

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146664.12 + phase: 0 /pseudo
(410 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
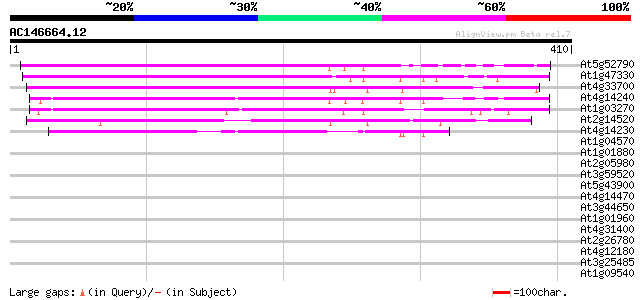
Score E
Sequences producing significant alignments: (bits) Value
At5g52790 unknown protein 293 1e-79
At1g47330 unknown protein 285 3e-77
At4g33700 unknown protein 267 9e-72
At4g14240 unknown protein 238 4e-63
At1g03270 unknown protein 223 2e-58
At2g14520 hypothetical protein 220 1e-57
At4g14230 hypothetical protein 197 9e-51
At1g04570 unknown protein 32 0.83
At1g01880 hypothetical protein 31 1.1
At2g05980 putative non-LTR retroelement reverse transcriptase 31 1.4
At3g59520 unknown protein 30 1.8
At5g43900 myosin heavy chain MYA2 (pir||S51824) 30 2.4
At4g14470 reverse transcriptase like protein 30 3.2
At3g44650 putative protein 30 3.2
At1g01960 hypothetical protein 29 4.1
At4g31400 unknown protein 29 5.4
At2g26780 unknown protein 29 5.4
At4g12180 putative reverse transcriptase 28 7.0
At3g25485 unknown protein 28 7.0
At1g09540 putative transcription factor (MYB61) 28 7.0
>At5g52790 unknown protein
Length = 500
Score = 293 bits (750), Expect = 1e-79
Identities = 183/418 (43%), Positives = 252/418 (59%), Gaps = 59/418 (14%)
Query: 9 DDEHCCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKI 68
+D CC FWV L+C + FA + S L LGL+S S V+LEV++KAG+PH +KNA KI
Sbjct: 4 NDVPCCETMFWVYLLVCVALVVFAGLMSGLTLGLMSLSIVELEVMIKAGEPHDRKNAEKI 63
Query: 69 MSIVKNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSR 128
+ +VKN+HL+LCTLL+ +LA+E + + ++ + P W A+L++ LI E+IPQA+ SR
Sbjct: 64 LPLVKNQHLLLCTLLIGNALAMEALPIFVDSLLPAWGAILISVTLILAFGEIIPQAVCSR 123
Query: 129 YGLRFGATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAG 188
YGL GA +S VR+++++FFP +YP+SKLLD LLGK H+ LLGR ELK+LV +H NEAG
Sbjct: 124 YGLSIGAKLSFLVRLIIIVFFPLSYPISKLLDLLLGKRHSTLLGRAELKSLVYMHGNEAG 183
Query: 189 KGGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLD-----------MSRI* 237
KGGELT ETTII+GALD++ K+AKDAMTP+S+ FSLDIN KLD SRI
Sbjct: 184 KGGELTHDETTIISGALDMSQKSAKDAMTPVSQIFSLDINFKLDEKTMGLIASAGHSRIP 243
Query: 238 CSAVRK--------MKHQLSL*L*GEFP------------GENWPLYDILNQFKKGQSHM 277
+V +K+ + + E N PLYDILN F+ G+SHM
Sbjct: 244 IYSVNPNVIIGFILVKNLIKVRPEDETSIRDLPIRRMPKVDLNLPLYDILNIFQTGRSHM 303
Query: 278 AVVLKSKENIRTAATNTEGFGPFLPHDYISISTEASNWQSEGSEYYSATLKNAMLQESKD 337
A V+ +K + TNT P SI+ + + + + S N+ E+
Sbjct: 304 AAVVGTKNH-----TNTN-----TPVHEKSINGSPN---KDANVFLSIPALNS--SETSH 348
Query: 338 SDPLHRSKQHDTSISLENMESLLGEEEVVGIITLEDVMEELLQVSLTSYHSTIIFVHL 395
