: 947 VEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVF 1006
+++ + +F+DD V+ + ++ +L +V+E+ + L KC F RE LGH+V
Sbjct: 611 LDEFVIIFIDDILVYSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVS 670
Query: 1007 DRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADF 1066
G+ VD KIE I+ PT+ EIRSFLG AG+YRRF+K F+S+ +P+T L KD F
Sbjct: 671 AEGVSVDPEKIEAIRDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPF 730
Query: 1067 TFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYY 1126
+ C + F LKE L + P++ P+ P+ + DAS +G VL QR I Y
Sbjct: 731 VWSPECEEGFVSLKEMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRG----KVIAY 786
Query: 1127 ASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRL 1186
AS+ L + NY T + E+ AV++A+ +R YL G + V+TDH ++KY+ + + R
Sbjct: 787 ASRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQ 846
Query: 1187 IRWILLLQEFDLEIKDKKGVENVVADHLSRLR 1218
+W+ L+ ++DLEI G NVVAD LS R
Sbjct: 847 RQWMELVADYDLEIAYHPGKANVVADALSHKR 878
Score = 100 bits (248), Expect = 9e-21
Identities = 80/296 (27%), Positives = 138/296 (46%), Gaps = 25/296 (8%)
Query: 1340 KDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEI----FDVWGIDFMGPFPSSFGNQYI 1395
+DV ++++CD CQ + +++P + + I +D IDF+ P S I
Sbjct: 941 EDVANWVAECDVCQL---VKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTKDAI 997
Query: 1396 LVAVDYVSKWVEAIA-SPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLL 1454
V VD ++K +A T+ A V+ K + I GVP ++SD S F S +
Sbjct: 998 WVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQ 1057
Query: 1455 QKLGVRHKIATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKT 1514
++G + +++T YHPQT GQ E + + ++ +L V W++ L +AY +Y
Sbjct: 1058 AEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPA 1117
Query: 1515 PIGMTPFKLVYGKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYEN 1574
IGM PF+ +Y + C P+ L +I + R L+LN E
Sbjct: 1118 SIGMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEA-------- 1169
Query: 1575 ARIYKERTKTWHDKKIIKRHFKSGDLVLLFNSRLK-----LFPGKLRSRWSGPFQV 1625
++R +++ DK+ + F+ GD V L + L+ + KL R+ GPF++
Sbjct: 1170 ----QDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRI 1221
>At1g20390 hypothetical protein
Length = 1791
Score = 269 bits (688), Expect = 9e-72
Identities = 168/520 (32%), Positives = 270/520 (51%), Gaps = 24/520 (4%)
Query: 703 PKEAPKVELKTLPSNL-RYEFLGPNSTYP---VIVNASLDEVETEKLLYVLKKYPKAIGY 758
PKEA S L R E + + + P V V A + +L+ +LK+ K +
Sbjct: 693 PKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAW 752
Query: 759 TIDDIKGINPSLCMHRILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDS 818
+I+D+KGI+P++ H + ++ +KP + +R+L P V +EV KLL AG I +
Sbjct: 753 SIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYP 812
Query: 819 KWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQM 878
+W++ VV KK G WR+C+DY LNKA KD +PLP ID++
Sbjct: 813 EWLANPVVVKKKNGK------------------WRVCVDYTDLNKACPKDSYPLPHIDRL 854
Query: 879 LERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRC 938
+E + + ++D +SG+ QI +H +DQEKT+F GT+ Y+ M FGL NA AT+QR
Sbjct: 855 VEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQRF 914
Query: 939 MMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREG 998
+ + +D + + +EV++DD V +D + +L K + + LN KC F V G
Sbjct: 915 VNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVTSG 974
Query: 999 IVLGHLVFDRGIEVDRAKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTS 1058
