

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.2 - phase: 0
(384 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
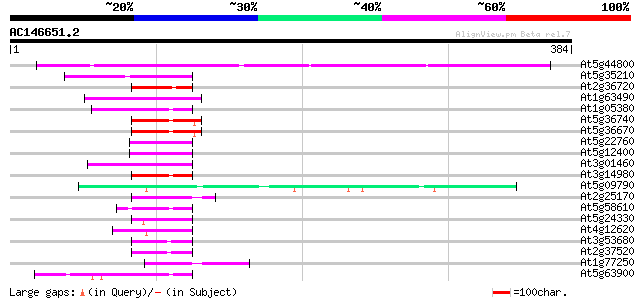
Score E
Sequences producing significant alignments: (bits) Value
At5g44800 helicase-like protein 191 6e-49
At5g35210 putative protein 60 2e-09
At2g36720 putative PHD-type zinc finger protein 59 3e-09
At1g63490 RB-binding protein -like 59 6e-09
At1g05380 unknown protein 59 6e-09
At5g36740 putative protein 58 8e-09
At5g36670 putative protein 58 8e-09
At5g22760 putative protein 57 1e-08
At5g12400 putative protein 56 3e-08
At3g01460 unknown protein 55 5e-08
At3g14980 PHD-finger protein, putative 54 1e-07
At5g09790 unknown protein 54 2e-07
At2g25170 putative chromodomain-helicase-DNA-binding protein 53 3e-07
At5g58610 putative protein 50 2e-06
At5g24330 putative protein 48 8e-06
At4g12620 origin recognition complex subunit 1 -like protein 47 2e-05
At3g53680 putative protein 47 2e-05
At2g37520 unknown protein 47 2e-05
At1g77250 unknown protein 47 2e-05
At5g63900 putative protein 45 5e-05
>At5g44800 helicase-like protein
Length = 2228
Score = 191 bits (485), Expect = 6e-49
Identities = 128/354 (36%), Positives = 189/354 (53%), Gaps = 9/354 (2%)
Query: 19 LKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHD 78
+K+KRRKLP D K S+ D+ + SS+ + K+ LKT+ + SSK+KG+D
Sbjct: 1 MKQKRRKLPSILDILDQKVDSSMAFDSPEYTSSSKPS--KQRLKTDSTPERNSSKRKGND 58
Query: 79 GYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNL 138
G ++ECVICDLGG+LLCCDSCPRTYH CL+PPLKRIP GKW CP C ++ LKP+N L
Sbjct: 59 GNYFECVICDLGGDLLCCDSCPRTYHTACLNPPLKRIPNGKWICPKCSPNSEALKPVNRL 118
Query: 139 DSISRRARTKTVPVKSKAGVNPVNLEKVSGIFGNKHISKKRST-KAKSISTMGGKFFGMK 197
D+I++RARTKT SKAG E+ S I+ + IS ++S+ K KSIS K G +
Sbjct: 119 DAIAKRARTKTKKRNSKAG---PKCERASQIYCSSIISGEQSSEKGKSISAEESKSTGKE 175
Query: 198 PVLSPVDATCSDKPMDPSLESCMEGTSCADADEKNLNLSPTV-APMDRMSVSPDKEVLSP 256
SP+D CS + + +S D+ + PT P D E LS
Sbjct: 176 VYSSPMDG-CSTAELGHASADDRPDSSSHGEDDLGKPVIPTADLPSDAGLTLLSCEDLSE 234
Query: 257 SKITNLDANDDLLEEKPDLSCDKIPFRKTLVLAITVGGEEMGKRKHKVIGDNANQKKRRT 316
SK+++ + + EK + + +I KT V + G + KRK ++ + ++ +
Sbjct: 235 SKLSDTEKTHEAPVEKLEHASSEIVENKT-VAEMETGKGKRKKRKRELNDGESLERCKTD 293
Query: 317 EKGKKVVITPIKSKSGNNKVQTKQKSKTHKISISASKGDVGKKKSDAQQKDKVL 370
+K K ++ + S S K K K K ++ + K++ +K K L
Sbjct: 294 KKRAKKSLSKVGSSSQTTKSPESSKKKKKKNRVTLKSLSKPQSKTETPEKVKKL 347
>At5g35210 putative protein
Length = 1606
Score = 60.1 bits (144), Expect = 2e-09
Identities = 29/88 (32%), Positives = 42/88 (46%), Gaps = 3/88 (3%)
Query: 38 QSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCD 97
