

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146681.9 + phase: 2 /pseudo
(249 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
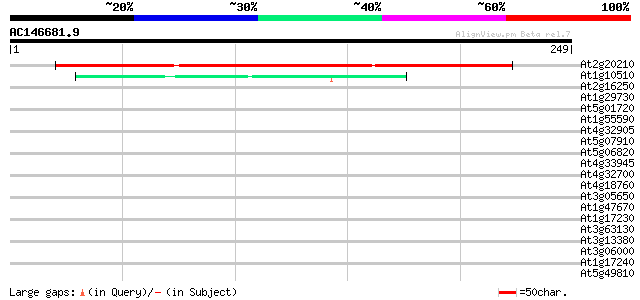
Score E
Sequences producing significant alignments: (bits) Value
At2g20210 hypothetical protein 177 5e-45
At1g10510 unknown protein 41 7e-04
At2g16250 putative LRR receptor protein kinase 39 0.003
At1g29730 Putative receptor-like serine/threonine kinase 33 0.14
At5g01720 putative F-box protein family, AtFBL3 33 0.19
At1g55590 unknown protein 32 0.24
At4g32905 unknown protein 31 0.54
At5g07910 putative protein 31 0.71
At5g06820 SRF2 (LRR receptor protein kinase like) 30 0.93
At4g33945 unknown protein 30 1.6
At4g32700 putative protein 30 1.6
At4g18760 predicted GPI-anchored protein 30 1.6
At3g05650 putative disease resistance protein 30 1.6
At1g47670 unknown protein 30 1.6
At1g17230 putative leucine-rich receptor protein kinase 30 1.6
At3g63130 RAN GTPase like protein 29 2.1
At3g13380 brassinosteroid receptor kinase, putative 29 2.1
At3g06000 hypothetical protein 29 2.1
At1g17240 putative receptor protein kinase, predicted GPI-anchor... 29 2.7
At5g49810 methionine S-methyltransferase (gb|AAD49574.1) 28 4.6
>At2g20210 hypothetical protein
Length = 271
Score = 177 bits (449), Expect = 5e-45
Identities = 104/203 (51%), Positives = 134/203 (65%), Gaps = 3/203 (1%)
Query: 21 FLI*FLSHRGNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMC 80
F+ +S RGN L + DAENL +A +MP LE LD+S NPIED G+R LI YF T
Sbjct: 22 FVSFLMSVRGNELDRYDAENLAHALLHMPGLESLDLSGNPIEDSGIRSLISYF--TKNPD 79
Query: 81 SRLACLKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTPIEV 140
SRLA L LE C+LSC V LD+L +L LK LS+A+N LGS+VA A+ + IE
Sbjct: 80 SRLADLNLENCELSCCGVIEFLDTLSMLEKPLKFLSVADNALGSEVAEAVVNSFTISIES 139
Query: 141 LDASGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVN 200
L+ GI L P GFL L L + L+ INISKNRGG++TARFLSKL+P AP+L+ ++
Sbjct: 140 LNIMGIGLGPLGFLALGRKLE-KVSKKLLSINISKNRGGLETARFLSKLIPLAPKLISID 198
Query: 201 ASSNCMPIESLSIISSALKFAKG 223
AS N MP E+L ++ +L+ AKG
Sbjct: 199 ASYNLMPPEALLMLCDSLRTAKG 221
>At1g10510 unknown protein
Length = 605
Score = 40.8 bits (94), Expect = 7e-04
Identities = 35/150 (23%), Positives = 60/150 (39%), Gaps = 8/150 (5%)
Query: 30 GNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLE 89
GNN+ + + A + L++ NPI +G + L + + + LKL
Sbjct: 432 GNNIHAEGVNAIAQALKDNAIITTLEVGYNPIGPDGAKAL----SEILKFHGNVKTLKLG 487
Query: 90 ACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTPIEVL---DASGI 146
C ++ H+ D L N T+ L + N L + A L R L E L D
Sbjct: 488 WCQIAAKGAEHVADMLR-YNNTISVLDLRANGLRDEGASCLARSLKVVNEALTSVDLGFN 546
Query: 147 DLLPSGFLELQNMLTIEEELSLVIINISKN 176
++ G + L E++++ IN+ N
Sbjct: 547 EIRDDGAFAIAQALKANEDVTVTSINLGNN 576
Score = 30.8 bits (68), Expect = 0.71
Identities = 39/173 (22%), Positives = 67/173 (38%), Gaps = 33/173 (19%)
