

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.12 - phase: 0
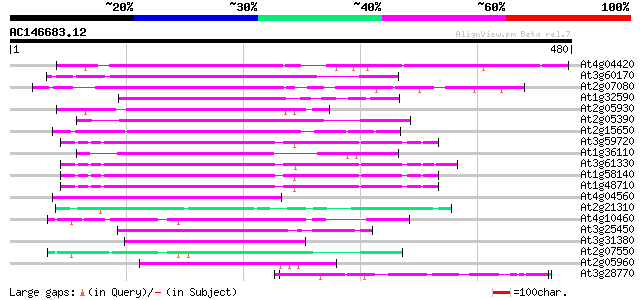
(480 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g04420 putative transposon protein 111 1e-24
At3g60170 putative protein 104 1e-22
At2g07080 putative gag-protease polyprotein 93 3e-19
At1g32590 hypothetical protein, 5' partial 89 4e-18
At2g05930 copia-like retroelement pol polyprotein 86 6e-17
At2g05390 putative retroelement pol polyprotein 84 1e-16
At2g15650 putative retroelement pol polyprotein 81 1e-15
At3g59720 copia-type reverse transcriptase-like protein 80 2e-15
At1g36110 hypothetical protein 80 2e-15
At3g61330 copia-type polyprotein 79 4e-15
At1g58140 hypothetical protein 79 7e-15
At1g48710 hypothetical protein 79 7e-15
At4g04560 putative transposon protein 70 2e-12
At2g21310 putative retroelement pol polyprotein 69 7e-12
At4g10460 putative retrotransposon 67 2e-11
At3g25450 hypothetical protein 67 2e-11
At3g31380 hypothetical protein 65 8e-11
At2g07550 putative retroelement pol polyprotein 60 3e-09
At2g05960 putative retroelement pol polyprotein 57 2e-08
At3g28770 hypothetical protein 55 1e-07
>At4g04420 putative transposon protein
Length = 1008
Score = 111 bits (277), Expect = 1e-24
Identities = 114/474 (24%), Positives = 204/474 (42%), Gaps = 68/474 (14%)
Query: 41 FSWWKTNMYSFIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRRIHTPAQK 89
+ WK M + I GL +E W EDG D L ++ A++
Sbjct: 21 YGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTKDQW--------NDAEE 72
Query: 90 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 149
K + + +I + + ++ ++ + +AK + L +EG+ VK ++ ML Q+
Sbjct: 73 AKAKANSRALSLIFNFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 132
Query: 150 ELFRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAK 209
E M++ +IEE + + S L K Y V K+LR LPSR+ K TA+ +
Sbjct: 133 ENLSMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 192
Query: 210 DLNTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDG 269
D +++ E++V L+ +E+ + + SK +AL + K
Sbjct: 193 DTDSIDFEEVVGMLQAYELEITSGK-GGYSKGLALAASAK-------------------- 231
Query: 270 DSDEDQSVK--MAMLSNKLEYLARK--QKKFLSKRGSYK---NSKKEDQKGCFNCKKPGH 322
+E Q +K M+M++ R+ +K F +G+ + S K D+ C C+ GH
Sbjct: 232 -KNEIQELKDTMSMMAKDFSRAMRRVEKKGFGRNQGTDRYRDRSSKRDEIQCHECQGYGH 290
Query: 323 FIADCPDLQKEKYKGKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAA 382
A+CP L+++ K S+ + +KF KS +SES S+ +++D K
Sbjct: 291 IKAECPSLKRKDLK-CSECNGLGHTKFDCVGSKSKPDKSCSSESESDSNDGDSEDYIKGF 349
Query: 383 MRLVATV-SSEAVSEAESDSEDE----------------NEVYSKIPRQELVDSLKELLS 425
+ V + + S++E+D EDE NE + K+ L+ S +++
Sbjct: 350 VSFVGIIEEKDESSDSEADGEDEDNSADEDSDIEKDVNINEEFRKLYDSWLMLSKEKVAW 409
Query: 426 LFEH-RTNELTDLKEKYVDLMKQQKTTLLELKASEEELKGFNLISTTYEDRLKI 478
L E + ELT+ + + Q+ + L++ K S E K L + R I
Sbjct: 410 LEEKLKVQELTEKLKGELTAANQKNSELIQ-KCSVAEEKNRELSQELSDTRKNI 462
>At3g60170 putative protein
Length = 1339
Score = 104 bits (259), Expect = 1e-22
Identities = 76/301 (25%), Positives = 135/301 (44%), Gaps = 43/301 (14%)
Query: 32 PKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKL 91
P+F+G + +W M +F+ ELW ++E+G+ + + + +R A ++
Sbjct: 13 PRFDG---YYDFWSMTMENFLRS--RELWRLVEEGIPAIVVGTTPVSEAQR---SAVEEA 64
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
K K++ + +I R + DKST+KA++ S+ ++GS KVK A+ L ++EL
Sbjct: 65 KLKDLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFEL 124
Query: 152 FRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
MK+ I+ R T+V+ ++ + S VSKILRSL ++ V +IEE+ DL