P+ ++ SIS E +EEV+GIITLEDVMEEL+Q + Y T +V L
Sbjct: 349 QSPI----RYIDSISDE-------DEEVIGIITLEDVMEELIQEEI--YDETDQYVEL 393
>At1g47330 unknown protein
Length = 527
Score = 285 bits (729), Expect = 3e-77
Identities = 174/445 (39%), Positives = 249/445 (55%), Gaps = 65/445 (14%)
Query: 10 DEHCCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIM 69
D CCG F + ++ + FA + + L LGL+S VDLEVL+K+G+P + NA KI
Sbjct: 4 DIPCCGTTFSLYVVIIIALVAFAGLMAGLTLGLMSLGLVDLEVLIKSGRPQDRINAGKIF 63
Query: 70 SIVKNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRY 129
+VKN+HL+LCTLL+ S+A+E + + ++K+ P W+A+LL+ LI + E++PQA+ +RY
Sbjct: 64 PVVKNQHLLLCTLLIGNSMAMEALPIFLDKIVPPWLAILLSVTLILVFGEIMPQAVCTRY 123
Query: 130 GLRFGATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGK 189
GL+ GA M+PFVRVLL+LFFP +YP+SK+LD +LGKGH LL R ELKT VN H NEAGK
Sbjct: 124 GLKVGAIMAPFVRVLLVLFFPISYPISKVLDWMLGKGHGVLLRRAELKTFVNFHGNEAGK 183
Query: 190 GGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLDMSRI*CSAVRKMKHQ-- 247
GG+LT ET+II GAL+LT KTAKDAMTP+S FSL++++ L++ + + + + H
Sbjct: 184 GGDLTTDETSIITGALELTEKTAKDAMTPISNAFSLELDTPLNLETL--NTIMSVGHSRV 241
Query: 248 -------------------LSL*L*GEFP------------GENWPLYDILNQFKKGQSH 276
L++ E P E PLYDILN+F+KG SH
Sbjct: 242 PVYFRNPTHIIGLILVKNLLAVDARKEVPLRKMSMRKIPRVSETMPLYDILNEFQKGHSH 301
Query: 277 MAVVLKSK--------------ENIRTAATNTEGFGPFL--PHDYISIST---------E 311
+AVV K E + T E F P +S +
Sbjct: 302 IAVVYKDLDEQEQSPETSENGIERRKNKKTKDELFKDSCRKPKAQFEVSEKEVFKIETGD 361
Query: 312 ASNWQSEGSEYYSATLKNAMLQESKDSDPLHRSKQHDTSISLEN--MESLLGEEEVVGII 369
A + +SE E + K ++L + + HR + +EN + EEVVG+I
Sbjct: 362 AKSGKSENGEEQQGSGKTSLL--AAPAKKRHRGCSF-CILDIENTPIPDFPTNEEVVGVI 418
Query: 370 TLEDVMEELLQVSLTSYHSTIIFVH 394
T+EDV+EELLQ + + +H
Sbjct: 419 TMEDVIEELLQEEILDETDEYVNIH 443
>At4g33700 unknown protein
Length = 424
Score = 267 bits (682), Expect = 9e-72
Identities = 168/420 (40%), Positives = 240/420 (57%), Gaps = 51/420 (12%)
Query: 13 CCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIV 72
CC +F++ + + FA + S L LGL+S S VDLEVL K+G P +K AAKI+ +V
Sbjct: 7 CCSPNFFIHIAVIVFLVLFAGLMSGLTLGLMSLSLVDLEVLAKSGTPEHRKYAAKILPVV 66
Query: 73 KNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLR 132
KN+HL+L TLL+ + A+E + + ++ + W A+L++ LI + E+IPQ++ SRYGL
Sbjct: 67 KNQHLLLVTLLICNAAAMETLPIFLDGLVTAWGAILISVTLILLFGEIIPQSICSRYGLA 126
Query: 133 FGATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKGGE 192
GAT++PFVRVL+ + P A+P+SKLLD LLG AL R ELKTLV+ H NEAGKGGE
Sbjct: 127 IGATVAPFVRVLVFICLPVAWPISKLLDFLLGHRRAALFRRAELKTLVDFHGNEAGKGGE 186
