

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.4 - phase: 0
(320 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
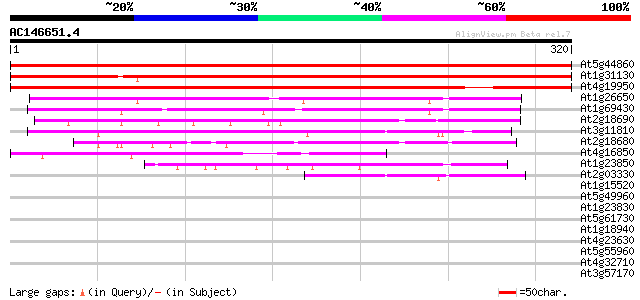
Score E
Sequences producing significant alignments: (bits) Value
At5g44860 unknown protein 466 e-132
At1g31130 unknown protein 435 e-122
At4g19950 putative protein 430 e-121
At1g26650 unknown protein 100 8e-22
At1g69430 hypothetical protein 100 1e-21
At2g18690 unknown protein 80 2e-15
At3g11810 unknown protein 66 3e-11
At2g18680 unknown protein 65 4e-11
At4g16850 hypothetical protein 50 2e-06
At1g23850 unknown protein 44 2e-04
At2g03330 unknown protein 42 6e-04
At1g15520 hypothetical protein 35 0.042
At5g49960 putative protein 35 0.054
At1g23830 unknown protein 34 0.093
At5g61730 ABC transport protein - like 32 0.35
At1g18940 hypothetical protein 31 1.0
At4g23630 unknown protein 30 1.3
At5g55960 unknown protein 30 1.7
At4g32710 putative protein kinase 30 1.7
At3g57170 unknown protein 30 1.7
>At5g44860 unknown protein
Length = 321
Score = 466 bits (1200), Expect = e-132
Identities = 232/321 (72%), Positives = 275/321 (85%), Gaps = 1/321 (0%)
Query: 1 MDLAPEELQFLNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHI 60
MDLA EELQFLNI IL+ES +IPK SPKTFYLITL LIFPLSFAILAHSLFT P+++ +
Sbjct: 1 MDLAAEELQFLNIQGILRESTTIPKFSPKTFYLITLTLIFPLSFAILAHSLFTQPILAQL 60
Query: 61 ES-PFTDPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTIS 119
++ P +D ++T+H+WTLLLI QF Y+IFLFAFSLLSTAAVVFTVASLYT KPVSFS+T+S
Sbjct: 61 DATPPSDQSKTNHEWTLLLIYQFIYVIFLFAFSLLSTAAVVFTVASLYTGKPVSFSSTMS 120
Query: 120 AIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVV 179
AIP V KRLFITFLWV+L+M YN VF+L LV++++A+D + +L FS+V+IFVLFL V
Sbjct: 121 AIPLVLKRLFITFLWVSLMMLVYNSVFLLFLVVLIVAIDLQSVILAVFSMVVIFVLFLGV 180
Query: 180 HVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMFSV 239
HVY+TA WHLASVVSVLEP+YG AAMKKSYELL GR A +V YL LC + +G+F
Sbjct: 181 HVYMTAWWHLASVVSVLEPIYGIAAMKKSYELLNGRTNMACSMVFMYLALCGITAGVFGG 240
Query: 240 IVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQEIDKSALHDHL 299
+VVHGGD + +F++I +GGFLVG+LVIVNLVGLLVQSVFYYVCKS+HHQ IDKSALHDHL
Sbjct: 241 VVVHGGDDFGLFTKIVVGGFLVGILVIVNLVGLLVQSVFYYVCKSFHHQPIDKSALHDHL 300
Query: 300 GGYLGEYVPLKSSIQMENLDI 320
GGYLG+YVPLKSSIQMEN DI
Sbjct: 301 GGYLGDYVPLKSSIQMENFDI 321
>At1g31130 unknown protein
Length = 321
Score = 435 bits (1118), Expect = e-122
