

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146664.11 + phase: 0
(248 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
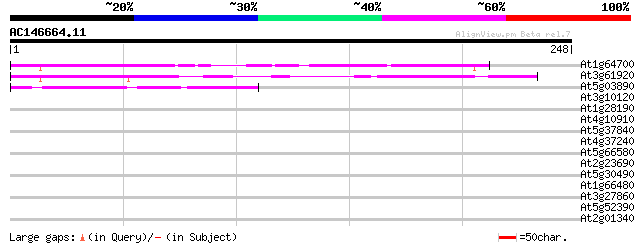
Score E
Sequences producing significant alignments: (bits) Value
At1g64700 unknown protein 134 5e-32
At3g61920 unknown protein 111 4e-25
At5g03890 putative protein 58 4e-09
At3g10120 unknown protein 39 0.003
At1g28190 unknown protein 36 0.017
At4g10910 unknown protein 35 0.029
At5g37840 putative protein 35 0.037
At4g37240 unknown protein 33 0.11
At5g66580 unknown protein 31 0.54
At2g23690 unknown protein 30 0.92
At5g30490 bcnt-like protein 30 1.6
At1g66480 hypothetical protein 30 1.6
At3g27860 hypothetical protein 28 3.5
At5g52390 photoassimilate-responsive protein PAR-like protein 28 6.0
At2g01340 unknown protein 28 6.0
>At1g64700 unknown protein
Length = 203
Score = 134 bits (337), Expect = 5e-32
Identities = 92/217 (42%), Positives = 121/217 (55%), Gaps = 28/217 (12%)
Query: 1 MGNCLLGANSDP--DTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLC 58
MGNCL G D D LIKVI DG ++EFY P+T F+++ F GHA+F D KPL
Sbjct: 1 MGNCLFGGLGDEEEDLLIKVIKSDGGVLEFYSPVTAGFVSHGFSGHALFSAVDLLWKPLA 60
Query: 59 QFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPA 118
LV GQSYYL P ++S K + +C HVRS S + +
Sbjct: 61 HDHLLVPGQSYYLFP-NIVSDELK-TFVGSC---------------HVRSNSESLSAI-T 102
Query: 119 PYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCIT 178
PYRMSLDY + +LKR+ T+ FS ++ + K + RR+S K IWKV L I
Sbjct: 103 PYRMSLDYNH----RVLKRSYTDVFSRNSHIRTRQKEKKTRRRRTSSKG-AIWKVNLIIN 157
Query: 179 PEKLLEILSQEVSTKELIESVRIVAK---CGVTAASS 212
E+LL+ILS++ T ELIESVR VAK +T++SS
Sbjct: 158 TEELLQILSEDGRTNELIESVRAVAKGETSSITSSSS 194
>At3g61920 unknown protein
Length = 187
Score = 111 bits (277), Expect = 4e-25
Identities = 85/242 (35%), Positives = 111/242 (45%), Gaps = 70/242 (28%)
Query: 1 MGNCLLGANSDP-------DTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTND-- 51
MGNC+ N D+LIKV+T +G +ME +PPI FITNEF GH I +
Sbjct: 1 MGNCVFKGNGGSRKLYDKDDSLIKVVTPNGGVMELHPPIFAEFITNEFPGHVIHDSLSLR 60
Query: 52 QSSKPLCQFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSV 111
SS PL +EL G YYLLP++ S ATA+ Q
Sbjct: 61 HSSPPLLHGEELFPGNIYYLLPLS----------SSAAATAQLDSSDQ------------ 98
Query: 112 PTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIW 171
L PYRMS K+ I ++ S G+W
Sbjct: 99 ----LSTPYRMSF----------------------------GKTPIMAA--LSGGGCGVW 124
Query: 172 KVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLS 231
KV+L I+PE+L EIL+++V T+ L+ESVR VAKCG GCG + SDQ S++
Sbjct: 125 KVRLVISPEQLAEILAEDVETEALVESVRTVAKCG-----GYGCGGGVHSRANSDQLSVT 179
Query: 232 SS 233
SS
Sbjct: 180 SS 181
>At5g03890 putative protein
Length = 179
Score = 58.2 bits (139), Expect = 4e-09
Identities = 36/110 (32%), Positives = 59/110 (52%), Gaps = 10/110 (9%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQF 60
MGNCL+ +IK++ DG ++E+ PI+V+ I +F GH+I N L