Query: 193 LTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLDM-----------SRI----- 236
LT ETTIIAGAL+L+ K KDAMTP+S+ F +DIN+KLD SR+
Sbjct: 187 LTHDETTIIAGALELSEKMVKDAMTPISDIFVIDINAKLDRDLMNLILEKGHSRVPVYYE 246
Query: 237 ---*CSAVRKMKHQLSL*L*GEFPGEN------------WPLYDILNQFKKGQSHMAVVL 281
+ +K+ L++ E P +N PLYDILN+F+KG SHMAVV+
Sbjct: 247 QPTNIIGLVLVKNLLTINPDEEIPVKNVTIRRIPRVPEILPLYDILNEFQKGLSHMAVVV 306
Query: 282 KSKE----------NIRTAATNTEGFGPFLPHD-YISISTEASNWQSEGSEYYSATLKNA 330
+ + +++ A + + G P + + W+S + S +
Sbjct: 307 RQCDKIHPLPSKNGSVKEARVDVDSEGTPTPQERMLRTKRSLQKWKSFPNRASSFKGGSK 366
Query: 331 MLQESKDSDPLHRSKQHDTSISLENMESLLGEEEVVGIITLEDVMEELLQVSL---TSYH 387
+ SKD+D ++ + L EEE VGIIT+EDV+EELLQ + T +H
Sbjct: 367 SKKWSKDND------ADILQLNGNPLPKLAEEEEAVGIITMEDVIEELLQEEIFDETDHH 420
>At4g14240 unknown protein
Length = 494
Score = 238 bits (607), Expect = 4e-63
Identities = 159/429 (37%), Positives = 236/429 (54%), Gaps = 75/429 (17%)
Query: 15 GNHFWVL--ALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIV 72
G+ W+ + C++ +F A I S L LGL+S V+LE+L ++G P+ +K AA I +V
Sbjct: 31 GSFEWITYAGISCFLVLF-AGIMSGLTLGLMSLGLVELEILQRSGTPNEKKQAAAIFPVV 89
Query: 73 KNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLR 132
+ +H +L TLL+ ++A+EG+ + ++K+F E+VA++L+ + EVIPQA+ +RYGL
Sbjct: 90 QKQHQLLVTLLLCNAMAMEGLPIYLDKLFNEYVAIILSVTFVLAYGEVIPQAICTRYGLA 149
Query: 133 FGATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKGGE 192
GA VR+L+ L +P A+P+ K+LD +LG + AL R +LK LV++H+ EAGKGGE
Sbjct: 150 VGANFVWLVRILMTLCYPIAFPIGKILDLVLGH-NDALFRRAQLKALVSIHSQEAGKGGE 208
Query: 193 LTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLD---MSRI*CSAVRKM----- 244
LT ETTII+GALDLT KTA++AMTP+ TFSLD+NSKLD M +I ++
Sbjct: 209 LTHDETTIISGALDLTEKTAQEAMTPIESTFSLDVNSKLDWEAMGKILARGHSRVPVYSG 268
Query: 245 --KHQLSL*L*GEF---------------------PGENWPLYDILNQFKKGQSHMAVVL 281
K+ + L L + PLYDILN+F+KG SHMA V+
Sbjct: 269 NPKNVIGLLLVKSLLTVRPETETLVSAVCIRRIPRVPADMPLYDILNEFQKGSSHMAAVV 328
Query: 282 KSK-----------ENIRTAATNTEGFGPFL-----PHDYISISTEASNWQSEGSEYYSA 325
K K E + +++ P L HD + ++ + +N QS
Sbjct: 329 KVKGKSKVPPSTLLEEHTDESNDSDLTAPLLLKREGNHDNVIVTIDKANGQS-------- 380
Query: 326 TLKNAMLQESKDSDPLHRSKQHDTSISLENMESLLGEEEVVGIITLEDVMEELLQVSLTS 385
++ +S P H S + E +E + EV+GIITLEDV EELLQ +
Sbjct: 381 ------FFQNNESGP------HGFSHTSEAIE----DGEVIGIITLEDVFEELLQEEIVD 424
Query: 386 YHSTIIFVH 394
+ VH
Sbjct: 425 ETDEYVDVH 433
>At1g03270 unknown protein
Length = 514
Score = 223 bits (567), Expect = 2e-58
Identities = 155/443 (34%), Positives = 232/443 (51%), Gaps = 80/443 (18%)