Identities = 220/323 (68%), Positives = 263/323 (81%), Gaps = 5/323 (1%)
Query: 1 MDLAPEELQFLNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHI 60
MDL PEELQFL IP +L+ESISI K SP+TFYLITL IFPLSFAILAHSLFT P+++ +
Sbjct: 1 MDLQPEELQFLTIPQLLQESISIKKRSPRTFYLITLSFIFPLSFAILAHSLFTQPILAKL 60
Query: 61 ESPFTDPAQTS---HDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNT 117
+ +DP + HDWT+LLI QF YLIFLFAFSLLSTAAVVFTVASLYT KPVSFS+T
Sbjct: 61 DK--SDPPNSDRSRHDWTVLLIFQFSYLIFLFAFSLLSTAAVVFTVASLYTGKPVSFSST 118
Query: 118 ISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFL 177
+SAIP VFKRLFITFLWV LLMF YN VF + LV++++A+D ++ L + V+I VL+
Sbjct: 119 LSAIPKVFKRLFITFLWVALLMFAYNAVFFVFLVMLLVALDLNSLGLAIVAGVIISVLYF 178
Query: 178 VVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMF 237
VHVY TALWHL SV+SVLEP+YG AAM+K+YELLKG+ + A L+ YLFLC +I +F
Sbjct: 179 GVHVYFTALWHLGSVISVLEPVYGIAAMRKAYELLKGKTKMAMGLIFVYLFLCGLIGVVF 238
Query: 238 SVIVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQEIDKSALHD 297
+VVHGG Y F+R +GG LVGVLV+VNLVGLLVQSVFYYVCKSYHHQ IDK+AL+D
Sbjct: 239 GAVVVHGGGKYGTFTRTLVGGLLVGVLVMVNLVGLLVQSVFYYVCKSYHHQTIDKTALYD 298
Query: 298 HLGGYLGEYVPLKSSIQMENLDI 320
LGGYLG+YVPLKS+IQ+E+LDI
Sbjct: 299 QLGGYLGDYVPLKSNIQLEDLDI 321
>At4g19950 putative protein
Length = 306
Score = 430 bits (1105), Expect = e-121
Identities = 221/321 (68%), Positives = 256/321 (78%), Gaps = 16/321 (4%)
Query: 1 MDLAPEELQFLNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHI 60
MDLAPEELQFLN IL+ES SIP+ S KTFYLITL LIFPLSFAILAHSLFT P+++ I
Sbjct: 1 MDLAPEELQFLNKRGILRESTSIPQYSLKTFYLITLTLIFPLSFAILAHSLFTQPILAQI 60
Query: 61 ES-PFTDPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTIS 119
++ P D +Q H+WT+LL+ QF Y+IFLFAFSLLSTAAVVFTVASLYT KPVSFS+T+S
Sbjct: 61 DTYPQADQSQLQHEWTVLLVFQFCYIIFLFAFSLLSTAAVVFTVASLYTGKPVSFSSTMS 120
Query: 120 AIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVV 179
AIP V KRLFITFLWV+LLM YN VF++ LV +++AVD N VL FS+V+IFVLFLVV
Sbjct: 121 AIPLVLKRLFITFLWVSLLMLAYNTVFLIFLVTLIVAVDLQNVVLAVFSLVVIFVLFLVV 180
Query: 180 HVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMFSV 239
HVY+TALWHLASVVSVLEP+YG AAMKKSYELLKG+ A +V YL C I+G+F
Sbjct: 181 HVYMTALWHLASVVSVLEPIYGLAAMKKSYELLKGKTLMACSMVFIYLVHCGFIAGVFGA 240
Query: 240 IVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQEIDKSALHDHL 299
+VV GGD Y +F+RI GGFL SVFYYVCKS+HHQEIDKSALHDHL
Sbjct: 241 VVVRGGDDYGIFARIVAGGFL---------------SVFYYVCKSFHHQEIDKSALHDHL 285
