

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.1 - phase: 0
(935 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
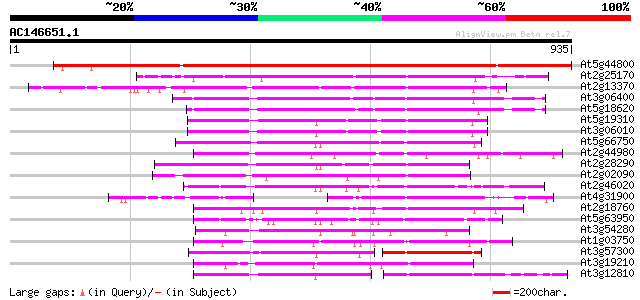
Score E
Sequences producing significant alignments: (bits) Value
At5g44800 helicase-like protein 1239 0.0
At2g25170 putative chromodomain-helicase-DNA-binding protein 515 e-146
At2g13370 pseudogene 423 e-118
At3g06400 putative ATPase (ISW2-like) 370 e-102
At5g18620 chromatin remodelling complex ATPase chain ISWI -like ... 369 e-102
At5g19310 homeotic gene regulator - like protein 341 1e-93
At3g06010 putative transcriptional regulator 341 1e-93
At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1) 317 2e-86
At2g44980 putative transcription activator 317 3e-86
At2g28290 putative SNF2 subfamily transcription regulator 297 2e-80
At2g02090 putative helicase 282 7e-76
At2g46020 putative SNF2 subfamily transcriptional activator 280 2e-75
At4g31900 putative protein 254 2e-67
At2g18760 putative SNF2/RAD54 family DNA repair and recombinatio... 246 3e-65
At5g63950 unknown protein 226 4e-59
At3g54280 TATA box binding protein (TBP) associated factor (TAF)... 213 3e-55
At1g03750 hypothetical protein 204 2e-52
At3g57300 helicase-like protein 197 2e-50
At3g19210 DNA repair protein, putative 185 1e-46
At3g12810 Snf2-related CBP activator protein, putative 180 3e-45
>At5g44800 helicase-like protein
Length = 2228
Score = 1239 bits (3205), Expect = 0.0
Identities = 624/876 (71%), Positives = 728/876 (82%), Gaps = 21/876 (2%)
Query: 74 ATDDLGSCARDGT----DQDDYAVSDEQLEKENDKLETEENLNVVLRGDGNSKLPNNCEM 129
++ + G +RD D DD A+ E + + +++ +E+ N L + + E
Sbjct: 433 SSSETGKSSRDSRLRDKDMDDSALGTEGMVEVKEEMLSEDISNATLSRHVDDEDMKVSET 492
Query: 130 HDSLE----------TKQKEVVLEKGMGSSGDNKVQDSIGEEVSYEFLVKWVGKSHIHNS 179
H S+E T +K V ++ + K D IGE VSYEFLVKWV KS+IHN+
Sbjct: 493 HVSVERELLEEAHQETGEKSTVADEEIEEPVAAKTSDLIGETVSYEFLVKWVDKSNIHNT 552
Query: 180 WISESHLKVIAKRKLENYKAKYGTATINICEEQWKNPERLLAIRTSKQGTSEAFVKWTGK 239
WISE+ LK +AKRKLENYKAKYGTA INICE++WK P+R++A+R SK+G EA+VKWTG
Sbjct: 553 WISEAELKGLAKRKLENYKAKYGTAVINICEDKWKQPQRIVALRVSKEGNQEAYVKWTGL 612
Query: 240 PYNECTWESLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQNDIVNLLEQP 299
Y+ECTWESL+EP+L++SSHLI F+ +E TLER SK N T++ + ++V L EQP
Sbjct: 613 AYDECTWESLEEPILKHSSHLIDLFHQYEQKTLERN-SKGNPTRE----RGEVVTLTEQP 667
Query: 300 KELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPC 359
+ELRGG+LF HQLEALNWLR+CW+KS+NVILADEMGLGKT+SA AF+SSLYFEF V+RPC
Sbjct: 668 QELRGGALFAHQLEALNWLRRCWHKSKNVILADEMGLGKTVSASAFLSSLYFEFGVARPC 727
Query: 360 LVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKF 419
LVLVPL TM NWL+EF+LWAP +NVV+YHG AK RAIIR YEWHA + +G KK +YKF
Sbjct: 728 LVLVPLSTMPNWLSEFSLWAPLLNVVEYHGSAKGRAIIRDYEWHAKNSTGTTKKPTSYKF 787
Query: 420 NVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPL 479
NVLLT+YEMVLAD SH RGVPWEVL+VDEGHRLKNSESKLFSLLN+ SFQHRVLLTGTPL
Sbjct: 788 NVLLTTYEMVLADSSHLRGVPWEVLVVDEGHRLKNSESKLFSLLNTFSFQHRVLLTGTPL 847
Query: 480 QNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQN 539
QNN+GEMYNLLNFLQP+SFPSLS+FEERF+DLTSAEKV+ELKKLV+PHMLRRLKKDAMQN
Sbjct: 848 QNNIGEMYNLLNFLQPSSFPSLSSFEERFHDLTSAEKVEELKKLVAPHMLRRLKKDAMQN 907
Query: 540 IPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYL 599