Sbjct: 1 MGNCLVMEKK----VIKIVRDDGKVLEYREPISVHHILTQFSGHSISHNNTH----LLPD 52
Query: 61 DELVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQS 110
+L++G+ YYLLP T+ K++ T A E G +++R+ S+S
Sbjct: 53 AKLLSGRLYYLLPTTM--TKKKVNKKVTFANPEVEGDERLLREEEDSSES 100
>At3g10120 unknown protein
Length = 173
Score = 38.9 bits (89), Expect = 0.003
Identities = 23/73 (31%), Positives = 39/73 (52%), Gaps = 4/73 (5%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQF 60
MGNCL+ +IK++ DG ++E+ P+ V+ I +F H + ++ L
Sbjct: 1 MGNCLVMEKK----VIKIMRNDGKVVEYRGPMKVHHILTQFSPHYSLFDSLTNNCHLHPQ 56
Query: 61 DELVAGQSYYLLP 73
+L+ G+ YYLLP
Sbjct: 57 AKLLCGRLYYLLP 69
>At1g28190 unknown protein
Length = 266
Score = 36.2 bits (82), Expect = 0.017
Identities = 22/85 (25%), Positives = 41/85 (47%), Gaps = 4/85 (4%)
Query: 16 IKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKPLCQFDELVAGQSYYL 71
IK++ DG++ + P+ V+ +T +F H I ++ Q + L + + L G +Y+L
Sbjct: 39 IKLVRSDGSLQVYDRPVVVSELTKDFPKHKICRSDLLYIGQKTPVLSETETLKLGLNYFL 98
Query: 72 LPMTVLSPNNKIDYPSTCATAETGG 96
LP + +T T + GG
Sbjct: 99 LPSDFFKNDLSFLTIATLKTPQNGG 123
>At4g10910 unknown protein
Length = 136
Score = 35.4 bits (80), Expect = 0.029
Identities = 17/50 (34%), Positives = 31/50 (62%)
Query: 163 SSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASS 212
S ++ G+WK K+ I ++L EIL+ E +T LI+ +R A + +++S
Sbjct: 69 SPQRRNGVWKAKVVIGSKQLEEILAVEGNTHALIDQLRFAAAEALVSSTS 118
>At5g37840 putative protein
Length = 214
Score = 35.0 bits (79), Expect = 0.037
Identities = 22/90 (24%), Positives = 40/90 (44%), Gaps = 9/90 (10%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQS-----SK 55
MGN ++ + +KV+ DG+I P+T + T E+ G + + +K
Sbjct: 1 MGNTIMVRRNK----VKVMKIDGDIFRLKTPVTASDATKEYPGFVLLDSETVKRLGVRAK 56
Query: 56 PLCQFDELVAGQSYYLLPMTVLSPNNKIDY 85
PL L +Y+L+ + + NK+ Y
Sbjct: 57 PLEPNQILKPNHTYFLVDLPPVDKRNKLPY 86
>At4g37240 unknown protein
Length = 168
Score = 33.5 bits (75), Expect = 0.11
Identities = 29/107 (27%), Positives = 48/107 (44%), Gaps = 10/107 (9%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKP 56
MG C ++ T K+I DG +MEF P+ V ++ ++ I ++ D +
Sbjct: 1 MGICSSSESTQVATA-KLILQDGRMMEFANPVKVGYVLLKYPMCFICNSDDMDFDDAVAA 59
Query: 57 LCQFDELVAGQSYYLLPMTVLSPNNKIDYPSTCATAET-----GGGG 98
+ +EL GQ Y+ LP+ L K + + A + GGGG
Sbjct: 60 ISADEELQLGQIYFALPLCWLRQPLKAEEMAALAVKASSALMRGGGG 106
>At5g66580 unknown protein
Length = 156
Score = 31.2 bits (69), Expect = 0.54
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 5/82 (6%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN----DQSSKP 56
MG C + D+ K+I DG + EF P+ V I + + ++ D +
Sbjct: 1 MGACASRESLRSDSA-KLILLDGTLQEFSSPVKVWQILQKNPTSFVCNSDEMDFDDAVSA 59
Query: 57 LCQFDELVAGQSYYLLPMTVLS 78
+ +EL +GQ Y++LP+T L+
Sbjct: 60 VAGNEELRSGQLYFVLPLTWLN 81
>At2g23690 unknown protein
Length = 278
Score = 30.4 bits (67), Expect = 0.92
Identities = 19/72 (26%), Positives = 34/72 (46%), Gaps = 4/72 (5%)