Query: 15 GNHFW--VLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIV 72
G+ +W V+ + C++ +F A I S L LGL+S V+LE+L ++G +K AA I+ +V
Sbjct: 29 GSPWWFVVVGVACFLVLF-AGIMSGLTLGLMSLGLVELEILQQSGSSAEKKQAAAILPVV 87
Query: 73 KNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLR 132
K +H +L TLL+ + A+E + + ++K+F +VAVLL+ + E+IPQA+ SRYGL
Sbjct: 88 KKQHQLLVTLLLCNAAAMEALPICLDKIFHPFVAVLLSVTFVLAFGEIIPQAICSRYGLA 147
Query: 133 FGATMSPFVRVLLLLFFPFAYPVSK---------------LLDCLLGKGHTALLGREELK 177
GA VR+L+++ +P AYP+ K +LD ++G T L R +LK
Sbjct: 148 VGANFLWLVRILMIICYPIAYPIGKVMLLCLLLSTFYMPQVLDAVIGHNDT-LFRRAQLK 206
Query: 178 TLVNLHANEAGKGGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLDMSRI* 237
LV++H+ EAGKGGELT ET II+GALDL+ KTA++AMTP+ TFSLD+N+KLD I
Sbjct: 207 ALVSIHSQEAGKGGELTHEETMIISGALDLSQKTAEEAMTPIESTFSLDVNTKLDWETIG 266
Query: 238 CSAVR-------------------KMKHQLSL*L*GEFP------------GENWPLYDI 266
R +K L++ E P + PLYDI
Sbjct: 267 KILSRGHSRIPVYLGNPKNIIGLLLVKSLLTVRAETEAPVSSVSIRKIPRVPSDMPLYDI 326
Query: 267 LNQFKKGQSHMAVVLKSKENIRTAATNTEGFGPFLPHDYISISTEASNWQSEGSEYYSAT 326
LN+F+KG SHMA V+K K+ + + + + + + Y S+
Sbjct: 327 LNEFQKGSSHMAAVVKVKDKDK--------------KNNMQLLSNGETPKENMKFYQSSN 372
Query: 327 LKNAMLQESK-------DSDPLH-----RSKQHDTSISLENMESLLGEE---EVVGIITL 371
L +L+ D P H R+ Q + +++ ++ LL + EV+GIITL
Sbjct: 373 LTAPLLKHESHDVVVDIDKVPKHVKNRGRNFQQNGTVT-RDLPCLLEDNEDAEVIGIITL 431
Query: 372 EDVMEELLQVSLTSYHSTIIFVH 394
EDV EELLQ + I VH
Sbjct: 432 EDVFEELLQAEIVDETDVYIDVH 454
>At2g14520 hypothetical protein
Length = 408
Score = 220 bits (560), Expect = 1e-57
Identities = 149/420 (35%), Positives = 220/420 (51%), Gaps = 79/420 (18%)
Query: 13 CCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNA------- 65
CCG F++ + + + FA + S L LGL+S S VDLEVL K+G P + +A
Sbjct: 7 CCGTSFFIHIAVIVLLVLFAGLMSGLTLGLMSMSLVDLEVLAKSGTPRDRIHAVGFGFDA 66
Query: 66 AKIMSIVKNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQAL 125
AKI+ +VKN+HL+LCTLL+ + A+E + + ++ + W A+L++ LI + E+IPQ++
Sbjct: 67 AKILPVVKNQHLLLCTLLICNAAAMEALPIFLDALVTAWGAILISVTLILLFGEIIPQSV 126
Query: 126 NSRYGLRFGATMSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHAN 185
SR+GL GAT++PFVRVL+ + P A+P+SK V
Sbjct: 127 CSRHGLAIGATVAPFVRVLVWICLPVAWPISK-------------------PNNVACQFF 167
Query: 186 EAGKGGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSKLDM------------ 233
+AGKGGELT ETTIIAGAL+L+ K AKDAMTP+S+TF +DIN+KLD
Sbjct: 168 QAGKGGELTHDETTIIAGALELSEKMAKDAMTPISDTFVIDINAKLDRDLMNLILDKGHS 227
Query: 234 -------SRI*CSAVRKMKHQLSL*L*GEFPGEN------------WPLYDILNQFKKGQ 274