Sbjct: 125 LAMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDL 184
Query: 212 NTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+TLS+++L SL HE LN H +++ + + + ++ S
Sbjct: 185 STLSIDELHGSLLVHEQRLNGHVQEEQALKVTHEERPSQGRGRGVFRGSRGR-------- 236
Query: 272 DEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQ 331
RG ++ C+ C GHF +CP+ +
Sbjct: 237 ---------------------------GRGRGRSGTNRAIVECYKCHNLGHFQYECPEWE 269
Query: 332 K 332
K
Sbjct: 270 K 270
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 93.2 bits (230), Expect = 3e-19
Identities = 105/446 (23%), Positives = 188/446 (41%), Gaps = 75/446 (16%)
Query: 20 DTADKKTDSGKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAI 79
+ D T +GK D + + +WK M I G E+G I
Sbjct: 2 EPTDVSTGTGKVLLL--DTKRYGYWKVCMTQIIRG------------------QEDGFKI 41
Query: 80 DR-RIHTPAQKKLYKKHH-KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKV 137
+ + + A++KL K + + I + E+ + +AK + +L + EG+ V
Sbjct: 42 TKPKANWTAEEKLQSKFNARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSHEGTSSV 101
Query: 138 KEAKALMLVHQYELFRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSR 197
K + + Q+E +M+ D I + S+ L + +++ K+Y V K+LR LP +
Sbjct: 102 KRTRLDHIATQFEYLKMEPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPK 161
Query: 198 WRPKVTAIEEAKDLNTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAY 257
+ + A + + +S DLV LK EM ++ + K SK+IA A
Sbjct: 162 FAAHKAVMRVAGNTDKISFVDLVGMLKLEEMKADQDKV-KPSKNIAF----------NAD 210
Query: 258 KASESEEESPDGDSDEDQSVKMAMLSNKL-EYLARKQKKFLSKRGSYKNSKKEDQK---- 312
+ SE +E DG MA+L+ + L R + RG + S+ +D +
Sbjct: 211 QGSEQFQEIKDG---------MALLARNFGKALKRVEIDGERSRGRFSRSENDDLRKKKE 261
Query: 313 -GCFNCKKPGHFIADCPDLQKEKYKGKSKKSSFSSSKF----RKQIKKSLMATWEDLDSE 367
C+ C GH +CP + K K K +KF + ++K+ + ++ D
Sbjct: 262 IQCYECGGFGHIKPECP-ITKRKEMKCLKCKGVGHTKFECPNKSKLKEKSLISFSD---- 316
Query: 368 SGSDKEEADDDAKAAMRLVATVSSEAVSE--AESDSEDENEVYSKIPRQELVDS------ 419
E+DD+ + + VA ++S S+ +++DS+ + E+ K + L DS
Sbjct: 317 -----SESDDEGEELLNFVAFMASSDSSKFMSDTDSDCDEELNPKDKYRVLYDSWVQLSK 371
Query: 420 -----LKELLSLFEHRTNELTDLKEK 440
+KE L+L N T+ K+K
Sbjct: 372 DKLKLVKEKLTLEAKLANVSTEDKQK 397
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 89.4 bits (220), Expect = 4e-18
Identities = 64/240 (26%), Positives = 110/240 (45%), Gaps = 32/240 (13%)
Query: 94 KHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFR 153
K HK++ + ASI +T + K T+K ++ S+ ++G+ +V+ A+ L +E+
Sbjct: 24 KDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSFEVLE 83
Query: 154 MKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNT 213
MK +I +SR + + ++ L + S V KILR+L ++ V AIEE+ ++
Sbjct: 84 MKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEESNNIKE 143
Query: 214 LSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDE 273
L+V+ L SSL HE +L+ H+ + + K+ ++ PDG
Sbjct: 144 LTVDGLQSSLMVHEQNLSRHDVEE-----------RVLKAETQWR--------PDGGRGR 184
Query: 274 DQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKE 333
S R + + + Y N D CF C K GH+ A+CP +KE
Sbjct: 185 GGSPSR----------GRGRGGYQGRGRGYVN---RDTVECFKCHKMGHYKAECPSWEKE 231
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 85.5 bits (210), Expect = 6e-17
Identities = 68/268 (25%), Positives = 116/268 (42%), Gaps = 47/268 (17%)
Query: 41 FSWWKTNMYSFIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRRIHTPAQK 89
+ WK M + I GL +E W EDG D L +++ + T +
Sbjct: 33 YGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTEDQWNDAEEAKATANSR 92
Query: 90 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 149
L +I S+ + ++ ++ + +AK + L +EG+ VK ++ ML Q+