Query: 300 GGYLGEYVPLKSSIQMENLDI 320
GGYLGEYVPLKS+IQMEN ++
Sbjct: 286 GGYLGEYVPLKSNIQMENFEV 306
>At1g26650 unknown protein
Length = 335
Score = 100 bits (250), Expect = 8e-22
Identities = 78/302 (25%), Positives = 145/302 (47%), Gaps = 29/302 (9%)
Query: 12 NIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIESPFTDPAQTS 71
N IL+E++ I + + L T +LI P+S +L + L L++ + A++S
Sbjct: 26 NALEILRETVRILRYNLGALMLTTAVLICPVSALLLPNFLVDQSLVNKLTVKLLLVAKSS 85
Query: 72 ------------HDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTIS 119
+ + SLLS AAVV++V Y+ + V S +
Sbjct: 86 GLPLQPFVKHSCQKFAETAVSSAMCFPVFITVSLLSKAAVVYSVDCSYSREVVDISKFLV 145
Query: 120 AIPNVFKRLFITFLWVTLLMF-CYNFVFILCLVLMVIAVDTDNSVLLF------FSVVLI 172
+ +++R+ T++W+ +L+ C+ F +L ++A+ + SVL F + +L+
Sbjct: 146 ILQKIWRRVVFTYVWICILIVGCFTFFCVL-----LVAICSSFSVLGFSPDFNVYGAMLV 200
Query: 173 FVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAV 232
+ F VV + + A V+SVLE + G A+ ++ +L+KG+++ L+ G A
Sbjct: 201 GLAFSVVFANAIIICNTAIVISVLEDVSGLGALMRASDLIKGQIQVGLLMFLGSTLGLAF 260
Query: 233 ISGMFS--VIVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQEI 290
+ G+F V V GDG SR++ G LV + V L+ ++ +VFY+ C+ Y+ E
Sbjct: 261 VEGLFDHRVKKVSYGDG---SSRLWEGPLLVLMYSFVTLIDSMMSAVFYFSCRVYYSMEA 317
Query: 291 DK 292
+
Sbjct: 318 SR 319
>At1g69430 hypothetical protein
Length = 350
Score = 100 bits (249), Expect = 1e-21
Identities = 79/302 (26%), Positives = 145/302 (47%), Gaps = 29/302 (9%)
Query: 11 LNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIES-------- 62
+N IL+E++ I + + F LI L+LI P+S +L + L +++ +
Sbjct: 41 MNALEILRETVRILRYNLGAFMLIALLLICPVSAILLPNLLVDQSVVNSLTVRLLLVSKS 100
Query: 63 ------PFTDPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSN 116
PF + T + F L SLLS AAVV++V Y+ K V +
Sbjct: 101 SGLPLLPFVRNSCQKFSETAVSSAMCFPLFI--TLSLLSRAAVVYSVDCTYSRKKVVVTK 158
Query: 117 TISAIPNVFKRLFITFLWV-TLLMFCYN----FVFILCLVLMVIAVDTDNSVLLFFSVVL 171
+ + ++KRL IT+LW+ T+++ C F+ +C V+ D + + +L
Sbjct: 159 FVVIMQRLWKRLVITYLWICTVIVVCLTSFCVFLVAVCSSFYVLGFSPDFNA---YGAIL 215
Query: 172 IFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCA 231
+ ++F VV + + V+S+LE + G A+ ++ +L+KG+ + L+ G
Sbjct: 216 VGLVFSVVFANAIIICNTTIVISILEDVSGPGALVRASDLIKGQTQVGLLIFLGSTIGLT 275
Query: 232 VISGMFS--VIVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQE 289
+ G+F V + GDG SR++ G LV + V L+ ++ +VFY+ C+SY +
Sbjct: 276 FVEGLFEHRVKSLSYGDG---SSRLWEGPLLVVMYSFVVLIDTMMSAVFYFSCRSYSMEA 332