IPPKTERMVPVEL+SIQAEYYRAMLTKNYQILRNIGKG+AQQSMLNIVMQLRKVCNHPYL
Sbjct: 908 IPPKTERMVPVELTSIQAEYYRAMLTKNYQILRNIGKGVAQQSMLNIVMQLRKVCNHPYL 967
Query: 600 IPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLN 659
IPGTEP+SGS+EFLH+MRIKASAKLTLLHSMLK+L+KEGHRVLIFSQMTKLLDILEDYLN
Sbjct: 968 IPGTEPESGSLEFLHDMRIKASAKLTLLHSMLKVLHKEGHRVLIFSQMTKLLDILEDYLN 1027
Query: 660 IEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDS 719
IEFGPKT+ERVDGSV+V DRQ AIARFNQDK+RFVFLLSTR+CGLGINLATADTVIIYDS
Sbjct: 1028 IEFGPKTFERVDGSVAVADRQAAIARFNQDKNRFVFLLSTRACGLGINLATADTVIIYDS 1087
Query: 720 DFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQK 779
DFNPHADIQAMNRAHRIGQS RLLVYRLVVRASVEERILQLAKKKLMLDQLF KSGSQK
Sbjct: 1088 DFNPHADIQAMNRAHRIGQSKRLLVYRLVVRASVEERILQLAKKKLMLDQLFVNKSGSQK 1147
Query: 780 EVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCT 839
E EDIL+WGTEELFNDS N KDT+E+N + + + ++E K RK+ GGLGDVY+DKCT
Sbjct: 1148 EFEDILRWGTEELFNDSAGENKKDTAESNGNL--DVIMDLESKSRKKGGGLGDVYQDKCT 1205
Query: 840 DNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKALEWNDEPTEEHVEGES 899
+ + KI+WD+ AI+KLLDRSNLQ ASTD A+ + +NDMLGS+K +EWN+E EE V ES
Sbjct: 1206 EGNGKIVWDDIAIMKLLDRSNLQSASTDAADTELDNDMLGSVKPVEWNEETAEEQVGAES 1265
Query: 900 PPHGADDMCTQNSEKKEDNAVIGGEENEWDRLLRLR 935
P DD +SE+K+D+ V EENEWDRLLR+R
Sbjct: 1266 PALVTDDTGEPSSERKDDDVVNFTEENEWDRLLRMR 1301
>At2g25170 putative chromodomain-helicase-DNA-binding protein
Length = 1359
Score = 515 bits (1326), Expect = e-146
Identities = 307/715 (42%), Positives = 434/715 (59%), Gaps = 83/715 (11%)
Query: 212 QWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMFETLT 271
+W +R+LA R + G E VK+ Y+EC WES E + + I RF + T
Sbjct: 177 EWTTVDRILACR-EEDGELEYLVKYKELSYDECYWES--ESDISTFQNEIQRFKDVNSRT 233
Query: 272 LEREASKENSTKKSSDRQNDIVNLLEQPKELRG----------GSLFPHQLEALNWLRKC 321
SK+ K++ D P+ L+ G L P+QLE LN+LR
Sbjct: 234 ---RRSKDVDHKRNP---RDFQQFDHTPEFLKDVMIYLFPAIEGLLHPYQLEGLNFLRFS 287
Query: 322 WYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPD 381
W K +VILADEMGLGKTI + A ++SL+ E + P LV+ PL T+ NW EFA WAP
Sbjct: 288 WSKQTHVILADEMGLGKTIQSIALLASLFEENLI--PHLVIAPLSTLRNWEREFATWAPQ 345
Query: 382 VNVVQYHGCAKARAIIRQYEWHAS-DPSGLNKKTEAY----------KFNVLLTSYEMVL 430
+NVV Y G A+ARA+IR++E++ S D + KK KF+VLLTSYEM+
Sbjct: 346 MNVVMYFGTAQARAVIREHEFYLSKDQKKIKKKKSGQISSESKQKRIKFDVLLTSYEMIN 405
Query: 431 ADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLL 490
D + + + WE +IVDEGHRLKN +SKLFS L S HR+LLTGTPLQNNL E++ L+
Sbjct: 406 LDSAVLKPIKWECMIVDEGHRLKNKDSKLFSSLTQYSSNHRILLTGTPLQNNLDELFMLM 465
Query: 491 NFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPV 550
+FL F SL F+E F D+ E++ L K+++PH+LRR+KKD M+++PPK E ++ V
Sbjct: 466 HFLDAGKFGSLEEFQEEFKDINQEEQISRLHKMLAPHLLRRVKKDVMKDMPPKKELILRV 525
Query: 551 ELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSV 610
+LSS+Q EYY+A+ T+NYQ+L KG AQ S+ NI+M+LRKVC HPY++ G EP
Sbjct: 526 DLSSLQKEYYKAIFTRNYQVLTK--KGGAQISLNNIMMELRKVCCHPYMLEGVEPVIHDA 583
Query: 611 EFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERV 670
+ +++ KL LL M+ L ++GHRVLI++Q +LD+LEDY + YER+
Sbjct: 584 NEAFKQLLESCGKLQLLDKMMVKLKEQGHRVLIYTQFQHMLDLLEDYCTHK--KWQYERI 641
Query: 671 DGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQA 729
DG V +RQ I RFN ++ ++F FLLSTR+ GLGINLATADTVIIYDSD+NPHAD+QA