Sbjct: 93 AL--------SLIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 144
Query: 150 ELFRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAK 209
E M++ +IEE + + S L K Y V K+LR LPSR+ K TA+ +
Sbjct: 145 ENLTMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 204
Query: 210 DLNTLSVEDLVSSLKGHEMSLNEHE----------TSKKSKSI--------------ALP 245
D N++ E++V + +E+ + + S K K + +
Sbjct: 205 DTNSIDFEEVVGMFQAYELEITSGKGGYGHIKAECPSLKRKDLKCSECKGLGHIKFDCVG 264
Query: 246 SKGKTSKSSKAYKASESEEESPDGDSDE 273
SK K +S +SESE +S DGDS++
Sbjct: 265 SKSKPDRSC----SSESESDSNDGDSED 288
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 84.3 bits (207), Expect = 1e-16
Identities = 60/288 (20%), Positives = 126/288 (42%), Gaps = 54/288 (18%)
Query: 58 ELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDK 117
++W+ ++ G DD++ K+ R ++ S+P + +++
Sbjct: 9 KVWETIDPGSDDME----------------------KNDMARALLFQSVPESTILQVGKH 46
Query: 118 STAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEMYSRFQTLVSGLQIL 177
T+KAM+ ++ G+++VKEAK L+ +++ MKD+ +I+E R + + + L
Sbjct: 47 KTSKAMWEAIKTRNLGAERVKEAKLQTLMAEFDRLNMKDNETIDEFVGRISEISTKSESL 106
Query: 178 KKSYVASDHVSKILRSLP-SRWRPKVTAIEEAKDLNTLSVEDLVSSLKGHEMSLNEHETS 236
+ S V K L+SLP ++ + A+E+ DLNT ED+V +K +E + + + S
Sbjct: 107 GEEIEESKIVKKFLKSLPRKKYIHIIAALEQILDLNTTGFEDIVGRMKTYEDRVCDEDDS 166
Query: 237 KKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKF 296
+ + + + ++S ++ + S G
Sbjct: 167 PEEQGKLMYANSESSYDTRGGRGRGRGRSSGRG--------------------------- 199
Query: 297 LSKRGSYKNSKKEDQKG-CFNCKKPGHFIADCPDLQKEKYKGKSKKSS 343
RG Y +++ K C+ C K GH+ ++C D + K + ++ +
Sbjct: 200 ---RGGYGYQQRDKSKVICYRCDKTGHYASECLDRLLKLIKAQEQQQN 244
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 80.9 bits (198), Expect = 1e-15
Identities = 64/298 (21%), Positives = 135/298 (44%), Gaps = 16/298 (5%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHH 96
D E++ +W M + +LW ++E+GV + E R T ++ +
Sbjct: 13 DGEKYDFWSIKMATIFR--TRKLWSVVEEGVPVEPVQAEETPETARAKTLREEAVTNDTM 70
Query: 97 KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKD 156
++ I+ ++ + +++ S++K + L ++GS +V+ K L +YE +M D
Sbjct: 71 ALQ-ILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENLKMYD 129
Query: 157 DGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSV 216
+ +I+ + L L + + + KIL SLP+++ V+ +E+ +DL+ L++
Sbjct: 130 NDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLDALTM 189
Query: 217 EDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQS 276
+L+ LK E + E S K + + SKG+ ES + + ++ +Q
Sbjct: 190 SELLGILKAQEARVTAREESTKEGAFYVRSKGR-----------ESGFKQDNTNNRVNQD 238
Query: 277 VKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEK 334
K ++ ++ + K + +K+ + K C+ C K GH+ +C KE+
Sbjct: 239 KKWCGFHKSSKH-TEEECREKPKNDDHGKNKRSNIK-CYKCGKIGHYANECRSKNKER 294
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 80.1 bits (196), Expect = 2e-15
Identities = 70/327 (21%), Positives = 147/327 (44%), Gaps = 20/327 (6%)
Query: 44 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 103
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEM 163
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 223
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 224 KGHEMSLNEHETSKKSKSI---ALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMA 280
+ +E E KK + I L + ++ ++Y+ + G
Sbjct: 195 QAYE------EKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 281 MLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSK 340
+ + K G K+ + C+NC K GH+ ++C +K+K +
Sbjct: 249 PHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFK---E 304