Query: 290 ID 291
++
Sbjct: 333 VE 334
>At2g18690 unknown protein
Length = 322
Score = 79.7 bits (195), Expect = 2e-15
Identities = 73/302 (24%), Positives = 143/302 (47%), Gaps = 29/302 (9%)
Query: 15 NILKESISIPKISPKTFY----LITLILIFPLSFAILAHSLFTHPLISHIES-------- 62
N++K+ ++I S K F L+ +L+FPL L + L ++ I +
Sbjct: 8 NVVKDVVAILNESRKLFLKNKKLMFSVLVFPLLLNCLVYFLNIFVIVPEITNLILEASLL 67
Query: 63 PFTDPAQTSHDWTLLLIIQFF--YLIFLFAFSLLSTAAVVFTV-----ASLYTSKPVSFS 115
P TDP + L+ + F ++ + F+ +S+ +F+V AS T K +F+
Sbjct: 68 PSTDPTSPEYAARLMRVFTDFRQFVGSSYIFAAVSSIINLFSVLVIVHASAITLKDENFN 127
Query: 116 NTISAIPNV--FKRLFITFLWVTLLMFCYNFVF--ILCLVLM--VIAVDTDNSVLLFFSV 169
+ ++ +K +T+ ++ L + F+F ILC +L+ + + +N L
Sbjct: 128 IKDFPVLSLKSWKGPLVTYFYIALFSLGFGFLFFIILCPILLFSIKSGSVENIGFLAVEA 187
Query: 170 VLIFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFL 229
++ ++F V Y W+L+ V+S+LE YGF A+ K+ +++KG LL LF
Sbjct: 188 GVLLIIFTVSQSYFAIYWNLSMVISILEESYGFQALGKAAKIVKGMKTKLFLL---NLFF 244
Query: 230 CAVISGMFSVIVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQE 289
+ SG+ ++ + G + + G LV ++ V + L+ +V Y+ CKS ++
Sbjct: 245 GLLASGLAQILQLI-NMGRSLAVTLTTGFVLVCLVFAVRMFQLVTYTVAYFQCKSLQGRD 303
Query: 290 ID 291
++
Sbjct: 304 VE 305
>At3g11810 unknown protein
Length = 348
Score = 65.9 bits (159), Expect = 3e-11
Identities = 65/305 (21%), Positives = 128/305 (41%), Gaps = 34/305 (11%)
Query: 11 LNIPNILKESISIPKISPKTFYLITLILIFPLSFAILAH----------SLFTHPLISHI 60
LN+ IL ES I + F ++++ + PL F+I + S +H +S +
Sbjct: 13 LNLWIILSESKRIINAHSRHFLALSVLFLLPLCFSITVYPSVFLLITDQSSASHNTVSLL 72
Query: 61 ESPFTDPAQTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISA 120
+ D +++ Y++ + F+LL+ ++ ++V + +PV ++ + +
Sbjct: 73 RGGLHNNNDDDIDTKTTVLLVIGYIVVITVFNLLAIGSIAYSVFQGFYGRPVKLNSAVKS 132
Query: 121 IPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFS-------VVLIF 173
F L T L++ + + L+V ++T V ++ V L+
Sbjct: 133 SFASFLPLLATLTSSNLIVLGGFLIPGILAFLLVKLIETIPGVEFDYASSYFQGFVTLVT 192
Query: 174 VLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVI 233
++ + + V + W LA VV V+E +G +K+S L+KG ++ ASL + + I
Sbjct: 193 IISIAIAVKLYVNWILAWVVVVVESAWGITPLKRSKRLVKG-MKCASLSIIFFFASTESI 251
Query: 234 SGMFSVIVVH--------GG----DGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYV 281
S + + GG D + V + F L ++ L L +V Y
Sbjct: 252 LVWISTLAAYAQLNDNENGGKSWTDAFFVVQIVITSAF----LTLLTLYNLAATTVMYMY 307
Query: 282 CKSYH 286
CK+ H
Sbjct: 308 CKAVH 312