Sbjct: 642 DGKVGGAERQIRIDRFNAKNSNKFCFLLSTRAGGLGINLATADTVIIYDSDWNPHADLQA 701
Query: 730 MNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGK----SGSQKEVEDIL 785
M RAHR+GQ+N++++YRL+ R ++EER++QL KKK++L+ L GK + +Q+E++DI+
Sbjct: 702 MARAHRLGQTNKVMIYRLINRGTIEERMMQLTKKKMVLEHLVVGKLKTQNINQEELDDII 761
Query: 786 KWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKI 845
++G++ELF SE++ + K S KI
Sbjct: 762 RYGSKELF----------ASEDDEAGK-----------------------------SGKI 782
Query: 846 MWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA--LEWNDEPTEEHVEGE 898
+D+ AI KLLDR +L +A + + EN L + K E+ DE +E +
Sbjct: 783 HYDDAAIDKLLDR-DLVEAEEVSVDDEEENGFLKAFKVANFEYIDENEAAALEAQ 836
>At2g13370 pseudogene
Length = 1738
Score = 423 bits (1088), Expect = e-118
Identities = 300/872 (34%), Positives = 459/872 (52%), Gaps = 109/872 (12%)
Query: 32 QNVKSSDEGELHNTDRVEKIHVYRRSITKESKNGNLLNSLSKATDDLGSCAR----DGTD 87
Q+ S ++ E N + + RR T NG N++ ++++ S +
Sbjct: 337 QDDDSEEDSENDNDEGFRSLA--RRGTTLRQNNGRSTNTIGQSSEVRSSTRSVRKVSYVE 394
Query: 88 QDDYAVSDEQLEKENDKLETEENLNVVLRGDGNSKLPNNCEMHDSLETKQKEVVLEKGMG 147
+D D+ ++N K + EE V+ +L M + ++T K V
Sbjct: 395 SEDSEDIDDGKNRKNQKDDIEEEDADVIEKVLWHQLKG---MGEDVQTNNKSTVPVLV-- 449
Query: 148 SSGDNKVQDSIGEEVSYEFLVKWVGKSHIHNSWISESHLKVIAK-RKLENYKAK------ 200
+++ D+ + EFL+KW G+SH+H W + S L+ ++ +K+ NY K
Sbjct: 450 ----SQLFDTEPDWNEMEFLIKWKGQSHLHCQWKTLSDLQNLSGFKKVLNYTKKVTEEIR 505
Query: 201 YGTATI-------NICEE-------QWKNPERLLAIRTSKQGTS----EAFVKWTGKPYN 242
Y TA ++ +E Q ER++A R SK G E VKW G Y
Sbjct: 506 YRTALSREEIEVNDVSKEMDLDIIKQNSQVERIIADRISKDGLGDVVPEYLVKWQGLSYA 565
Query: 243 ECTWE----------SLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ--- 289
E TWE ++DE + S + + +E++ +K S+ +
Sbjct: 566 EATWEKDVDIAFAQVAIDEYKAREVSIAV------QGKMVEQQRTKGKGENSFSNAELWL 619
Query: 290 ----NDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAF 345
+ L EQP+ L GG+L +QLE LN+L W NVILADEMGLGKT+ + +
Sbjct: 620 LFSVASLRKLDEQPEWLIGGTLRDYQLEGLNFLVNSWLNDTNVILADEMGLGKTVQSVSM 679
Query: 346 ISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHAS 405
+ L ++ P LV+VPL T+ NW EF W P +N++ Y G +R + +
Sbjct: 680 LGFLQNTQQIPGPFLVVVPLSTLANWAKEFRKWLPGMNIIVYVGTRASREV-------RN 732
Query: 406 DPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNS 465
+ ++K KFN LLT+YE+VL D + + W L+VDE HRLKNSE++L++ L
Sbjct: 733 KTNDVHKVGRPIKFNALLTTYEVVLKDKAVLSKIKWIYLMVDEAHRLKNSEAQLYTALLE 792
Query: 466 ISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKL-- 523
S ++++L+TGTPLQN++ E++ LL+FL P F + F E + +L+S + EL L
Sbjct: 793 FSTKNKLLITGTPLQNSVEELWALLHFLDPGKFKNKDEFVENYKNLSSFNE-SELANLHL 851
Query: 524 -VSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIA--Q 580
+ PH+LRR+ KD +++PPK ER++ VE+S +Q +YY+ +L +N+ ++ KG+ Q
Sbjct: 852 ELRPHILRRVIKDVEKSLPPKIERILRVEMSPLQKQYYKWILERNFH---DLNKGVRGNQ 908
Query: 581 QSMLNIVMQLRKVCNHPYLIPGTEPDSG---SVEFLHEMRIKASAKLTLLHSMLKILYKE 637
S+LNIV++L+K CNHP+L + G + + I +S KL +L +L L +
Sbjct: 909 VSLLNIVVELKKCCNHPFLFESADHGYGGDINDNSKLDKIILSSGKLVILDKLLVRLRET 968
Query: 638 GHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSR-FVFL 696
HRVLIFSQM ++LDIL +YL++ ++R+DGS RQ A+ FN S F FL
Sbjct: 969 KHRVLIFSQMVRMLDILAEYLSLR--GFQFQRLDGSTKAELRQQAMDHFNAPASDDFCFL 1026