R + +K+ L++ E +N PLYDILN+F+KG
Sbjct: 228 RVPVYYEQRTNIIGLVLVKNLLTINPDEEIQVKNVTIRRIPRVPETLPLYDILNEFQKGH 287
Query: 275 SHMAVVLKSKENIRTAATNTEGFGPFLPHDYISISTEAS-------------NWQSEGSE 321
SHMAVV++ + I +N + + + + E S W+S +
Sbjct: 288 SHMAVVVRQCDKIHPLQSN-DAANETVNEVRVDVDYERSPQETKLKRRRSLQKWKSFPNR 346
Query: 322 YYSATLKNAMLQESKDSDPLHRSKQHDTSISLENMESLLGEEEVVGIITLEDVMEELLQV 381
S ++ + D+D L ++ + L EE+ VGIIT+EDV+EELLQV
Sbjct: 347 ANSLGSRSKRWSKDNDADIL--------QLNEHPLPKLDEEEDAVGIITMEDVIEELLQV 398
>At4g14230 hypothetical protein
Length = 408
Score = 197 bits (501), Expect = 9e-51
Identities = 126/313 (40%), Positives = 176/313 (55%), Gaps = 61/313 (19%)
Query: 29 MFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVKNEHLVLCTLLMAKSL 88
+ FA I S L LGL+S V+LE+L ++G P +K +A I +V+ +H +L TLL+ +L
Sbjct: 45 VLFAGIMSGLTLGLMSLGLVELEILQRSGTPKEKKQSAAIFPVVQKQHQLLVTLLLFNAL 104
Query: 89 ALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLRFGATMSPFVRVLLLLF 148
A+EG+ + ++K+F E+VA++L+ + + EVIPQA+ +RYGL GA +
Sbjct: 105 AMEGLPIYLDKIFNEYVAIILSVTFVLFVGEVIPQAICTRYGLAVGANL----------- 153
Query: 149 FPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKGGELTLHETTIIAGALDLT 208
V +LD +LG + L R +LK LV++H AGKGGELT ETTII+GALDLT
Sbjct: 154 ------VWLMLDWVLGH-NDPLFRRAQLKALVSIHGEAAGKGGELTHDETTIISGALDLT 206
Query: 209 MKTAKDAMTPLSETFSLDINSKLDMSRI*CSAVRKMKHQLSL*L*GEFPGENWPLYDILN 268
KTA++AMTP+ TFSLD+NSKLD P N PLYDILN
Sbjct: 207 EKTAQEAMTPIESTFSLDVNSKLD----------------------RVPA-NMPLYDILN 243
Query: 269 QFKKGQSHMAVVLKSK------------EN---IRTAATNTEGFGPFL-----PHDYISI 308
+F+KG SHMA V+K K EN ++ N+E P L HD + +
Sbjct: 244 EFQKGSSHMAAVVKVKGKSKGHPSTLHEENSGESNVSSNNSELTAPLLLKREGNHDSVIV 303
Query: 309 STEASNWQSEGSE 321
+ +N QS SE
Sbjct: 304 RIDKANGQSFISE 316
>At1g04570 unknown protein
Length = 542
Score = 31.6 bits (70), Expect = 0.83
Identities = 31/124 (25%), Positives = 53/124 (42%), Gaps = 4/124 (3%)
Query: 14 CGNHFWVLALLC--WVFMFFAAISS-ALALGLLSFSQVDLEVLVKAGQPH-IQKNAAKIM 69
CG +WV C W+ + F + S AL L Q + + A + + + I
Sbjct: 91 CGLGYWVQGSRCFPWLALNFHMVHSLALQPSTLQLVQYSCSLPMVAKPLYGVLSDVLYIG 150
Query: 70 SIVKNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRY 129
S + ++ + L + G+ ++ P VA +L + L + I EV AL + Y
Sbjct: 151 SGRRVPYIAIGVFLQVLAWGSMGIFQGAREVLPSLVACVLLSNLGASITEVAKDALVAEY 210
Query: 130 GLRF 133
GLR+
Sbjct: 211 GLRY 214
>At1g01880 hypothetical protein
Length = 570
Score = 31.2 bits (69), Expect = 1.1
Identities = 19/54 (35%), Positives = 27/54 (49%), Gaps = 4/54 (7%)