+++ ++ V S+ + T G+ + SS G+ EC IC + G LLCCD
Sbjct: 371 KTSAYKEKEVTDSSTNESKDLDSRCTNGGSNEVSSDLDGNSD---ECRICGMDGTLLCCD 427
Query: 98 SCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
CP YH +C+ IP G W CP C
Sbjct: 428 GCPLAYHSRCIGVVKMYIPDGPWFCPEC 455
>At2g36720 putative PHD-type zinc finger protein
Length = 958
Score = 59.3 bits (142), Expect = 3e-09
Identities = 24/42 (57%), Positives = 28/42 (66%), Gaps = 2/42 (4%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
CVIC GGNLL CDSCPR +H++C+ P IP G W C C
Sbjct: 580 CVICADGGNLLLCDSCPRAFHIECVSLP--SIPRGNWHCKYC 619
Score = 36.2 bits (82), Expect = 0.031
Identities = 17/48 (35%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Query: 93 LLCCDSCPRTYHLQCLDP----PLKRIPMGKWQCP-SCFEENDQLKPL 135
++ CD C + YH+ CL LK +P G W C C N L+ L
Sbjct: 675 IIICDQCEKEYHIGCLSSQNIVDLKELPKGNWFCSMDCTRINSTLQKL 722
>At1g63490 RB-binding protein -like
Length = 1458
Score = 58.5 bits (140), Expect = 6e-09
Identities = 27/80 (33%), Positives = 41/80 (50%)
Query: 52 SRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPP 111
S S S+KR + + ++G D +C G +L CDSC + +H+ CL PP
Sbjct: 209 SESRSSKRRKRNADVKNPKVENEEGVDQACEQCKSDKHGEVMLLCDSCNKGWHIYCLSPP 268
Query: 112 LKRIPMGKWQCPSCFEENDQ 131
LK IP+G W C C +++
Sbjct: 269 LKHIPLGNWYCLECLNTDEE 288
>At1g05380 unknown protein
Length = 1138
Score = 58.5 bits (140), Expect = 6e-09
Identities = 28/69 (40%), Positives = 34/69 (48%), Gaps = 2/69 (2%)
Query: 57 AKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIP 116
A M K A G D C IC GG+L+CCD CP TYH CL ++ +P
Sbjct: 601 AWNMQKDATNLALHQVDTDGDDPNDDACGICGDGGDLICCDGCPSTYHQNCLG--MQVLP 658
Query: 117 MGKWQCPSC 125
G W CP+C
Sbjct: 659 SGDWHCPNC 667
>At5g36740 putative protein
Length = 960
Score = 58.2 bits (139), Expect = 8e-09
Identities = 25/51 (49%), Positives = 32/51 (62%), Gaps = 5/51 (9%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC---FEENDQ 131
C IC GG+L+CCD CP T+H CLD +K+ P G W C +C F E D+
Sbjct: 653 CGICGDGGDLICCDGCPSTFHQSCLD--IKKFPSGAWYCYNCSCKFCEKDE 701
>At5g36670 putative protein
Length = 1030
Score = 58.2 bits (139), Expect = 8e-09
Identities = 25/51 (49%), Positives = 32/51 (62%), Gaps = 5/51 (9%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC---FEENDQ 131
C IC GG+L+CCD CP T+H CLD +K+ P G W C +C F E D+
Sbjct: 500 CGICGDGGDLICCDGCPSTFHQSCLD--IKKFPSGAWYCYNCSCKFCEKDE 548
>At5g22760 putative protein
Length = 1516
Score = 57.4 bits (137), Expect = 1e-08
Identities = 21/43 (48%), Positives = 25/43 (57%)
Query: 83 ECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
EC +C + G LLCCD CP YH +C+ IP G W CP C