Query: 697 LSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEER 756
LSTR+ GLGINLATADTV+I+DSD+NP D+QAM+RAHRIGQ + +YR V SVEE
Sbjct: 1027 LSTRAGGLGINLATADTVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIYRFVTSKSVEEE 1086
Query: 757 ILQLAKKKLMLDQLF----------------KGKSGSQKEVEDILKWGTEELFNDSCALN 800
IL+ AK+K++LD L KG + + E+ IL++G EELF +
Sbjct: 1087 ILERAKRKMVLDHLVIQKLNAEGRLEKRETKKGSNFDKNELSAILRFGAEELFKED---K 1143
Query: 801 GKDTSENNNSNKD-----EAVAEVEHKHRKRT 827
+ S+ + D E +VE KH T
Sbjct: 1144 NDEESKKRLLSMDIDEILERAEQVEEKHTDET 1175
>At3g06400 putative ATPase (ISW2-like)
Length = 1057
Score = 370 bits (950), Expect = e-102
Identities = 233/637 (36%), Positives = 348/637 (54%), Gaps = 57/637 (8%)
Query: 272 LEREASKENSTKKSSDRQNDIVN--LLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVI 329
+ E E K+ D N LL QP ++G + +QL LNWL + + N I
Sbjct: 153 ITEEEEDEEYLKEEEDGLTGSGNTRLLTQPSCIQG-KMRDYQLAGLNWLIRLYENGINGI 211
Query: 330 LADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHG 389
LADEMGLGKT+ + ++ L+ ++ P +V+ P T+GNW+ E + P + V++ G
Sbjct: 212 LADEMGLGKTLQTISLLAYLHEYRGINGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLG 271
Query: 390 CAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEG 449
+ R IR+ A KF++ +TS+EM + + + R W +I+DE
Sbjct: 272 NPEERRHIRE------------DLLVAGKFDICVTSFEMAIKEKTALRRFSWRYIIIDEA 319
Query: 450 HRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFN 509
HR+KN S L + S +R+L+TGTPLQNNL E++ LLNFL P F S F+E F
Sbjct: 320 HRIKNENSLLSKTMRLFSTNYRLLITGTPLQNNLHELWALLNFLLPEIFSSAETFDEWFQ 379
Query: 510 ---DLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTK 566
+ E V +L K++ P +LRRLK D + +PPK E ++ V +S +Q +YY+A+L K
Sbjct: 380 ISGENDQQEVVQQLHKVLRPFLLRRLKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQK 439
Query: 567 NYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTL 626
+ L + G ++ +LNI MQLRK CNHPYL G EP G + I + K+ L
Sbjct: 440 D---LEAVNAGGERKRLLNIAMQLRKCCNHPYLFQGAEP--GPPYTTGDHLITNAGKMVL 494
Query: 627 LHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARF 686
L +L L + RVLIFSQMT+LLDILEDYL + Y R+DG+ +R +I +
Sbjct: 495 LDKLLPKLKERDSRVLIFSQMTRLLDILEDYLM--YRGYLYCRIDGNTGGDERDASIEAY 552
Query: 687 NQDKS-RFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVY 745
N+ S +FVFLLSTR+ GLGINLATAD VI+YDSD+NP D+QA +RAHRIGQ + V+
Sbjct: 553 NKPGSEKFVFLLSTRAGGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVF 612
Query: 746 RLVVRASVEERILQLAKKKLMLDQLF---------KGKSGSQKEVEDILKWGTEELFNDS 796
R +++EE++++ A KKL LD L K KS ++ E+ ++++G E +F+
Sbjct: 613 RFCTESAIEEKVIERAYKKLALDALVIQQGRLAEQKSKSVNKDELLQMVRYGAEMVFSSK 672
Query: 797 CALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLL 856
+ + + + +EA AE++ K + K T+++ + D++A
Sbjct: 673 DSTITDEDIDRIIAKGEEATAELDAKMK------------KFTEDAIQFKMDDSADFYDF 720
Query: 857 DRSNLQDASTDIAEGDSENDMLGSMKALEWNDEPTEE 893
D N + D + S+N WND P E
Sbjct: 721 DDDNKDENKLDFKKIVSDN----------WNDPPKRE 747
>At5g18620 chromatin remodelling complex ATPase chain ISWI -like
protein
Length = 1069
Score = 369 bits (946), Expect = e-102
Identities = 227/610 (37%), Positives = 342/610 (55%), Gaps = 53/610 (8%)
Query: 295 LLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFK 354
LL QP ++G L +QL LNWL + + N ILADEMGLGKT+ + ++ L+