Query: 341 KSSFSSSKFRKQIKKSLMATWEDLDSE 367
K+++ K +++ LMA+++ + E
Sbjct: 305 KANYVEEKIQEE-DMLLMASYKKDEQE 330
>At1g36110 hypothetical protein
Length = 745
Score = 80.1 bits (196), Expect = 2e-15
Identities = 67/283 (23%), Positives = 118/283 (41%), Gaps = 66/283 (23%)
Query: 58 ELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDK 117
++W+ +E G+DD K+ RG+I SIP + +++
Sbjct: 42 KMWETIEPGIDD----------------------GYKNTMARGLIFQSIPESLTLQVGTL 79
Query: 118 STAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEMYSRFQTLVSGLQIL 177
+TAK ++ S+ + G+ +VKEA+ L+ ++E +MK+ I+ R L + L
Sbjct: 80 ATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMKMKESEKIDVFAGRLAELATRSDAL 139
Query: 178 KKSYVASDHVSKILRSLPSR-WRPKVTAIEEAKDLNTLSVEDLVSSLKGHEMSLNEHETS 236
+ S V K L +LP R + + ++E+ DLN S ED+V +K +
Sbjct: 140 GSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLNNTSFEDIVGRIKVY---------- 189
Query: 237 KKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLE---YLARKQ 293
EE DG+ ED K+ + + Y +R +
Sbjct: 190 --------------------------EERVWDGEEQEDDQGKLMYANTDTQDSWYASRGR 223
Query: 294 KK---FLSK-RGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQK 332
+ F + RG + S+ + C+ C K GH+ ++CPD K
Sbjct: 224 GQGGGFNGRGRGRGRGSRDTSKVTCYRCDKLGHYASNCPDSVK 266
>At3g61330 copia-type polyprotein
Length = 1352
Score = 79.3 bits (194), Expect = 4e-15
Identities = 74/343 (21%), Positives = 152/343 (43%), Gaps = 21/343 (6%)
Query: 44 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 103
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEM 163
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 223
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 224 KGHEMSLNEHETSKKSKSIA---LPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMA 280
+ +E E KK + IA L + ++ ++Y+ + G
Sbjct: 195 QAYE------EKKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 281 MLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSK 340
+ + K G K+ + C+NC K GH+ ++C +K++ +
Sbjct: 249 PHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFE---E 304
Query: 341 KSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAM 383
K+ + K +++ LMA+++ D + + K D A M
Sbjct: 305 KAHYVEEKIQEE-DMLLMASYKK-DEQKENHKWYLDSGASNHM 345
>At1g58140 hypothetical protein
Length = 1320
Score = 78.6 bits (192), Expect = 7e-15
Identities = 69/327 (21%), Positives = 147/327 (44%), Gaps = 20/327 (6%)
Query: 44 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 103
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEM 163
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 223
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 224 KGHEMSLNEHETSKKSKSI---ALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMA 280
+ +E E KK + I L + ++ ++Y+ + G
Sbjct: 195 QAYE------EKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 281 MLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSK 340
+ + K G K+ + C+NC K GH+ ++C +K++ +
Sbjct: 249 PHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFE---E 304
Query: 341 KSSFSSSKFRKQIKKSLMATWEDLDSE 367
K+++ K +++ LMA+++ + E
Sbjct: 305 KANYVEEKIQEE-DMLLMASYKKDEQE 330
>At1g48710 hypothetical protein
Length = 1352
Score = 78.6 bits (192), Expect = 7e-15
Identities = 69/327 (21%), Positives = 147/327 (44%), Gaps = 20/327 (6%)
Query: 44 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 103
W M + + D +W+I+E G ++ + EG+ + + K+ K +I
Sbjct: 21 WSLRMKAILGAHD--VWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEM 163
+ + K+ + ++AK + L +++G+ +VK+ + L ++E +MK+ + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 223