Sbjct: 415 ECRLCGMDGTLLCCDGCPLAYHSRCIGVVKMYIPDGPWYCPEC 457
>At5g12400 putative protein
Length = 1595
Score = 56.2 bits (134), Expect = 3e-08
Identities = 19/43 (44%), Positives = 25/43 (57%)
Query: 83 ECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
+C C + G+LLCCD CP YH +C+ +P G W CP C
Sbjct: 610 DCCFCKMDGSLLCCDGCPAAYHSKCVGLASHLLPEGDWYCPEC 652
>At3g01460 unknown protein
Length = 2176
Score = 55.5 bits (132), Expect = 5e-08
Identities = 27/72 (37%), Positives = 36/72 (49%)
Query: 54 SASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLK 113
SA K+ +K + K +G C + ++L CD+C YH CL+PPL
Sbjct: 1263 SAEMKKEIKDIVVSVNKLPKAPWDEGVCKVCGVDKDDDSVLLCDTCDAEYHTYCLNPPLI 1322
Query: 114 RIPMGKWQCPSC 125
RIP G W CPSC
Sbjct: 1323 RIPDGNWYCPSC 1334
Score = 36.6 bits (83), Expect = 0.024
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 2/45 (4%)
Query: 93 LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFE--ENDQLKPL 135
++ CD+C R +H+ C++ ++ P W C C E +L PL
Sbjct: 98 VVVCDACERGFHMSCVNDGVEAAPSADWMCSDCRTGGERSKLWPL 142
>At3g14980 PHD-finger protein, putative
Length = 1189
Score = 53.9 bits (128), Expect = 1e-07
Identities = 20/42 (47%), Positives = 27/42 (63%), Gaps = 2/42 (4%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C +C GG L+CCD+CP T+H CL ++ +P G W C SC
Sbjct: 729 CGVCGDGGELICCDNCPSTFHQACLS--MQVLPEGSWYCSSC 768
>At5g09790 unknown protein
Length = 352
Score = 53.5 bits (127), Expect = 2e-07
Identities = 67/321 (20%), Positives = 124/321 (37%), Gaps = 34/321 (10%)
Query: 48 ASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGN------LLCCDSCPR 101
+SES K M + + +++ D Y V C+ G+ LL CD C R
Sbjct: 28 SSESPPPRKMKSMAEIMAKSVPVVEQEEEEDEDSYSNVTCEKCGSGEGDDELLLCDKCDR 87
Query: 102 TYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLDSISRRARTKTVPVKSKAGVNPV 161
+H++CL P + R+P+G W C C ++ +P+ RR+ + TV + + + V
Sbjct: 88 GFHMKCLRPIVVRVPIGTWLCVDCSDQ----RPVRKETRKRRRSCSLTVKKRRRKLLPLV 143
Query: 162 NLEKVSGIFGNKHISKKRSTKAKSISTMGGKF---FGMKPVLSPVDATCSDKPMDPSLES 218
E + T A +++ +G K+ P ++P A S
Sbjct: 144 PSEDPDQRLA------QMGTLASALTALGIKYSDGLNYVPGMAPRSANQSKLEKGGMQVL 197
Query: 219 CMEGTSCADADE---KNLNLSPTVA---PMDRMSVSPDKEVLSPSKITNLDANDDLLEEK 272
C E + + + P V P++ +V D + + I + D L+ +
Sbjct: 198 CKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGDVDYLKNR 257
Query: 273 PDLSCDKIPFRKTLVLA------ITVGGEEMGKRKHKVIGDNANQKKRRTEKGKKVVITP 326
CD I TL+L+ + + ++ G + G N + + ++ K V
Sbjct: 258 EKDDCDSI---MTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKCVRYS 314
Query: 327 IKSKSGNNKVQTKQKSKTHKI 347
I + V T+ SK ++
Sbjct: 315 INGECRVLLVATRDISKGERL 335