Query: 51 LEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVNHLLDSLPVLNG 110
L+ L++S NPI DEG + L T S + L+L + D+ + + + L N
Sbjct: 228 LKILNLSGNPIGDEGAKTL----CATLMENSSIEILQLNSTDIGDEGAKEIAELLK-RNS 282
Query: 111 TLKSLSIAENCLGSKVAGALGRFLSTPIEVLDASGIDLLPSGFLELQNMLTIEEELSLVI 170
TL+ + + N ++D SG L LE +
Sbjct: 283 TLRIIELNNN-------------------MIDYSGFTSLAGALLENNTIRN--------- 314
Query: 171 INISKNRGGIQTARFLSKLLPQAPRLVDVNASSNCMPIESLSIISSALKFAKG 223
++++ N GG A L+K L L +++ N + E + + L KG
Sbjct: 315 LHLNGNYGGALGANALAKGLEGNKSLRELHLHGNSIGDEGTRALMAGLSSHKG 367
Score = 30.8 bits (68), Expect = 0.71
Identities = 31/126 (24%), Positives = 55/126 (43%), Gaps = 5/126 (3%)
Query: 109 NGTLKSLSIAENCLGSKVAGALGRFLST---PIEVLDASGIDLLPSGFLELQNMLTIEEE 165
N +L+ L + N +G + AL LS+ + +LD + G + I+
Sbjct: 337 NKSLRELHLHGNSIGDEGTRALMAGLSSHKGKVALLDLGNNSISAKGAFYVAEY--IKRS 394
Query: 166 LSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASSNCMPIESLSIISSALKFAKGIY 225
SLV +N+ N G + A ++ L Q + ++ N + E ++ I+ ALK I
Sbjct: 395 KSLVWLNLYMNDIGDEGAEKIADSLKQNRSIATIDLGGNNIHAEGVNAIAQALKDNAIIT 454
Query: 226 GVPVSY 231
+ V Y
Sbjct: 455 TLEVGY 460
>At2g16250 putative LRR receptor protein kinase
Length = 925
Score = 38.5 bits (88), Expect = 0.003
Identities = 32/123 (26%), Positives = 61/123 (49%), Gaps = 14/123 (11%)
Query: 83 LACLKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLS-TPIEVL 141
++ L LE DLS VN ++ +L++L++++N L S V +LG+ L+ + +++
Sbjct: 125 VSLLALEVLDLSSCSVNGVVPFTLGNLTSLRTLNLSQNSLTSLVPSSLGQLLNLSQLDLS 184
Query: 142 DASGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNA 201
S +LP F L+N+LT++ + + I G + +L+ +N
Sbjct: 185 RNSFTGVLPQSFSSLKNLLTLDVSSNYLTGPIPPGLGALS-------------KLIHLNF 231
Query: 202 SSN 204
SSN
Sbjct: 232 SSN 234
>At1g29730 Putative receptor-like serine/threonine kinase
Length = 966
Score = 33.1 bits (74), Expect = 0.14
Identities = 42/165 (25%), Positives = 79/165 (47%), Gaps = 29/165 (17%)
Query: 88 LEACDLSCDVVNHLLDSLPVLNGTL---KSLSIAENCLGSKVAGALGRFLSTPIEVLDAS 144
LE+ DL N+L S+P+ +L KS+S+ N L + LG+F++ + VL+A+
Sbjct: 124 LESIDL---YNNYLYGSIPMEWASLPYLKSISVCANRLSGDIPKGLGKFINLTLLVLEAN 180
Query: 145 GID-LLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASS 203
+P EL N++ ++ + +S N Q L K L + +L +++ S
Sbjct: 181 QFSGTIPK---ELGNLVNLQG------LGLSSN----QLVGGLPKTLAKLTKLTNLHLSD 227
Query: 204 NCMP------IESLSIISSALKFAKGIYG-VP--VSYIGDLLDLR 239
N + I L + +A G+ G +P + ++ +L+D+R
Sbjct: 228 NRLNGSIPEFIGKLPKLQRLELYASGLRGPIPDSIFHLENLIDVR 272
>At5g01720 putative F-box protein family, AtFBL3
Length = 665
Score = 32.7 bits (73), Expect = 0.19
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 8/41 (19%)
Query: 51 LEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEAC 91
LE+LD++DN I+DEGL+ + C L+ LKL C
Sbjct: 409 LEELDLTDNEIDDEGLKSI--------SSCLSLSSLKLGIC 441
>At1g55590 unknown protein
Length = 607
Score = 32.3 bits (72), Expect = 0.24