>At2g18680 unknown protein
Length = 287
Score = 65.5 bits (158), Expect = 4e-11
Identities = 73/275 (26%), Positives = 125/275 (44%), Gaps = 33/275 (12%)
Query: 37 ILIFPLSFAILAH---SLFTHPLISHI--ES---PFTDPAQTSHDWTLLLII----QFFY 84
+L+FPL L + ++ P I+++ ES P TDP L+ + QF
Sbjct: 4 VLVFPLLLNCLVYLFNAIAIKPEITNLILESSLLPMTDPNTPEFAAHLMRVFVDFRQFVN 63
Query: 85 LIFLFA-----FSLLSTAAVVFTVASLYTSKPVSFSNTISAIP----NVFKRLFITFLWV 135
+++F +LLST +V AS T K SF I P +K +T ++
Sbjct: 64 SLYIFIAVSSIINLLSTLVMVH--ASALTHKDDSFE--IKDFPILTLKYWKGPLVTNFYI 119
Query: 136 TLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVVHVYITALWHLASVVSV 195
L Y F+F + L +V +S L +F++F V Y+ +W+L+ V+S+
Sbjct: 120 VLFSLGYWFLFFIVLFSIVFFSTKLDS--LAAKSRALFIVFAVFESYLAIVWNLSMVISI 177
Query: 196 LEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMFSVI-VVHGGDGYDVFSRI 254
LE YG A+ K+ +++KG LL LF + G+ ++ +V + V +
Sbjct: 178 LEDTYGIQALGKAAKIVKGMKPKLFLL---NLFFGLLSFGLVQILRLVDWSSSFSV--TL 232
Query: 255 FIGGFLVGVLVIVNLVGLLVQSVFYYVCKSYHHQE 289
G L+ + +V + L+ +V Y+ CKS Q+
Sbjct: 233 TTGLVLMSSVFVVRMFQLVTYTVAYFQCKSLQGQD 267
>At4g16850 hypothetical protein
Length = 295
Score = 49.7 bits (117), Expect = 2e-06
Identities = 56/224 (25%), Positives = 98/224 (43%), Gaps = 32/224 (14%)
Query: 1 MDLAPEELQFLNIPNIL-----KESISIPKISPKTFYLITLILIFPL-SFAILAHSLFTH 54
+D+ +E QF+++ N + I +P + L L L L SF + H
Sbjct: 52 VDVNKDEKQFMHLWNSFVRKQRNKHILLPIFAFIAIPLAALHLSLTLTSFRLKNHVFRLE 111
Query: 55 PLISHIESPFTDPA---QTSHDWTLLLIIQFFYLIFLFAFSLLSTAAVVFTVASLYTSKP 111
L + + + F ++ D LL ++F Y + F S +++ V+ + + +
Sbjct: 112 ALANVVHTRFEARQIWQESREDAVSLLHLKFRYFVPSFILSCMASITVITSTSFSHQGIN 171
Query: 112 VSFSNTISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVL 171
S ++ +++ + + R+ T + +V A+ L + L
Sbjct: 172 PSLKSSFASVKSSWMRVTATSI-------------------IVYALVGYTPTLRY----L 208
Query: 172 IFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGR 215
I +LFL V VYI A+ L VVSVLE YGF A+K+ L+KGR
Sbjct: 209 ITILFLGVEVYIMAITGLGFVVSVLEERYGFDAIKEGTALMKGR 252
>At1g23850 unknown protein
Length = 354
Score = 43.5 bits (101), Expect = 2e-04
Identities = 60/256 (23%), Positives = 107/256 (41%), Gaps = 54/256 (21%)
Query: 78 LIIQFFYLIFLFAFSLL---STAAVVFTVASLYTSK--PVSFSN----TISAIPNVFKRL 128
L+IQ F L++LF + L+ +T +V +++TS+ P+ F T+ N +
Sbjct: 84 LLIQTF-LLYLFPYGLIDLFTTTTIVSASWTVHTSEEEPLRFGQLVRRTVEICQNRLEGC 142
Query: 129 FITFLWVTLLM--FCYNFVFILCLVLMVIAV-------------------------DTDN 161