>At2g25170 putative chromodomain-helicase-DNA-binding protein
Length = 1359
Score = 52.8 bits (125), Expect = 3e-07
Identities = 22/58 (37%), Positives = 31/58 (52%), Gaps = 6/58 (10%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLDSI 141
C C NL+ C++C +H +CL PPLK + W+CP C + PLN +D I
Sbjct: 52 CQACGESTNLVSCNTCTYAFHAKCLVPPLKDASVENWRCPEC------VSPLNEIDKI 103
>At5g58610 putative protein
Length = 1065
Score = 50.1 bits (118), Expect = 2e-06
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 4/52 (7%)
Query: 74 KKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
++G + F C +C GG L+ CD CP +H CL L+ +P G W C SC
Sbjct: 689 RQGENDVF--CSVCHYGGKLILCDGCPSAFHANCLG--LEDVPDGDWFCQSC 736
>At5g24330 putative protein
Length = 349
Score = 48.1 bits (113), Expect = 8e-06
Identities = 21/45 (46%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Query: 84 CVICDLG---GNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C C G LL CD C + +HL CL P L +P G W CPSC
Sbjct: 35 CEECSSGKQPAKLLLCDKCDKGFHLFCLRPILVSVPKGSWFCPSC 79
>At4g12620 origin recognition complex subunit 1 -like protein
Length = 813
Score = 47.0 bits (110), Expect = 2e-05
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 2/57 (3%)
Query: 71 SSKKKGHDGYFYECVICDLGGN--LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
S+ + D +C IC ++ CD C +HL+CL PPLK +P G W C C
Sbjct: 156 SNSDEEEDPEIEDCQICFKSDTNIMIECDDCLGGFHLKCLKPPLKEVPEGDWICQFC 212
>At3g53680 putative protein
Length = 839
Score = 47.0 bits (110), Expect = 2e-05
Identities = 19/42 (45%), Positives = 24/42 (56%), Gaps = 2/42 (4%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C IC GG+LL C CP+ +H CL + +P G W C SC
Sbjct: 489 CSICGNGGDLLLCAGCPQAFHTACL--KFQSMPEGTWYCSSC 528
Score = 36.2 bits (82), Expect = 0.031
Identities = 28/91 (30%), Positives = 38/91 (40%), Gaps = 18/91 (19%)
Query: 93 LLCCDSCPRTYHLQCLDP----PLKRIPMGKWQC-----------PSCFEENDQLKPLNN 137
++ CD C + YH+ CL LK IP KW C S Q P
Sbjct: 582 VILCDQCEKEYHVGCLRENELCDLKGIPQDKWFCCSDCSRIHRVLQSSASCGPQTIPTLL 641
Query: 138 LDSISRRARTKTVPVKSKAGVNPVNLEKVSG 168
LD+ISR+ R K + + + N V +SG
Sbjct: 642 LDTISRKYREKGIYIDNG---NTVEWRMLSG 669
>At2g37520 unknown protein
Length = 825
Score = 46.6 bits (109), Expect = 2e-05
Identities = 19/42 (45%), Positives = 24/42 (56%), Gaps = 2/42 (4%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C IC GG+LL C CP+ +H CL + +P G W C SC
Sbjct: 467 CSICGDGGDLLLCAGCPQAFHTACL--KFQSMPEGTWYCSSC 506
Score = 34.3 bits (77), Expect = 0.12
Identities = 23/73 (31%), Positives = 29/73 (39%), Gaps = 15/73 (20%)
Query: 93 LLCCDSCPRTYHLQCLDP----PLKRIPMGKWQCPSCFEE-----------NDQLKPLNN 137