Identities = 34/138 (24%), Positives = 59/138 (42%), Gaps = 8/138 (5%)
Query: 48 MPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVNHLLDSLPV 107
+P+L LD+S +DE L +++ G S + L CL+L A + + HL + L +
Sbjct: 41 LPSLTSLDLSVFSPDDETLNHVLRGCIGLSSL--TLNCLRLNAASVRGVLGPHLRE-LHL 97
Query: 108 LNGTLKSLSIAENCLGSKVAGALGRFLSTPIEVLDASGIDLLPSGFLELQNMLTIEEELS 167
L +L S ++ + + +E+ D D+ S ++ N E L
Sbjct: 98 LRCSLLSSTVL-----TYIGTLCPNLRVLTLEMADLDSPDVFQSNLTQMLNGCPYLESLQ 152
Query: 168 LVIINISKNRGGIQTARF 185
L I I + Q+ RF
Sbjct: 153 LNIRGILVDATAFQSVRF 170
>At4g32905 unknown protein
Length = 716
Score = 31.2 bits (69), Expect = 0.54
Identities = 22/78 (28%), Positives = 34/78 (43%), Gaps = 6/78 (7%)
Query: 64 EGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLG 123
EGLR ++ G ++ CL+ C+ V+HLL P G S+A+ C+
Sbjct: 336 EGLRNMLKIMIGNAD------CLRAATCNWMELFVSHLLYLRPFTKGLDGMHSLAQKCVQ 389
Query: 124 SKVAGALGRFLSTPIEVL 141
SK + L I +L
Sbjct: 390 SKPVNTSHKLLRLLIGIL 407
>At5g07910 putative protein
Length = 268
Score = 30.8 bits (68), Expect = 0.71
Identities = 30/107 (28%), Positives = 51/107 (47%), Gaps = 20/107 (18%)
Query: 112 LKSLSIAENCLGSKVAGALGRFLSTPIEVLDASGIDLLPSGFLEL---------QNML-- 160
++ L IA+N L ++ G LG+ S + +LD + I LP +L +NML
Sbjct: 70 MQRLLIADN-LVERLPGNLGKLQSLKVLMLDGNRISCLPDELGQLVRLEQLSISRNMLIY 128
Query: 161 ---TIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASSN 204
TI +L+++N+S NR + L + + L +V A+ N
Sbjct: 129 LPDTIGSLRNLLLLNVSNNR-----LKSLPESVGSCASLEEVQANDN 170
>At5g06820 SRF2 (LRR receptor protein kinase like)
Length = 735
Score = 30.4 bits (67), Expect = 0.93
Identities = 37/130 (28%), Positives = 60/130 (45%), Gaps = 29/130 (22%)
Query: 40 NLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVN 99
+LG ++ NL+ LD+S N +E E IP+ + +A N
Sbjct: 86 SLGNQLQHLHNLKILDVSFNNLEGE-----IPFGLPPNATHINMA-------------YN 127
Query: 100 HLLDSLPV---LNGTLKSLSIAENCLGSKVAGALGR-FLSTPIEVLDASGIDL---LPSG 152
+L S+P L +L+SL+++ N L +G LG F I+ +D S +L LPS
Sbjct: 128 NLTQSIPFSLPLMTSLQSLNLSHNSL----SGPLGNVFSGLQIKEMDLSFNNLTGDLPSS 183
Query: 153 FLELQNMLTI 162
F L N+ ++
Sbjct: 184 FGTLMNLTSL 193
>At4g33945 unknown protein
Length = 464
Score = 29.6 bits (65), Expect = 1.6
Identities = 33/143 (23%), Positives = 57/143 (39%), Gaps = 33/143 (23%)
Query: 58 DNPIEDEGLRYLIPYFAGTSEMCSR------LACLKLEACDLSCDVVNHL---------- 101
D P+ DE + F +E+CS K A DL+C + + +
Sbjct: 78 DGPLRDEVFDEISSLFKNLNELCSSQESGNAAIATKHGAVDLTCSICSKIKISTRSNRVL 137
Query: 102 ---LDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLSTPIEVLD---------ASGIDLL 149
+L VL ++S NC G + L S+ ++LD A+G +++
Sbjct: 138 VPCFKALAVLIRDIQSTERFRNCTGPNIVVDLLNDSSSDSDLLDAGFAVVAAAATGNEVV 197
Query: 150 PSGFLELQNMLTIEEELSLVIIN 172
F+EL+ +EL L ++N
Sbjct: 198 KQLFMELK-----IDELILQVLN 215
>At4g32700 putative protein
Length = 1548
Score = 29.6 bits (65), Expect = 1.6
Identities = 28/89 (31%), Positives = 41/89 (45%), Gaps = 5/89 (5%)