IT L+V LL + F+F+ +I++ ++
Sbjct: 143 LITSLYVLLLSTPVFFGFLFVATNYFHIISLTGSGENSYYYSINIEEDAEGYYRSSSVNS 202
Query: 162 SVLLFFSVVL------IFVLFLVVHVYITALWHLASVVSVLEP------LYGFAAMKKSY 209
V + F VL IF+ L + +A W++ VVSVLE +YG A+ S
Sbjct: 203 PVKMLFDAVLAMFHGAIFLGLLAMFSKWSAGWNMGLVVSVLEEEENGQSIYGTDALTLSS 262
Query: 210 ELLKGRVRYASLLVCGYL-FLCAVISGMFSVIVVHGGDGYDVFSRIFIGGFLVGVLVIVN 268
KG + ++ +L F A+ F +G +R+ F VG++ + N
Sbjct: 263 NYGKGHEKRGLQVMLVFLVFAIAMRMPCFCFKCTESSNG----NRVLYTSFYVGLICVGN 318
Query: 269 LVGLLVQSVFYYVCKS 284
++ + VFY C++
Sbjct: 319 MIKWVACVVFYEDCRT 334
>At2g03330 unknown protein
Length = 385
Score = 41.6 bits (96), Expect = 6e-04
Identities = 35/135 (25%), Positives = 63/135 (45%), Gaps = 11/135 (8%)
Query: 169 VVLIFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLF 228
V L+ ++ + V V + W LA VV V+E +G +K S L+KG ++ ASL + +
Sbjct: 225 VTLVTIISIAVAVKLYVNWILAWVVVVVESAWGITPLKTSKRLVKG-MKCASLSIIFFFA 283
Query: 229 LCAVISGMFSVIVVH--------GGDGYDVFSRIFIGGFLV-GVLVIVNLVGLLVQSVFY 279
I S + + GG + S F+ ++ +L ++ L L+ +V Y
Sbjct: 284 STESILVWISTLAAYALLNDNENGGKSWK-GSFFFVQNWITSALLTLLMLYNLVATTVMY 342
Query: 280 YVCKSYHHQEIDKSA 294
CK+ H + + + A
Sbjct: 343 MYCKAVHGELVGEIA 357
>At1g15520 hypothetical protein
Length = 1423
Score = 35.4 bits (80), Expect = 0.042
Identities = 49/188 (26%), Positives = 78/188 (41%), Gaps = 26/188 (13%)
Query: 119 SAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLV 178
SA+P F ++FI +V + Y + + AV + + L F + +
Sbjct: 1244 SAMPYAFAQVFIEIPYVLVQAIVYGLIVYAMIGFEWTAVKFFWYLFFMYGSFLTFTFYGM 1303
Query: 179 VHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKG-RVRYASLLVC--GYLFLCAVISG 235
+ V +T H+ASVVS YG + L G + S+ V Y +LC V
Sbjct: 1304 MAVAMTPNHHIASVVS--SAFYGI------WNLFSGFLIPRPSMPVWWEWYYWLCPVAWT 1355
Query: 236 MFSVIVVHGGD----------GYDVFSRIFIG---GFLVGVLVIVNLV-GLLVQSVFYYV 281
++ +I GD F R F G GFL GV+ +N++ LL +F
Sbjct: 1356 LYGLIASQFGDITEPMADSNMSVKQFIREFYGYREGFL-GVVAAMNVIFPLLFAVIFAIG 1414
Query: 282 CKSYHHQE 289
KS++ Q+
Sbjct: 1415 IKSFNFQK 1422
>At5g49960 putative protein
Length = 824
Score = 35.0 bits (79), Expect = 0.054
Identities = 26/91 (28%), Positives = 50/91 (54%), Gaps = 11/91 (12%)
Query: 158 DTDNSVLLFFSVVLIFVLFLVVHVYITALWHLASVVSVLEPLYGFAAMKKSYELLKGRVR 217
+T++ ++FFSV++ FVL ++++Y+ L H+ +++ KK LK R+
Sbjct: 130 ETNSRAVVFFSVIITFVLPFLLYMYLDDLSHVKNLLRRTN-------QKKEDVPLKKRLA 182
Query: 218 YASLLVCGYLFLCAVISGMF---SVIVVHGG 245
Y SL VC ++ A + + V++V+GG