Sbjct: 183 LLTQPACIQG-KLRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLAYLHEYRG 241
Query: 355 VSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKT 414
++ P +V+ P T+GNW+ E + P + V++ G + R IR+ +
Sbjct: 242 INGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLGNPEERRHIRE------------ELL 289
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
A KF++ +TS+EM + + + R W +I+DE HR+KN S L + S +R+L+
Sbjct: 290 VAGKFDICVTSFEMAIKEKTTLRRFSWRYIIIDEAHRIKNENSLLSKTMRLFSTNYRLLI 349
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFN---DLTSAEKVDELKKLVSPHMLRR 531
TGTPLQNNL E++ LLNFL P F S F+E F + E V +L K++ P +LRR
Sbjct: 350 TGTPLQNNLHELWALLNFLLPEVFSSAETFDEWFQISGENDQQEVVQQLHKVLRPFLLRR 409
Query: 532 LKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLR 591
LK D + +PPK E ++ V +S +Q +YY+A+L K+ +++ G+ ++ +LNI MQLR
Sbjct: 410 LKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEVVNGGGE---RKRLLNIAMQLR 466
Query: 592 KVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
K CNHPYL G EP G + + + K+ LL +L L RVLIFSQMT+LL
Sbjct: 467 KCCNHPYLFQGAEP--GPPYTTGDHLVTNAGKMVLLDKLLPKLKDRDSRVLIFSQMTRLL 524
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKS-RFVFLLSTRSCGLGINLAT 710
DILEDYL + Y R+DG+ +R +I +N+ S +FVFLLSTR+ GLGINLAT
Sbjct: 525 DILEDYLM--YRGYQYCRIDGNTGGDERDASIEAYNKPGSEKFVFLLSTRAGGLGINLAT 582
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
AD VI+YDSD+NP D+QA +RAHRIGQ + V+R ++E ++++ A KKL LD L
Sbjct: 583 ADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTENAIEAKVIERAYKKLALDAL 642
Query: 771 F--KGKSGSQK-----EVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
+G+ QK E+ ++++G E +F+ + + + + +EA AE++ K
Sbjct: 643 VIQQGRLAEQKTVNKDELLQMVRYGAEMVFSSKDSTITDEDIDRIIAKGEEATAELDAKM 702
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
+ K T+++ + D++A D N ++ D + SEN
Sbjct: 703 K------------KFTEDAIQFKMDDSADFYDFDDDNKDESKVDFKKIVSEN-------- 742
Query: 884 LEWNDEPTEE 893
WND P E
Sbjct: 743 --WNDPPKRE 750
>At5g19310 homeotic gene regulator - like protein
Length = 1041
Score = 341 bits (874), Expect = 1e-93
Identities = 211/522 (40%), Positives = 301/522 (57%), Gaps = 44/522 (8%)
Query: 297 EQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVS 356
+QP L+GG L +QLE L W+ + N ILADEMGLGKTI A I+ L +
Sbjct: 353 KQPSLLQGGELRSYQLEGLQWMVSLYNNDYNGILADEMGLGKTIQTIALIAYLLESKDLH 412
Query: 357 RPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEA 416
P L+L P + NW EFALWAP ++ Y G + R IR +
Sbjct: 413 GPHLILAPKAVLPNWENEFALWAPSISAFLYDGSKEKRTEIRA-------------RIAG 459
Query: 417 YKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNS-ISFQHRVLLT 475
KFNVL+T Y++++ D + + + W +IVDEGHRLKN E L L + + R+LLT
Sbjct: 460 GKFNVLITHYDLIMRDKAFLKKIDWNYMIVDEGHRLKNHECALAKTLGTGYRIKRRLLLT 519
Query: 476 GTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFN---------DLTSAEK---VDELKKL 523
GTP+QN+L E+++LLNFL P F S+ FEE FN LT E+ ++ L +
Sbjct: 520 GTPIQNSLQELWSLLNFLLPHIFNSIHNFEEWFNTPFAECGSASLTDEEELLIINRLHHV 579
Query: 524 VSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSM 583
+ P +LRR K + + +P KT+ ++ ++S+ Q YY+ +T ++ + G G +S+
Sbjct: 580 IRPFLLRRKKSEVEKFLPGKTQVILKCDMSAWQKLYYK-QVTDVGRVGLHSGNG-KSKSL 637
Query: 584 LNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLI 643
N+ MQLRK CNHPYL G + + + ++AS K LL +L L K GHR+L+