Query: 122 LGSKVAGALGRFLSTPIEVLDASGIDLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQ 181
+GS + + T + D G D P +EL N + +E + V+I S +G
Sbjct: 189 VGSTIYNKKMEVVRTIPKAADMGGKD--PDHIVELCN--EVVQEGNSVLIFCSSRKGCES 244
Query: 182 TARFLSKLLPQAPRLVDVNASSNCMPIES 210
TAR +SKL+ P VD +S M I S
Sbjct: 245 TARHISKLIKNVPVNVD-GENSEFMDIRS 272
>At4g18760 predicted GPI-anchored protein
Length = 431
Score = 29.6 bits (65), Expect = 1.6
Identities = 24/104 (23%), Positives = 44/104 (42%), Gaps = 10/104 (9%)
Query: 42 GYAFAYMPNLEDLDISDNPIEDEGLRYLIPYF----------AGTSEMCSRLACLKLEAC 91
G A + NL DL +S P+ GL ++ A S + L
Sbjct: 158 GVFLARLVNLTDLTVSSVPVSTSGLFVILGNMHEIVSLTISHANLSGNIPKSFHSNLTFI 217
Query: 92 DLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLS 135
DLS +++ + + L LKSL++++N + + ++G +S
Sbjct: 218 DLSDNLLKGSIPTSITLLSNLKSLNLSKNTISGDIPDSIGDLIS 261
>At3g05650 putative disease resistance protein
Length = 868
Score = 29.6 bits (65), Expect = 1.6
Identities = 27/104 (25%), Positives = 44/104 (41%), Gaps = 18/104 (17%)
Query: 77 SEMCSRLACLKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLST 136
S +C+ + + L+ D N+L S+P G LKS N +++ G L R +
Sbjct: 484 SFICALRSLITLDLSD------NNLNGSIPPCMGNLKSTLSFLNLRQNRLGGGLPRSIFK 537
Query: 137 PIEVLDASGIDL---LPSGFLELQNMLTIEEELSLVIINISKNR 177
+ LD L LP F+ L +L ++N+ NR
Sbjct: 538 SLRSLDVGHNQLVGKLPRSFIRLS---------ALEVLNVENNR 572
>At1g47670 unknown protein
Length = 519
Score = 29.6 bits (65), Expect = 1.6
Identities = 22/106 (20%), Positives = 47/106 (43%), Gaps = 9/106 (8%)
Query: 150 PSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASSNCMPIE 209
P+ F+ + + I + V++N+ +G ++ L+KL PQ L + +
Sbjct: 45 PTSFISPRFLSPIGTPMKRVLVNM---KGYLEEVGHLTKLNPQDAWLPITESRNGNAHYA 101
Query: 210 SLSIISSALKFAKGIYGVPVSYIG------DLLDLRCWQVVTSWLL 249
+ +++ + F + V +++G L CWQ+ T W+L
Sbjct: 102 AFHNLNAGVGFQALVLPVAFAFLGWSWGILSLTIAYCWQLYTLWIL 147
>At1g17230 putative leucine-rich receptor protein kinase
Length = 1133
Score = 29.6 bits (65), Expect = 1.6
Identities = 43/170 (25%), Positives = 67/170 (39%), Gaps = 25/170 (14%)
Query: 47 YMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLEACDLSCDVVNHLLDSLP 106
++P L DL + DN +E + IP G S + L + A LS + H
Sbjct: 377 FLPYLVDLQLFDNQLEGK-----IPPLIG---FYSNFSVLDMSANSLSGPIPAHFCRF-- 426
Query: 107 VLNGTLKSLSIAENCLGSKVAGALGRFLS-TPIEVLDASGIDLLPSGFLELQNMLTIEEE 165
TL LS+ N L + L S T + + D LP LQN+ +E
Sbjct: 427 ---QTLILLSLGSNKLSGNIPRDLKTCKSLTKLMLGDNQLTGSLPIELFNLQNLTALELH 483
Query: 166 LSLVIINISKNRGGIQTA--------RFLSKLLPQ---APRLVDVNASSN 204
+ + NIS + G ++ F ++ P+ ++V N SSN
Sbjct: 484 QNWLSGNISADLGKLKNLERLRLANNNFTGEIPPEIGNLTKIVGFNISSN 533
>At3g63130 RAN GTPase like protein
Length = 535
Score = 29.3 bits (64), Expect = 2.1
Identities = 48/178 (26%), Positives = 76/178 (41%), Gaps = 11/178 (6%)
Query: 31 NNLRKDD-AENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLE 89
NN+ D+ A + P+LED S I EG L A E CS L L L