+SR T+ + L+ + + K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 224 KGHEMSLNEHETSKKSKSI---ALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMA 280
+ +E E KK + I L + ++ ++Y+ + G
Sbjct: 195 QAYE------EKKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 281 MLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSK 340
+ + K G K+ + C+NC K GH+ ++C +K++ +
Sbjct: 249 PHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFE---E 304
Query: 341 KSSFSSSKFRKQIKKSLMATWEDLDSE 367
K+++ K +++ LMA+++ + E
Sbjct: 305 KANYVEEKIQEE-DMLLMASYKKDEQE 330
>At4g04560 putative transposon protein
Length = 590
Score = 70.5 bits (171), Expect = 2e-12
Identities = 45/199 (22%), Positives = 96/199 (47%), Gaps = 3/199 (1%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDG-VDDLDLDEEGAAIDR-RIHTPAQKKLYKK 94
D E + +WK + I +D + W +EDG + D + + + R A +K
Sbjct: 349 DAEHYGYWKVLIKRSIQSIDMDAWFAVEDGWMPPTTKDAKRDIVSKSRTEWIADEKTAAN 408
Query: 95 HH-KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFR 153
H+ + +I S+ R ++ ++ +AK ++ L +FE + VK + ML ++E
Sbjct: 409 HNSQALSVIFGSLLRNKFTQVQGCLSAKEVWEILQVSFECTNNVKRTRLDMLASEFENLT 468
Query: 154 MKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNT 213
M+ + S+++ + ++ +L K+Y V K LRSLP +++ +AI+ + + +
Sbjct: 469 MEAEESVDDFNGKLSSITQEAVVLGKTYKDKKMVKKFLRSLPDKFQSHKSAIDVSLNSDQ 528
Query: 214 LSVEDLVSSLKGHEMSLNE 232
L + +V ++ ++ E
Sbjct: 529 LKFDQVVGMMQAYDTDKEE 547
Score = 38.5 bits (88), Expect = 0.008
Identities = 54/264 (20%), Positives = 105/264 (39%), Gaps = 43/264 (16%)
Query: 32 PKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTP--AQK 89
P FN E + +W+ M + ++LW+I+++GV + + H+P Q+
Sbjct: 8 PIFN--KENYGFWRIKMKTIFQ--TKKLWEIVDEGVPKPPAEGD--------HSPEAVQQ 55
Query: 90 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 149
K + ++ + I +T +SD + AS AL +Y
Sbjct: 56 KTRCEAASLKDLTALQILQTA---VSDSIFPRIAPAS--------------SALGKPWEY 98
Query: 150 ELFRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAK 209
E +MK+ +I ++ + + L++ + V KIL SLP R+ V +++ K
Sbjct: 99 ENLKMKESDNINTFMTKLIEMGNQLRVHGEEKSDYQIVQKILISLPKRFDIIVAMMKQTK 158
Query: 210 DLNTLSVEDL--VSSLKGHEMS----------LNEHETSKKSKSIALPSKGKTSKSSKAY 257
DL +LS V K H S L++ + + + G +K+ +
Sbjct: 159 DLTSLSAGKWCDVCERKNHNESDCWMKKNKGVLSQQVGNNERRCFVCNKPGHLAKNCRLR 218
Query: 258 KASESEEESPDGDSDEDQSVKMAM 281
+ + + + DED + A+
Sbjct: 219 RTERVDLSLEETNDDEDHMLFSAV 242
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 68.6 bits (166), Expect = 7e-12
Identities = 78/341 (22%), Positives = 138/341 (39%), Gaps = 54/341 (15%)
Query: 40 EFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEG--AAIDRRIHTPAQKKLYKKHHK 97
++ WK + + I EL +LE +D ++EE A D + K L +K K
Sbjct: 16 DYVLWKEKLLAHI-----ELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKEKRGK 70
Query: 98 IRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDD 157
R ++ S+ K+ + TA M L F ++ Y +M D
Sbjct: 71 ARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGY---KMSDS 127
Query: 158 GSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVE 217
+IEE + F L+S L+ +K S D +L SLP ++ ++ K TL+++
Sbjct: 128 MTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGK--TTLALD 185
Query: 218 DLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSV 277
++ ++ +SK + L + GK K+S S++ G S+
Sbjct: 186 EITGAI--------------RSKVLELGASGKMLKNS-----SDALFVQDRGRSE----- 221
Query: 278 KMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKG 337
K+ K + S SK ++K C+ C K GHF C +++ KG
Sbjct: 222 --------------KRDKSSERNKSQSRSKSREKKVCWVCGKEGHFKKQCYVWKEKNKKG 267