++ CD C + YH+ CL LK IP KW C S Q P
Sbjct: 564 VILCDQCEKEYHVGCLRENGFCDLKEIPQEKWFCCSNCSRIHTAVQNSVSCGPQTLPTPL 623
Query: 138 LDSISRRARTKTV 150
LD I R+ R K +
Sbjct: 624 LDMICRKDREKGI 636
>At1g77250 unknown protein
Length = 522
Score = 46.6 bits (109), Expect = 2e-05
Identities = 19/72 (26%), Positives = 34/72 (46%), Gaps = 6/72 (8%)
Query: 93 LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLDSISRRARTKTVPV 152
++ CD C YH+ C+ PP + +P G+W C +C + + + K V
Sbjct: 417 IVLCDGCDDAYHIYCMRPPCESVPNGEWFCTAC------KAAILKVQKARKAFEKKMETV 470
Query: 153 KSKAGVNPVNLE 164
+ + G+ P NL+
Sbjct: 471 QKQKGIKPKNLQ 482
Score = 36.6 bits (83), Expect = 0.024
Identities = 17/46 (36%), Positives = 20/46 (42%), Gaps = 6/46 (13%)
Query: 86 ICDLGG------NLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
IC L G + L CD C YH+ C P K +P W C C
Sbjct: 242 ICKLCGEKAEARDCLACDHCEDMYHVSCAQPGGKGMPTHSWYCLDC 287
Score = 33.1 bits (74), Expect = 0.26
Identities = 16/45 (35%), Positives = 23/45 (50%), Gaps = 3/45 (6%)
Query: 88 DLGGNLLCCDS--CP-RTYHLQCLDPPLKRIPMGKWQCPSCFEEN 129
D GG + CD CP + YH++CL ++ +W C SC N
Sbjct: 363 DSGGKYITCDHPFCPHKYYHIRCLTSRQIKLHGVRWYCSSCLCRN 407
>At5g63900 putative protein
Length = 557
Score = 45.4 bits (106), Expect = 5e-05
Identities = 38/123 (30%), Positives = 52/123 (41%), Gaps = 19/123 (15%)
Query: 18 VLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSA----SAKRML----------KT 63
V + K+ IG + EQ K+ +ASE R S K++L K
Sbjct: 181 VFNGDQEKIRIGLNVDVIHEQQ--KKRRKMASEEIRRPKMVKSLKKVLQVMEKKQQKNKH 238
Query: 64 EEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIP-MGKWQC 122
E+ + +F K D C +C GG+LL CD CP +H CL L +P W C
Sbjct: 239 EKESLRFCRKDCSPDMNCDVCCVCHWGGDLLLCDGCPSAFHHACLG--LSSLPEEDLWFC 296
Query: 123 PSC 125
P C
Sbjct: 297 PCC 299
Score = 34.7 bits (78), Expect = 0.090
Identities = 17/55 (30%), Positives = 24/55 (42%), Gaps = 8/55 (14%)
Query: 78 DGYFYECVICDLGGN--------LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPS 124
D +F C CD+ G+ L+ C+ C R +HL CL + W C S
Sbjct: 292 DLWFCPCCCCDICGSMESPANSKLMACEQCQRRFHLTCLKEDSCIVSSRGWFCSS 346
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,108,064
Number of Sequences: 26719
Number of extensions: 417506
Number of successful extensions: 1642
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 1479
Number of HSP's gapped (non-prelim): 198
length of query: 384
length of database: 11,318,596
effective HSP length: 101
effective length of query: 283
effective length of database: 8,619,977
effective search space: 2439453491
effective search space used: 2439453491
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146651.2