Sbjct: 183 Y-SLDVCFSVYPYAKLLALLLATVVLIVYGG 212
>At1g23830 unknown protein
Length = 345
Score = 34.3 bits (77), Expect = 0.093
Identities = 54/242 (22%), Positives = 94/242 (38%), Gaps = 48/242 (19%)
Query: 84 YLIFLFAFS---LLSTAAVVFTVASLYTSK--PVSF----SNTISAIPNVFKRLFITFLW 134
+L++ F ++ LL+T +V + +YTSK P+ +I N IT L+
Sbjct: 94 FLLYFFPYTILDLLTTTTIVAASSIVYTSKEEPLGLLYLVERSIKICQNRVGGCLITSLY 153
Query: 135 VTLLMFCYNFVFILCLVLMVIAVDTDNSVLLF---------------FSVVLIFVLFLVV 179
V L F L L ++ T+ + + F+VV+ L +
Sbjct: 154 VLLWSTSVFLFFFLFFFLQFLSGSTNYVSIPYLSREYKGFHYQPTGLFNVVVPLTLLMQC 213
Query: 180 HVYITAL---------WHLASVVSVLEP------LYGFAAMKKSYELLKGRVRYASLLVC 224
++I W++ VVSVLE +YG A+ S KG + L+
Sbjct: 214 TLFIVLTAKYSKWSSGWNMGLVVSVLEEDEDGQGIYGGDALSLSGWYRKGHEKRDLWLML 273
Query: 225 GYLF--LCAVISGMFSVIVVHGGDGYDVFSRIFIGGFLVGVLVIVNLVGLLVQSVFYYVC 282
+L L + ++S G + GF VG++ + NL+ + Y+ C
Sbjct: 274 MFLVFGLATRMPCLYSKCSASGNG-------VMYTGFYVGLICVGNLLKWVTCLACYHDC 326
Query: 283 KS 284
K+
Sbjct: 327 KT 328
>At5g61730 ABC transport protein - like
Length = 940
Score = 32.3 bits (72), Expect = 0.35
Identities = 30/121 (24%), Positives = 58/121 (47%), Gaps = 12/121 (9%)
Query: 78 LIIQFFYL---IFLFAFSLLSTAAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFITFLW 134
+II FYL +F AFS+ V + S+ T K + ++ + V++ + W
Sbjct: 222 VIISAFYLMGPVFFLAFSMFG---FVLQLGSVVTEKELKLREAMTTM-GVYESAY----W 273
Query: 135 VTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVVHVYITAL-WHLASVV 193
++ L++ F+ L L++ + L S VL+F+LF + + L + L+S++
Sbjct: 274 LSWLIWEGILTFVSSLFLVLFGMMFQFEFFLKNSFVLVFLLFFLFQFNMIGLAFALSSII 333
Query: 194 S 194
S
Sbjct: 334 S 334
>At1g18940 hypothetical protein
Length = 526
Score = 30.8 bits (68), Expect = 1.0
Identities = 19/71 (26%), Positives = 36/71 (49%), Gaps = 1/71 (1%)
Query: 10 FLNIPNILKESISIPK-ISPKTFYLITLILIFPLSFAILAHSLFTHPLISHIESPFTDPA 68
+L I ILK ++S+P + T ++ ++L PL A+ AH +S + SP D
Sbjct: 230 YLMITIILKSTLSLPSWANAVTLAVLLVLLSSPLLVAVRAHRDSIEKPLSSVYSPLVDNL 289
Query: 69 QTSHDWTLLLI 79
+ + +L++
Sbjct: 290 EATTSGEILML 300
>At4g23630 unknown protein
Length = 275
Score = 30.4 bits (67), Expect = 1.3
Identities = 27/117 (23%), Positives = 56/117 (47%), Gaps = 14/117 (11%)
Query: 131 TFLWVTLLMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFV-----LFLVVHVYITA 185
T WV + Y+ + +LC V++V+ +VL +S +F+ VH+
Sbjct: 109 TAAWVVFELMEYHLLTLLCHVMIVVL-----AVLFLWSNATMFINKSPPKIPEVHIPEEP 163