Sbjct: 274 NNMTGDEGATAIAEIVRECPSLEDFRCSSTRIGSEGGVAL----AEALEHCSHLKKLDLR 329
Query: 90 ACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFL--STP-IEVLDASGI 146
+ L +L VL L + ++ L + AL L S P +EVL+ +G
Sbjct: 330 DNMFGVEGGIALAKTLSVLTH-LTEIYMSYLNLEDEGTEALSEALLKSAPSLEVLELAGN 388
Query: 147 DLLPSGFLELQNMLTIEEELSLVIINISKNRGGIQTARFLSKLLPQAPRLVDVNASSN 204
D+ L + ++ SL +N+S+N + ++K + +LV+V+ S+N
Sbjct: 389 DITVKSTGNLAACIASKQ--SLAKLNLSENELKDEGTILIAKAVEGHDQLVEVDLSTN 444
>At3g13380 brassinosteroid receptor kinase, putative
Length = 1164
Score = 29.3 bits (64), Expect = 2.1
Identities = 31/110 (28%), Positives = 44/110 (39%), Gaps = 26/110 (23%)
Query: 83 LACLKLEACDLSCDVVNHLLDSLPVLNGTLKSLSIAENCLG----SKVAGALGRFL---- 134
L C LE DLS + + L G+L+SL++ N L S V L R
Sbjct: 299 LLCRTLEVLDLSGNSLTGQLPQSFTSCGSLQSLNLGNNKLSGDFLSTVVSKLSRITNLYL 358
Query: 135 -------STPIEVLDASGIDLL-----------PSGFLELQNMLTIEEEL 166
S PI + + S + +L PSGF LQ+ +E+ L
Sbjct: 359 PFNNISGSVPISLTNCSNLRVLDLSSNEFTGEVPSGFCSLQSSSVLEKLL 408
>At3g06000 hypothetical protein
Length = 211
Score = 29.3 bits (64), Expect = 2.1
Identities = 25/100 (25%), Positives = 45/100 (45%), Gaps = 14/100 (14%)
Query: 30 GNNLRKDDAENLGYAFAYMPNLEDLDISDNPIEDEGLRYLIPYFAGTSEMCSRLACLKLE 89
GNN+ + A + A +L+ L++S+N ++DE G E+ + +LE
Sbjct: 75 GNNITYEAATAIAVCLAAKRHLKKLNLSENDLKDE----------GCVEIVKSMEDWELE 124
Query: 90 ACDLSCDVVNH----LLDSLPVLNGTLKSLSIAENCLGSK 125
D+S + + L + V G+ K L+I N + K
Sbjct: 125 YVDMSYNDLRREGALRLARVVVKKGSFKMLNIDGNMISLK 164
>At1g17240 putative receptor protein kinase, predicted GPI-anchored
protein
Length = 729
Score = 28.9 bits (63), Expect = 2.7
Identities = 25/63 (39%), Positives = 35/63 (54%), Gaps = 3/63 (4%)
Query: 99 NHLLDSLPVLNGTLKSLSIAENCLGSKVAGALGRFLS--TPIEVLDASGIDLLPSGFLEL 156
N+L S+PV G LK L I E LG+ ++G++ LS T +E LD S +L S L
Sbjct: 586 NNLTGSIPVEVGQLKVLHILE-LLGNNLSGSIPDELSNLTNLERLDLSNNNLSGSIPWSL 644
Query: 157 QNM 159
N+
Sbjct: 645 TNL 647
>At5g49810 methionine S-methyltransferase (gb|AAD49574.1)
Length = 1071
Score = 28.1 bits (61), Expect = 4.6
Identities = 22/53 (41%), Positives = 25/53 (46%), Gaps = 3/53 (5%)
Query: 76 TSEMCSRLACLKLEACDLSCDVVNHL-LDSLPVLNGTLKSLSIAENCLGSKVA 127
+S L K C L D+ +H L SLP NG LK L AEN L S A
Sbjct: 538 SSSFLHLLEVTKEIGCRLFLDISDHFELSSLPASNGVLKYL--AENQLPSHAA 588
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.330 0.146 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,447,771
Number of Sequences: 26719
Number of extensions: 227958
Number of successful extensions: 848
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 830
Number of HSP's gapped (non-prelim): 38
length of query: 249
length of database: 11,318,596
effective HSP length: 97
effective length of query: 152
effective length of database: 8,726,853
effective search space: 1326481656
effective search space used: 1326481656
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146681.9