LG++V RGIE + +I I ++ P + +E++ G RFI + P +
Sbjct: 975 EFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDKCLPFYN 1034
Query: 1059 LLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRND 1118
LL + A F +D +AF +LK+ L T PI+ P+ + SD+AV +VL + +
Sbjct: 1035 LLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHAVSSVLVREDR 1094
Query: 1119 KKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLN 1178
+ I+Y SK+L A+ Y EK LAVV + K R Y I V TD ++ L+
Sbjct: 1095 GEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLTD-QPLRVALH 1153
Query: 1179 KKDAKPRLIRWILLLQEFDLEIKDKKGVEN-VVADHLSRL 1217
R+ +W + L E+D++ + + +++ V+AD L L
Sbjct: 1154 SPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLADFLIEL 1193
Score = 200 bits (508), Expect = 7e-51
Identities = 123/416 (29%), Positives = 205/416 (48%), Gaps = 13/416 (3%)
Query: 1246 WYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPENEVSS 1305
W + ++L+ G LP + + ++ K+ E +L + + G C+ E++
Sbjct: 1380 WRVEIRDYLSDGTLPSD-KWTARRLRIKAAKYTLMKE-HLLKVSAFGAMLNCLHGTEINE 1437
Query: 1306 ILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMP 1365
I+ H + G H+ + + K+ GF+WP++ D F +KC++CQR +
Sbjct: 1438 IMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTEL 1497
Query: 1366 LNNILEVEIFDVWGIDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKK 1425
L + F W +D +GP P+S ++ILV DY +KWVEA + T A V K
Sbjct: 1498 LRAGVAPYPFMRWAMDIVGPMPASRQKRFILVMTDYFTKWVEAESYATIRANDVQNFVWK 1557
Query: 1426 VIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIATPYHPQTSGQVEVSNRQIKAI 1485
I R G+P +I+D GS FIS FE +R +TP +PQ +GQ E +N+ I +
Sbjct: 1558 FIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNKTILSG 1617
Query: 1486 LEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVYGKSCHLPVELEHKAY---WA 1542
L+K + + W+++LD LW+YRT ++ TPF YG P E+ + +
Sbjct: 1618 LKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYSSLRRSMM 1677
Query: 1543 IRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTWHDKKIIKRHFKSGDLVL 1602
++N L+ + D+ L++LEE+R A + Y+ +++K+ RHF GDLVL
Sbjct: 1678 VKNPELNDRMMLDR----LDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGDLVL 1733
Query: 1603 --LFNSRLKLFPGKLRSRWSGPFQVRTVYPYGAIEIFSEETGSF--TVNGQRLKIY 1654
+F + ++ GKL + W G +QV + G E+ + + T N LK Y
Sbjct: 1734 RKVFENTAEINAGKLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTWNSMHLKRY 1789
Score = 44.3 bits (103), Expect = 6e-04
Identities = 26/80 (32%), Positives = 39/80 (48%)
Query: 71 DDQETVRLYLFPFSLKDKASNWFNSLKPGSITSWDQLRREFLCRFFPPSKTAQLRGKLYQ 130
D+ + R +F L A NWF+ LKP SI S+ QL FL + P + L+
Sbjct: 202 DNFDAGRCQIFIEHLTGPAHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWS 261
Query: 131 FTQKNEESLFDVWEHFKEIL 150
+Q +ESL + FK ++
Sbjct: 262 ISQGAKESLRSFVDRFKLVV 281
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 255 bits (652), Expect = 1e-67
Identities = 154/406 (37%), Positives = 219/406 (53%), Gaps = 25/406 (6%)
Query: 775 ILLEEDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLT 834
I LE P + R+ P +KK++ +LL G I P S S W +PV V KK G
Sbjct: 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRP-SSSPWGAPVLFVKKKDG-- 202
Query: 835 VIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGY 894
+R+CIDYR LNK T K+ +PLP ID+++++L F +D
Sbjct: 203 ----------------SFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLA 246