Query: 17 KVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQS----SKPLCQFDELVAGQSYYLL 72
K+I DG +MEF P+ V ++ + I ++D + +E GQ Y+ L
Sbjct: 16 KLILHDGRMMEFTSPVKVGYVLQKNPMCFICNSDDMDFDNVVSAISADEEFQLGQLYFAL 75
Query: 73 PMTVLSPNNKID 84
P++ L + K +
Sbjct: 76 PLSSLHHSLKAE 87
>At5g30490 bcnt-like protein
Length = 236
Score = 29.6 bits (65), Expect = 1.6
Identities = 38/152 (25%), Positives = 59/152 (38%), Gaps = 45/152 (29%)
Query: 113 TTPLPAPYRMSLDYQYYQGVGLLKRTSTES--------FSCRTNRSS--------VNKST 156
+T LPA + +++ Y GV + K+ + S SC S V +T
Sbjct: 40 STALPAKNSANSNWKSYLGVNVKKKDTCVSDVQKSVLNHSCSEEAKSIAAAALAAVRNAT 99
Query: 157 ISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVTAASSGGCG 216
++++ SSR I +VK + QE+ K L+E+ A G G
Sbjct: 100 VTAAAASSRGKIEITEVK---------DFAGQEIEVKRLVEAD------SKEALERGNKG 144
Query: 217 ISSTTSIVSDQWSLSSSGRSAPSKVDALVLDI 248
SSS +APS VDA++ I
Sbjct: 145 --------------SSSSSAAPSAVDAVLEQI 162
>At1g66480 hypothetical protein
Length = 223
Score = 29.6 bits (65), Expect = 1.6
Identities = 20/81 (24%), Positives = 34/81 (41%), Gaps = 12/81 (14%)
Query: 17 KVITFDGNIMEFYPPITVNFITNEFQGHAIFPTN-----DQSSKPLCQFDELVAGQSYYL 71
KV+ DG P+T +T ++ G+ + + SKPL L ++Y+L
Sbjct: 13 KVMKIDGETFRIKTPVTAREVTADYPGYVLLDSQAVKHFGVRSKPLEPNQTLKPKKTYFL 72
Query: 72 LPMTVLSP-------NNKIDY 85
+ + L P NK+ Y
Sbjct: 73 VELPKLPPETTAVDTENKLPY 93
>At3g27860 hypothetical protein
Length = 652
Score = 28.5 bits (62), Expect = 3.5
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 3/47 (6%)
Query: 143 FSCRTNR---SSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEIL 186
FSCR + S S I + R + +T I VKL TPE++L ++
Sbjct: 401 FSCRARKIKIESAESSQIQVAVRMTTPNTAIDVVKLGATPEEMLALI 447
>At5g52390 photoassimilate-responsive protein PAR-like protein
Length = 195
Score = 27.7 bits (60), Expect = 6.0
Identities = 15/50 (30%), Positives = 26/50 (52%)
Query: 135 LKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLE 184
+KR+ E ++CR++ NK T K+ G+ + L I+ + LLE
Sbjct: 53 MKRSGIEVYTCRSSEIEANKVTNIIESDECIKACGLDRKALGISSDALLE 102
>At2g01340 unknown protein
Length = 225
Score = 27.7 bits (60), Expect = 6.0
Identities = 14/46 (30%), Positives = 21/46 (45%), Gaps = 5/46 (10%)
Query: 1 MGNCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAI 46
MGN L G + KV+ DG + P+T + +F GH +
Sbjct: 1 MGNSLGGKKTT-----KVMKIDGETFKLKTPVTAEEVLKDFPGHVL 41
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.130 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,584,678
Number of Sequences: 26719
Number of extensions: 223831
Number of successful extensions: 507
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 495
Number of HSP's gapped (non-prelim): 16
length of query: 248
length of database: 11,318,596
effective HSP length: 97
effective length of query: 151
effective length of database: 8,726,853
effective search space: 1317754803
effective search space used: 1317754803
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146664.11