Sbjct: 638 QNLTMQLRKCCNHPYLFVGADYNMCKKPEI----VRASGKFELLDRLLPKLKKAGHRILL 693
Query: 644 FSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQ-DKSRFVFLLSTRSC 702
FSQMT+L+D+LE YL++ Y R+DGS R + +FN+ D F+FLLSTR+
Sbjct: 694 FSQMTRLIDLLEIYLSLN--DYMYLRLDGSTKTDQRGILLKQFNEPDSPYFMFLLSTRAG 751
Query: 703 GLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAK 762
GLG+NL TADT+II+DSD+NP D QA +RAHRIGQ + V+ LV S+EE IL+ AK
Sbjct: 752 GLGLNLQTADTIIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSIGSIEEVILERAK 811
Query: 763 KKLMLDQ------LFKGKSGSQ---KEVEDILKWGTEELFND 795
+K+ +D LF S +Q + +E+I+ GT L D
Sbjct: 812 QKMGIDAKVIQAGLFNTTSTAQDRREMLEEIMSKGTSSLGED 853
>At3g06010 putative transcriptional regulator
Length = 1132
Score = 341 bits (874), Expect = 1e-93
Identities = 213/522 (40%), Positives = 300/522 (56%), Gaps = 43/522 (8%)
Query: 297 EQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVS 356
EQP L GG L +QLE L W+ + + N ILADEMGLGKTI + I+ L V
Sbjct: 423 EQPSLLEGGELRSYQLEGLQWMVSLFNNNLNGILADEMGLGKTIQTISLIAYLLENKGVP 482
Query: 357 RPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEA 416
P L++ P + NW+ EFA W P + Y G + R IR+ K
Sbjct: 483 GPYLIVAPKAVLPNWVNEFATWVPSIAAFLYDGRLEERKAIRE------------KIAGE 530
Query: 417 YKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLF-SLLNSISFQHRVLLT 475
KFNVL+T Y++++ D + + + W +IVDEGHRLKN ES L +LL + R+LLT
Sbjct: 531 GKFNVLITHYDLIMRDKAFLKKIEWYYMIVDEGHRLKNHESALAKTLLTGYRIKRRLLLT 590
Query: 476 GTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFN---------DLTSAEK---VDELKKL 523
GTP+QN+L E+++LLNFL P F S+ FEE FN LT E+ + L +
Sbjct: 591 GTPIQNSLQELWSLLNFLLPHIFNSVQNFEEWFNAPFADRGNVSLTDEEELLIIHRLHHV 650
Query: 524 VSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSM 583
+ P +LRR K + + +P KT+ ++ ++S+ Q YY+ +T ++ G G +S+
Sbjct: 651 IRPFILRRKKDEVEKFLPGKTQVILKCDMSAWQKVYYK-QVTDMGRVGLQTGSG-KSKSL 708
Query: 584 LNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLI 643
N+ MQLRK CNHPYL G + + + ++AS K LL +L L K GHR+L+
Sbjct: 709 QNLTMQLRKCCNHPYLFVGGDYNMWKKPEI----VRASGKFELLDRLLPKLRKAGHRILL 764
Query: 644 FSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQ-DKSRFVFLLSTRSC 702
FSQMT+L+D+LE YL + Y R+DG+ R + +FN+ D F+FLLSTR+
Sbjct: 765 FSQMTRLIDVLEIYLTLN--DYKYLRLDGTTKTDQRGLLLKQFNEPDSPYFMFLLSTRAG 822
Query: 703 GLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAK 762
GLG+NL TADTVII+DSD+NP D QA +RAHRIGQ + V+ LV SVEE IL+ AK
Sbjct: 823 GLGLNLQTADTVIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSVGSVEEVILERAK 882
Query: 763 KKLMLDQ------LFKGKSGSQ---KEVEDILKWGTEELFND 795
+K+ +D LF S +Q + +E+I++ GT L D
Sbjct: 883 QKMGIDAKVIQAGLFNTTSTAQDRREMLEEIMRKGTSSLGTD 924
>At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1)
Length = 764
Score = 317 bits (813), Expect = 2e-86
Identities = 209/542 (38%), Positives = 298/542 (54%), Gaps = 48/542 (8%)
Query: 277 SKENSTKKSSD--RQNDIVNLL-EQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADE 333
SKE+ +SD + ++ L E L GG L +QL+ + WL W N ILAD+
Sbjct: 169 SKEDGETINSDLTEEETVIKLQNELCPLLTGGQLKSYQLKGVKWLISLWQNGLNGILADQ 228
Query: 334 MGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKA 393
MGLGKTI F+S L + P LV+ PL T+ NW E A + P +N + YHG
Sbjct: 229 MGLGKTIQTIGFLSHLKGN-GLDGPYLVIAPLSTLSNWFNEIARFTPSINAIIYHGDKNQ 287