Query: 186 LWHLASVVSVLEPLYGFAAMKKSYELLKGRVRYASLLVCGYLFLCAVISGMFSVIVV 242
+ LAS + + E GF++++ E+ GR L+ L++ +++ G F+ + +
Sbjct: 164 ILQLASGLRI-EINRGFSSLR---EIASGRDLKKFLIAIAGLWVLSILGGCFNFLTL 216
>At5g55960 unknown protein
Length = 648
Score = 30.0 bits (66), Expect = 1.7
Identities = 35/155 (22%), Positives = 67/155 (42%), Gaps = 30/155 (19%)
Query: 97 AAVVFTVASLYTSKPVSFSNTISAIPNVFKRLFITFLWVTLLMFCYNFVFILCLVLMVIA 156
A VF++ +L S F N IS + + F+WV ++ + V+ ++
Sbjct: 435 AKFVFSIGNLIISGAAEFFNFISQL--------MIFIWVLYILITSESGGVTEQVMNMLP 486
Query: 157 VDTD-------------NSVLL------FFSVVLIFVLFLVVHVYITALWHLASVVSVLE 197
++ + VLL FF L ++LF + +++ + + + +S L
Sbjct: 487 INASARNRCVEVLDLAISGVLLATAEIAFFQGCLTWLLFRLYNIHFLYMSTVLAFISALL 546
Query: 198 PL--YGFAAMKKSYEL-LKGRVRYASLLVCGYLFL 229
P+ Y FA + + +L L+GR A +L +L L
Sbjct: 547 PIFPYWFATIPAALQLVLEGRYIVAVILSVTHLVL 581
>At4g32710 putative protein kinase
Length = 731
Score = 30.0 bits (66), Expect = 1.7
Identities = 21/55 (38%), Positives = 28/55 (50%), Gaps = 10/55 (18%)
Query: 257 GGFLVGVLVIVNLVGLLVQSVFYYVCK-------SYHHQEIDKSALHDHLGGYLG 304
GG +VG L+++ L+GLL VFY + S HHQ S + H GG G
Sbjct: 284 GGVIVGALLLI-LLGLLF--VFYRATRNRNNNSSSAHHQSKTPSKVQHHRGGNAG 335
>At3g57170 unknown protein
Length = 560
Score = 30.0 bits (66), Expect = 1.7
Identities = 32/138 (23%), Positives = 59/138 (42%), Gaps = 21/138 (15%)
Query: 91 FSLLSTAAVVFTVASLYTSKPV-SFSNTISAIPNVFK------------RLFITFLWVTL 137
F+ S + VF A +T+ + S S + +P FK L + +W TL
Sbjct: 229 FNTESVCSFVFDFAKEFTNGILRSGSVWLMGVPAGFKLNTELAGVLGMVSLNVIQIWSTL 288
Query: 138 LMFCYNFVFILCLVLMVIAVDTDNSVLLFFSVVLIFVLFLVVHVYITALWHLASVVSVLE 197
+F +F+F L V+ ++ + +V F V+ + F +H+ + W + V S
Sbjct: 289 WVFMASFIFCLIRVIAILGITFGATVSAAF--VIDVITFATLHI-MALHWAITLVYS--- 342
Query: 198 PLYGFAAMKKSYELLKGR 215
+ A+ + L +GR
Sbjct: 343 --HQIQALAALWRLFRGR 358
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.332 0.146 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,747,679
Number of Sequences: 26719
Number of extensions: 272766
Number of successful extensions: 1029
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 994
Number of HSP's gapped (non-prelim): 38
length of query: 320
length of database: 11,318,596
effective HSP length: 99
effective length of query: 221
effective length of database: 8,673,415
effective search space: 1916824715
effective search space used: 1916824715
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146651.4