Query: 338 KSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDD 378
+ + SS+ + + +A E ES +D +E D++
Sbjct: 268 NNSEKGESSNVIGQAADAAALAVRE----ESNADNQEVDNE 304
>At4g10460 putative retrotransposon
Length = 1230
Score = 67.0 bits (162), Expect = 2e-11
Identities = 71/316 (22%), Positives = 131/316 (40%), Gaps = 55/316 (17%)
Query: 33 KFNGDPEEFSWWKTNMYSF--IMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKK 90
KF+G + ++ WK + + ++GL L + + D L+ +EEG ++ +
Sbjct: 10 KFDGHGD-YTLWKEKLMAHMDLLGLTVALRET-QSVSDPLESEEEGKESEKG---DKEAL 64
Query: 91 LYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKAL----MLV 146
+ +K K R IV S+ K + TA +M +L K+ +KAL L
Sbjct: 65 MEEKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEAL-------DKLYMSKALPNRIYLK 117
Query: 147 HQYELFRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIE 206
+ ++M+++ S+E F L++ L+ D +L SLP ++ ++
Sbjct: 118 QKLYSYKMQENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLK 177
Query: 207 EAKDLNTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
TLSV+++V+++ E+ L ++ S + ++ L K K SE +E+
Sbjct: 178 YGSGRTTLSVDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRG----MSEQKEKG 233
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIAD 326
G S S+ + KGC+ C + GHF
Sbjct: 234 NKGRS---------------------------------RSRSKGWKGCWICGEEGHFKTS 260
Query: 327 CPDLQKEKYKGKSKKS 342
CP+ K++ KGK + S
Sbjct: 261 CPNKGKQQNKGKDQAS 276
>At3g25450 hypothetical protein
Length = 1343
Score = 67.0 bits (162), Expect = 2e-11
Identities = 53/219 (24%), Positives = 105/219 (47%), Gaps = 19/219 (8%)
Query: 93 KKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELF 152
+K+ R ++ SIP + +++ + T+ A++ ++ + G+++VKEA+ L+ +++
Sbjct: 58 EKNDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMAEFDKL 117
Query: 153 RMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIEEAKDL 211
+MKD +I++ R + + L + S V K L+SLP ++ V A+E+ DL
Sbjct: 118 KMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALEQVLDL 177
Query: 212 NTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
T + ED+ +K +E + + + S + +GK +E EEE D
Sbjct: 178 KTTTFEDIAGRIKTYEDRVWDDDDSHE-------DQGKL--------MTEVEEEVVDDLE 222
Query: 272 DEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKED 310
+E++ V + K + R K R + K+ED
Sbjct: 223 EEEEEVINKEIKAKSHVIDRLLKLI---RLQEQKEKEED 258
>At3g31380 hypothetical protein
Length = 262
Score = 65.1 bits (157), Expect = 8e-11
Identities = 43/156 (27%), Positives = 76/156 (48%), Gaps = 1/156 (0%)
Query: 99 RGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDG 158
RG++V SIP +++ + TAK ++ S+ G+++VKEA+ L+ +E +MK+
Sbjct: 3 RGLLVQSIPEAFTLQVGNLQTAKEVWDSIKTRHVGAERVKEARVQTLMADFEKMKMKEAE 62
Query: 159 SIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIEEAKDLNTLSVE 217
I++ R L + L + V K L SLP ++ + ++E+ DLN + E
Sbjct: 63 KIDDFAGRLSELSTKSAPLGVNIEVPKLVKKFLNSLPRKKYIHIIASLEQVLDLNNTTFE 122
Query: 218 DLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKS 253
D+V +K +E + + E L SKS
Sbjct: 123 DIVGCMKVNEERVYDPEEETNEDQNKLMYTSSDSKS 158
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 60.1 bits (144), Expect = 3e-09
Identities = 64/310 (20%), Positives = 125/310 (39%), Gaps = 55/310 (17%)
Query: 33 KFNGDPEEFSWWKTNMYSF--IMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKK 90
KF+G + ++ WK + + I+GL+ L + G LDE + ++ +
Sbjct: 10 KFDGRGD-YTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEK--FEA 66
Query: 91 LYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKAL--MLVHQ 148
L +K K R IV S+ K+ +STA AM +L K+ +KAL + +