Query: 52 VLVKAGQPHIQKNAAKIMSIVKNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWV 105
V V G P K+ A+I ++ + C L + K +GVSV K+F EWV
Sbjct: 71 VFVVDGTPSPLKSQARISRFFRSSGIDTCNLPVIK----DGVSVERNKLFSEWV 120
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 30.8 bits (68), Expect = 1.4
Identities = 18/73 (24%), Positives = 36/73 (48%), Gaps = 11/73 (15%)
Query: 94 SVLMEKMFPEWVAVLLATALISI-IAEVIPQALNSRYGLRFGATMSPFVRVLLLLFFPFA 152
++ + +MF W+ + + TA S+ + + S GLR G ++SP++ V+ +
Sbjct: 765 AIHLPEMFIHWIELCIGTASFSVQVNGELSGFFRSERGLRQGCSLSPYLYVICM------ 818
Query: 153 YPVSKLLDCLLGK 165
+L C+L K
Sbjct: 819 ----NVLSCMLDK 827
>At3g59520 unknown protein
Length = 269
Score = 30.4 bits (67), Expect = 1.8
Identities = 14/47 (29%), Positives = 25/47 (52%), Gaps = 2/47 (4%)
Query: 16 NHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQ 62
N++W L +L W+ + F + S G FS +++E L A P ++
Sbjct: 204 NNYWALTMLGWIVVVF--VFSLKKSGAYDFSFLEIESLTDASLPSVR 248
>At5g43900 myosin heavy chain MYA2 (pir||S51824)
Length = 1505
Score = 30.0 bits (66), Expect = 2.4
Identities = 20/68 (29%), Positives = 39/68 (56%), Gaps = 3/68 (4%)
Query: 311 EASNWQSEGSEYYSATLKNAMLQESKDSDPLHRSKQHDTSI--SLENMESLLGEEEVVGI 368
++ N +S E S+ ++ M +ES D+D D+SI S++++ S + E++ VGI
Sbjct: 1431 DSYNTRSVSQEVISS-MRTLMTEESNDADSDSFLLDDDSSIPFSIDDISSSMEEKDFVGI 1489
Query: 369 ITLEDVME 376
E+++E
Sbjct: 1490 KPAEELLE 1497
>At4g14470 reverse transcriptase like protein
Length = 318
Score = 29.6 bits (65), Expect = 3.2
Identities = 14/49 (28%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Query: 99 KMFPEWVAVLLATALISI-IAEVIPQALNSRYGLRFGATMSPFVRVLLL 146
++F W+ + + TA S+ + + NS GLR G ++SP++ V+++
Sbjct: 59 QVFVHWIMLCVTTASFSVQVNGELAGYFNSSRGLRQGCSLSPYLFVIVM 107
>At3g44650 putative protein
Length = 762
Score = 29.6 bits (65), Expect = 3.2
Identities = 20/69 (28%), Positives = 35/69 (49%), Gaps = 7/69 (10%)
Query: 99 KMFPEWVAVLLATALISI-IAEVIPQALNSRYGLRFGATMSPFVRVLLLLFFPFAYPVSK 157
+MF W+ + + T S+ + + S GLR G +SP++ V+ + +SK
Sbjct: 176 EMFIHWIRLCITTPSFSVQVNGELAGFFQSSRGLRQGCALSPYLFVICM------DVLSK 229
Query: 158 LLDCLLGKG 166
LLD ++G G
Sbjct: 230 LLDKVVGIG 238
>At1g01960 hypothetical protein
Length = 1750
Score = 29.3 bits (64), Expect = 4.1
Identities = 20/68 (29%), Positives = 32/68 (46%)
Query: 337 DSDPLHRSKQHDTSISLENMESLLGEEEVVGIITLEDVMEELLQVSLTSYHSTIIFVHLQ 396
++D L RSK + SLE+ E+ L E T ++ L+ Y+ I HL
Sbjct: 1589 NADLLLRSKLQELGSSLESQEAPLLRLENESFQTCMTFLDNLISDQPVGYNEAEIESHLI 1648
Query: 397 TLCKMCVE 404
+LC+ +E
Sbjct: 1649 SLCREVLE 1656
>At4g31400 unknown protein
Length = 322