Query: 394 RAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHF-RGVPWEVLIVDEGHRL 452
R +R+ KT KF +++TSYE+ + D R PW+ +++DEGHRL
Sbjct: 288 RDELRRKHM---------PKTVGPKFPIVITSYEVAMNDAKRILRHYPWKYVVIDEGHRL 338
Query: 453 KNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERF---- 508
KN + KL L + +++LLTGTPLQNNL E+++LLNF+ P F S FE F
Sbjct: 339 KNHKCKLLRELKHLKMDNKLLLTGTPLQNNLSELWSLLNFILPDIFTSHDEFESWFDFSE 398
Query: 509 ---NDLTSAEK-------VDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAE 558
N+ T E+ V +L ++ P +LRR+K D ++P K E ++ ++ Q +
Sbjct: 399 KNKNEATKEEEEKRRAQVVSKLHGILRPFILRRMKCDVELSLPRKKEIIMYATMTDHQKK 458
Query: 559 YYRAMLTKNYQ--ILRNIGKGIAQQSMLN-IVMQLRKVCNHPYLIPGTEPDSGSVEFLHE 615
+ ++ + + N +G + LN +V+QLRK CNHP L+ G S + E
Sbjct: 459 FQEHLVNNTLEAHLGENAIRGQGWKGKLNNLVIQLRKNCNHPDLLQGQIDGSYLYPPVEE 518
Query: 616 MRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYE--RVDGS 673
+ + K LL +L L+ H+VLIFSQ TKLLDI++ Y F K +E R+DGS
Sbjct: 519 I-VGQCGKFRLLERLLVRLFANNHKVLIFSQWTKLLDIMDYY----FSEKGFEVCRIDGS 573
Query: 674 VSVTDRQTAIARFNQDKSRF-VFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNR 732
V + +R+ I F+ +KS +FLLSTR+ GLGINL ADT I+YDSD+NP D+QAM+R
Sbjct: 574 VKLDERRRQIKDFSDEKSSCSIFLLSTRAGGLGINLTAADTCILYDSDWNPQMDLQAMDR 633
Query: 733 AHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKG---------KSGSQKEVED 783
HRIGQ+ + VYRL S+E R+L+ A KL L+ + G KS + E ED
Sbjct: 634 CHRIGQTKPVHVYRLSTAQSIETRVLKRAYSKLKLEHVVIGQGQFHQERAKSSTPLEEED 693
Query: 784 IL 785
IL
Sbjct: 694 IL 695
>At2g44980 putative transcription activator
Length = 861
Score = 317 bits (811), Expect = 3e-86
Identities = 232/683 (33%), Positives = 342/683 (49%), Gaps = 82/683 (12%)
Query: 306 SLFPHQLEALNWLRKCWYKSRNVILA-DEMGLGKTISACAFISSLYFEFKVSRPCLVLVP 364
+L PHQ+E ++WL + + NV+L D+MGLGKT+ A +F+S L F + P LVL P
Sbjct: 50 TLKPHQVEGVSWLIQKYLLGVNVVLELDQMGLGKTLQAISFLSYLKFRQGLPGPFLVLCP 109
Query: 365 LVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLT 424
L W++E + P++ V++Y G R +R+ + F+VLLT
Sbjct: 110 LSVTDGWVSEINRFTPNLEVLRYVGDKYCRLDMRK---------SMYDHGHFLPFDVLLT 160
Query: 425 SYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFS-LLNSISFQHRVLLTGTPLQNNL 483
+Y++ L D +PW+ I+DE RLKN S L++ LL R+L+TGTP+QNNL
Sbjct: 161 TYDIALVDQDFLSQIPWQYAIIDEAQRLKNPNSVLYNVLLEQFLIPRRLLITGTPIQNNL 220
Query: 484 GEMYNLLNFLQPASFPSL----SAFEERFNDL---TSAEKVDELKKLVSPHMLRRLKKDA 536
E++ L++F P F +L SAF+E + L E LK ++ MLRR K
Sbjct: 221 TELWALMHFCMPLVFGTLDQFLSAFKETGDGLDVSNDKETYKSLKFILGAFMLRRTKSLL 280
Query: 537 MQN----IPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSML-NIVMQLR 591
+++ +PP TE V V L S+Q + Y ++L K L + G + + L NIV+QLR
Sbjct: 281 IESGNLVLPPLTELTVMVPLVSLQKKIYTSILRKELPGLLELSSGGSNHTSLQNIVIQLR 340
Query: 592 KVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
K C+HPYL PG EP+ E ++AS KL +L +LK L+ GHRVL+FSQMT L
Sbjct: 341 KACSHPYLFPGIEPEPFEEG---EHLVQASGKLLVLDQLLKRLHDSGHRVLLFSQMTSTL 397
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSR-----------FVFLLSTR 700
DIL+D++ E +YER+DGSV +R AI F+ R FVF++STR
Sbjct: 398 DILQDFM--ELRRYSYERLDGSVRAEERFAAIKNFSAKTERGLDSEVDGSNAFVFMISTR 455
Query: 701 SCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQL 760
+ G+G+NL ADTVI Y+ D+NP D QA+ RAHRIGQ + +L LV SVEE IL+
Sbjct: 456 AGGVGLNLVAADTVIFYEQDWNPQVDKQALQRAHRIGQISHVLSINLVTEHSVEEVILRR 515