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLAL-------DKLYMSKALPNRIYPK 119
Query: 149 YEL--FRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIE 206
+L F+M ++ S+E F +++ L+ + D +L +LP + ++
Sbjct: 120 QKLYSFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLK 179
Query: 207 EAKDLNTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
+ + L+++++ +++ E+ L + S K ++ L K K K +
Sbjct: 180 YSSGKSILTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENKGKGEQKG------ 233
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIAD 326
+G K K + + GC+ C + GHF +
Sbjct: 234 ---------------------------------KGKGKKGKSKKKPGCWTCGEEGHFRSS 260
Query: 327 CPDLQKEKYK 336
CP+ K ++K
Sbjct: 261 CPNQNKPQFK 270
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 57.0 bits (136), Expect = 2e-08
Identities = 48/199 (24%), Positives = 92/199 (46%), Gaps = 31/199 (15%)
Query: 112 MKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEMYSRFQTLV 171
+++ T+KA++ + + G+++VKEAK L+ +++ +MKD+ +I+E R +
Sbjct: 35 LQVGKLKTSKAVWDKIQSRNLGAERVKEAKLKTLMAEFDKLKMKDNETIDECAGRLSEIS 94
Query: 172 SGLQILKKSYVASDHVSKILRSLPS-RWRPKVTAIEEAKDLNTLSVEDLVSSLKGHEMSL 230
+ L + + V K L+SLP+ ++ V A+E+ DL + +D+V +K +E +
Sbjct: 95 TKSTSLGEDIEETKVVKKFLKSLPTKKYIHIVAALEQVLDLKNTTFKDIVGRIKTYEDKI 154
Query: 231 --------NEHETSKK-------------SKSIALP---------SKGKTSKSSKAYKAS 260
E E +K S+ I L + + K K
Sbjct: 155 WVLITCLKKEAEEEEKSVVGVEAEEELVISREITLRLLVIAVINYASNCPDRLLKLIKLQ 214
Query: 261 ESEEESPDGDSDEDQSVKM 279
E ++E+ D D DE +S+ M
Sbjct: 215 ERQQEAEDDDDDEVESLMM 233
>At3g28770 hypothetical protein
Length = 2081
Score = 54.7 bits (130), Expect = 1e-07
Identities = 62/238 (26%), Positives = 111/238 (46%), Gaps = 35/238 (14%)
Query: 227 EMSLNEHETSK-KSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNK 285
E +E SK + K K KT + +K K +++ + DS+E +S K S
Sbjct: 998 EKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRD 1057
Query: 286 LEYLARKQKKFLSKRGS--YKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSKKSS 343
L+ +K+++ K+ S +K+ KKED+K D ++KE+ K + KK
Sbjct: 1058 LK-AKKKEEETKEKKESENHKSKKKEDKKE----------HEDNKSMKKEEDKKEKKKHE 1106
Query: 344 FSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMRLVATVSSEAVSEAESDSED 403
S S+ +++ KK + E L+ ++ + K+E ++ K + + + + ESD ++
Sbjct: 1107 ESKSRKKEEDKKDM----EKLEDQNSNKKKEDKNEKKKSQHV-------KLVKKESDKKE 1155
Query: 404 ENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKTTLLELKASEEE 461
+ E K +E+ S + + NE+ D KEK QQK E+K SEE+
Sbjct: 1156 KKENEEKSETKEIESS--------KSQKNEV-DKKEK-KSSKDQQKKKEKEMKESEEK 1203
Score = 42.7 bits (99), Expect = 4e-04
Identities = 57/234 (24%), Positives = 89/234 (37%), Gaps = 52/234 (22%)
Query: 232 EHETSKKSKSIALPSKGKTSKSSKAYKASESEE--ESPDGDSDEDQSVKMAMLSNKLEYL 289
E E+SK K+ + K+SK + K E +E E ++ED+ + ++ NK +
Sbjct: 1167 EIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKE 1226
Query: 290 ARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSKKSSFSSSKF 349
+K+ KN K+D+K K G K+S S SK
Sbjct: 1227 TKKE----------KNKPKDDKK------------------NTTKQSGGKKESMESESKE 1258
Query: 350 RKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMRLVATVSSEAVSEAESDSEDENEVYS 409
+ +KS T D D +AD A S + S+A+SD E +NE+
Sbjct: 1259 AENQQKSQATTQADSDESKNEILMQADSQA----------DSHSDSQADSD-ESKNEILM 1307
Query: 410 KIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKTTLLELKASEEELK 463
+ Q R NE K+ V K+QK T E +++ K
Sbjct: 1308 QADSQATT-----------QRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKK 1350
Score = 40.0 bits (92), Expect = 0.003