Score = 28.9 bits (63), Expect = 5.4
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query: 312 ASNWQSEGSEYYSATLKNAMLQESKDSDPLHRSKQHDTSISLENMESLLGEEEVV 366
+ WQ+E + +KN ++ S++ P HR+K + ++ ME LGE+ ++
Sbjct: 143 SQGWQNEKAFTSPLFIKNRIVMVSENDSPAHRNKVQEV---VKMMEVELGEDWIL 194
>At2g26780 unknown protein
Length = 1732
Score = 28.9 bits (63), Expect = 5.4
Identities = 22/83 (26%), Positives = 38/83 (45%), Gaps = 20/83 (24%)
Query: 90 LEGVSVLMEKMFPEWVAVLLATALISI--IAEVIPQ---------ALNSRYGL------- 131
LE + + + K P W + L ++ I + ++IP+ LN+R G+
Sbjct: 1246 LENLRISISKGSPMWETLDLCINIVDIESLEQLIPRLTQLVRGGVGLNTRVGVASFISLL 1305
Query: 132 --RFGATMSPFVRVLLLLFFPFA 152
+ G+ + PF +LL L FP A
Sbjct: 1306 VQKVGSEIKPFTGMLLRLLFPVA 1328
>At4g12180 putative reverse transcriptase
Length = 662
Score = 28.5 bits (62), Expect = 7.0
Identities = 16/58 (27%), Positives = 33/58 (56%), Gaps = 5/58 (8%)
Query: 94 SVLMEKMFPE----WVAVLLATALISI-IAEVIPQALNSRYGLRFGATMSPFVRVLLL 146
+VL+ FP+ W+ + + TA S+ + + NS GLR G +++P++ V+++
Sbjct: 132 NVLLTLDFPQEFVHWIMLCVTTASFSVQVNRELAGYFNSLRGLRQGCSLTPYLFVIVM 189
>At3g25485 unknown protein
Length = 979
Score = 28.5 bits (62), Expect = 7.0
Identities = 20/72 (27%), Positives = 36/72 (49%), Gaps = 7/72 (9%)
Query: 94 SVLMEKMFPEWVAVLLATALISI-IAEVIPQALNSRYGLRFGATMSPFVRVLLLLFFPFA 152
S+ + F W+ + + TA S+ + + NS GLR G ++SP++ V+ +
Sbjct: 534 SLNFSQEFIHWIMLCITTASFSVQVNGELVGFFNSSRGLRQGCSLSPYLFVIAM------ 587
Query: 153 YPVSKLLDCLLG 164
+SK+LD G
Sbjct: 588 DVLSKMLDRAAG 599
>At1g09540 putative transcription factor (MYB61)
Length = 366
Score = 28.5 bits (62), Expect = 7.0
Identities = 23/98 (23%), Positives = 39/98 (39%), Gaps = 3/98 (3%)
Query: 296 GFGPFLPHDYISISTEASNWQSEG---SEYYSATLKNAMLQESKDSDPLHRSKQHDTSIS 352
GF +P+ S S N E SEY + +S+ S P++ + D +
Sbjct: 269 GFSWSIPNSSTSSSQVKPNHNFEEIKWSEYLNTPFFIGSTVQSQTSQPIYIKSETDYLAN 328
Query: 353 LENMESLLGEEEVVGIITLEDVMEELLQVSLTSYHSTI 390
+ NM + E +G DV + LQ S+ ++
Sbjct: 329 VSNMTDPWSQNENLGTTETSDVFSKDLQRMAVSFGQSL 366
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,223,490
Number of Sequences: 26719
Number of extensions: 311435
Number of successful extensions: 1220
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1192
Number of HSP's gapped (non-prelim): 32
length of query: 410
length of database: 11,318,596
effective HSP length: 102
effective length of query: 308
effective length of database: 8,593,258
effective search space: 2646723464
effective search space used: 2646723464
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146664.12