Query: 895 SGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVF 954
SG+ QIPI P D KT F +G F + MPFGL NAPA F + M +F DF+++ + +F
Sbjct: 247 SGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIF 306
Query: 955 MDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVFDRGIEVDR 1014
+DD VH +++ +L VLER + L K F R LGH++ D+G+ VD
Sbjct: 307 IDDILVHSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDP 366
Query: 1015 AKIEIIKKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTSLLLKDADFTFDDSCLQ 1074
KI IK+ P + EIRSFLG AG+YRRF+ F+S+ +PLT L KD F + D C +
Sbjct: 367 EKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEK 426
Query: 1075 AFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGAVLGQRNDKKMHAIYYASKTLDGA 1134
+F LK LI AP++ P+ P+ + DAS +G VL Q+ I YAS+ L
Sbjct: 427 SFLELKAMLINAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKH 482
Query: 1135 QVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKK 1180
+ NY T + E+ AVV+A+ +R YL+ ++ D + LN K
Sbjct: 483 EKNYPTHDLEMAAVVFALKIWRSYLI--RLAQERDEEIKGWTLNNK 526
Score = 108 bits (271), Expect = 2e-23
Identities = 87/350 (24%), Positives = 164/350 (46%), Gaps = 40/350 (11%)
Query: 1295 RRCIPENEV--SSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKC 1352
R C+P + IL H S + H + K ++ L + W + KDV +++KC C
Sbjct: 542 RVCVPNDRALKEEILREAHQSKFSIHPGSNKM-YRDLKRYYHWVGMRKDVARWVAKCPTC 600
Query: 1353 QRTGSITKRNEMPLNNILEVEI----FDVWGIDFMGPFPSSFGNQY--ILVAVDYVSKWV 1406
Q + +++P + + I +D +DF+ P+ +++ + V VD ++K
Sbjct: 601 QL---VKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSA 657
Query: 1407 EAIASPTND-AQVVIKMFKKVIFPRFGVPRVVISDGGSHFISRHFEKLLQKLGVRHKIAT 1465
+A D A+++ + + I G+P ++SD + F S+ + + LG R ++T
Sbjct: 658 HFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLST 717
Query: 1466 PYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWSNKLDDALWAYRTAYKTPIGMTPFKLVY 1525
YHPQT GQ E + + ++ +L V +W L +AY +++ IGM+P++ +Y
Sbjct: 718 AYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALY 777
Query: 1526 GKSCHLPVELEHKAYWAIRNLNLDPNLAGDKRKLQLNELEELRMDAYENARIYKERTKTW 1585
G++C P+ + + L D+ ++ L+ +A ++R K++
Sbjct: 778 GRACRTPL-----CWTPVGERRLFGPTIVDETTERMKFLKIKLKEA-------QDRQKSY 825
Query: 1586 HDKKIIKRHFKSGDLVLL----------FNSRLKLFPGKLRSRWSGPFQV 1625
+K+ + F+ GDLV L F SR KL P R+ GP++V
Sbjct: 826 ANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSP-----RYVGPYKV 870
Score = 32.0 bits (71), Expect = 3.1
Identities = 17/84 (20%), Positives = 41/84 (48%), Gaps = 3/84 (3%)
Query: 544 LQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDV 603
++ A YP ++ + V V V +PAD +++ +++ + ++LG +L ID
Sbjct: 2 VRAAGGQAMYPTRLVRGISVVVNGVNMPADLIIVPLKKHD---VILGMDWLGKYKGHIDC 58
Query: 604 KKGKLAFNVGKETVEFELAKLMKG 627
+ ++ F + ++F+ + G
Sbjct: 59 HRERIQFERDEGMLKFQGIRTTSG 82
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,860,132
Number of Sequences: 26719
Number of extensions: 1764596
Number of successful extensions: 5575
Number of sequences better than 10.0: 198
Number of HSP's better than 10.0 without gapping: 147
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 5039
Number of HSP's gapped (non-prelim): 404
length of query: 1672
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1559
effective length of database: 8,299,349
effective search space: 12938685091
effective search space used: 12938685091
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146683.16