Identities = 52/256 (20%), Positives = 104/256 (40%), Gaps = 18/256 (7%)
Query: 224 KGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLS 283
K E E +K + + + KGK K K +ES + + + + K ++
Sbjct: 917 KKDEKKEGNKEENKDTINTSSKQKGKDKKKKK--------KESKNSNMKKKEEDKKEYVN 968
Query: 284 NKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSKKSS 343
N+L +KQ+ + +NSK +++ KK D +EK + + KKS
Sbjct: 969 NEL----KKQEDNKKETTKSENSKLKEENKDNKEKKESE---DSASKNREKKEYEEKKSK 1021
Query: 344 FSSSKFRKQIKKSLMATWEDLDSESGSDKEEADD--DAKAAMRLVATVSSEAVSEAESDS 401
+ + +K+ KKS E+ DSE K+E ++ D KA + T + +S
Sbjct: 1022 -TKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKK 1080
Query: 402 EDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKTTLLELKASEEE 461
+++ + + + + KE E ++ + + K+ L Q E K +++
Sbjct: 1081 KEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKK 1140
Query: 462 LKGFNLISTTYEDRLK 477
+ L+ + + K
Sbjct: 1141 SQHVKLVKKESDKKEK 1156
Score = 40.0 bits (92), Expect = 0.003
Identities = 50/260 (19%), Positives = 107/260 (40%), Gaps = 37/260 (14%)
Query: 207 EAKDLNTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E K+L+ + ++ + K E++ N+ + +K+ + + G++ K+ E++E+
Sbjct: 553 EDKNLDNIGADEQKKNDKSVEVTTNDGDHTKEKREETQGNNGESVKNENL----ENKEDK 608
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIAD 326
+ DE K +N L K+++ + NSK D KG
Sbjct: 609 KELKDDESVGAK----TNNETSLEEKREQTQKGHDNSINSKIVDNKG------------G 652
Query: 327 CPDLQKEKYKGKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMRLV 386
D KEK ++ ++ + ++ K + D SE G + +E + D+ +L
Sbjct: 653 NADSNKEKEVHVGDSTNDNNMESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSMEDKKL- 711
Query: 387 ATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMK 446
E+++DS+D+ V K ++ E + ++ + K K + +
Sbjct: 712 ------ENKESQTDSKDDKSVDDKQEEAQIYGG--------ESKDDKSVEAKGKKKESKE 757
Query: 447 QQKTTLLE--LKASEEELKG 464
+KT E ++ EE ++G
Sbjct: 758 NKKTKTNENRVRNKEENVQG 777
Score = 39.3 bits (90), Expect = 0.005
Identities = 57/237 (24%), Positives = 103/237 (43%), Gaps = 32/237 (13%)
Query: 221 SSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMA 280
SS KG E N + S + K + SK K+ + E + G+S +D+SV+
Sbjct: 691 SSEKGEEGKENNKD-SMEDKKLENKESQTDSKDDKSVDDKQEEAQIYGGESKDDKSVEAK 749
Query: 281 MLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSK 340
K E +++ KK + +N K+E+ +G N K+ ++ KG+ K
Sbjct: 750 --GKKKE--SKENKKTKTNENRVRN-KEENVQG--NKKE-----------SEKVEKGEKK 791
Query: 341 KSSFSSSKFRKQIKK-SLMATWEDLDSESGSDKEEADDDAKAAMRLVATVSSEAVSEAES 399
+S + S K KK S ++ SG D +E +++K S EA + E+
Sbjct: 792 ESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQ------SVEAKEKNEN 845
Query: 400 DSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKTTLLELK 456
D N V +K ++L D E + N+ +K+K ++ + K++ E++
Sbjct: 846 GGVDTN-VGNKEDSKDLKDDRS-----VEVKANKEESMKKKREEVQRNDKSSTKEVR 896
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.126 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,783,818
Number of Sequences: 26719
Number of extensions: 486819
Number of successful extensions: 3454
Number of sequences better than 10.0: 340
Number of HSP's better than 10.0 without gapping: 96
Number of HSP's successfully gapped in prelim test: 250
Number of HSP's that attempted gapping in prelim test: 2904
Number of HSP's gapped (non-prelim): 642
length of query: 480
length of database: 11,318,596
effective HSP length: 103
effective length of query: 377
effective length of database: 8,566,539
effective search space: 3229585203
effective search space used: 3229585203
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146683.12