Query: 761 AKKKLMLDQLFKGKSGSQKE---------VEDILKWGTEELFND-----------SCALN 800
A++KL L G + +KE V + ++ EE+ N+ S A
Sbjct: 516 AERKLQLSHNVVGDNMEEKEEDGGDLRSLVFGLQRFDPEEIHNEESDNLKMVEISSLAEK 575
Query: 801 GKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDE----------- 849
+N +K+E E+ + G D D +S + W E
Sbjct: 576 VVAIRQNVEPDKEERRFEI-NSSDTLLGNTSSASLDSELDEASYLSWVEKLKEAARSSKD 634
Query: 850 NAILKLLDRSNLQDAST---DIAEGDSENDMLGSMKALEWNDEPTEEHVEGESPPHGAD- 905
I++L +R NL + + A +E L + A + EE + + +D
Sbjct: 635 EKIIELGNRKNLSEERNLRIEAARKKAEEKKLATWGAHGYQSLSVEEPILPDDVDSSSDA 694
Query: 906 -------DMCTQNSEKKEDNAVI 921
CT S + A+I
Sbjct: 695 GSVNFVFGDCTNPSTVSHEPAII 717
>At2g28290 putative SNF2 subfamily transcription regulator
Length = 1331
Score = 297 bits (761), Expect = 2e-80
Identities = 202/542 (37%), Positives = 285/542 (52%), Gaps = 42/542 (7%)
Query: 242 NECTWESLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQNDIV-NLLEQPK 300
+E E+ DE +S++ F++F + + S +K + I N+ EQP
Sbjct: 727 DETLIENEDESDQAKASYISAFFHLFLMVMIHTIQHYLESNEKYYLMAHSIKENINEQPS 786
Query: 301 ELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCL 360
L GG L +Q+ L WL + N ILADEMGLGKT+ + I L P L
Sbjct: 787 SLVGGKLREYQMNGLRWLVSLYNNHLNGILADEMGLGKTVQVISLICYLMETKNDRGPFL 846
Query: 361 VLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFN 420
V+VP + W +E WAP ++ + Y G R + ++ KFN
Sbjct: 847 VVVPSSVLPGWQSEINFWAPSIHKIVYCGTPDERRKL------------FKEQIVHQKFN 894
Query: 421 VLLTSYEMVLA--DYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTP 478
VLLT+YE ++ D + W +I+DEGHR+KN+ KL + L HR+LLTGTP
Sbjct: 895 VLLTTYEYLMNKHDRPKLSKIHWHYIIIDEGHRIKNASCKLNADLKHYVSSHRLLLTGTP 954
Query: 479 LQNNLGEMYNLLNFLQPASFPSLSAFEERFND----------LTSAEK----VDELKKLV 524
LQNNL E++ LLNFL P F S F + FN L S E+ ++ L +++
Sbjct: 955 LQNNLEELWALLNFLLPNIFNSSEDFSQWFNKPFQSNGESSALLSEEENLLIINRLHQVL 1014
Query: 525 SPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSML 584
P +LRRLK +P K ER++ E S+ Y + ++ + L +IG +++
Sbjct: 1015 RPFVLRRLKHKVENELPEKIERLIRCEASA----YQKLLMKRVEDNLGSIGNA-KSRAVH 1069
Query: 585 NIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIF 644
N VM+LR +CNHPYL S E ++ KL +L ML L HRVL F
Sbjct: 1070 NSVMELRNICNHPYL-----SQLHSEEHFLPPIVRLCGKLEMLDRMLPKLKATDHRVLFF 1124
Query: 645 SQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRF-VFLLSTRSCG 703
S MT+LLD++EDYL ++ G K Y R+DG S DR I FN+ S F +FLLS R+ G
Sbjct: 1125 STMTRLLDVMEDYLTLK-GYK-YLRLDGQTSGGDRGALIDGFNKSGSPFFIFLLSIRAGG 1182
Query: 704 LGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKK 763
+G+NL ADTVI++D+D+NP D+QA RAHRIGQ +LV R SVEE++ A+
Sbjct: 1183 VGVNLQAADTVILFDTDWNPQVDLQAQARAHRIGQKKDVLVLRFETVNSVEEQVRASAEH 1242
Query: 764 KL 765
KL
Sbjct: 1243 KL 1244
>At2g02090 putative helicase
Length = 763
Score = 282 bits (721), Expect = 7e-76
Identities = 201/601 (33%), Positives = 293/601 (48%), Gaps = 92/601 (15%)
Query: 238 GKPYNECTWESLD--EPVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQNDIVNL 295
GK +C S D + + SS + R++ ET T+ QNDI +
Sbjct: 154 GKALQKCAKISADLRKELYGTSSGVTDRYSEVETSTVRIVT------------QNDIDDA 201
Query: 296 LEQPKELRGGSLFPHQLEALNWLRKCWYKS-RNVILADEMGLGKTISACAFISSLYFEFK 354
+ L P+QL +N+L + K ILADEMGLGKTI A +++ L
Sbjct: 202 CKAEDSDFQPILKPYQLVGVNFLLLLYKKGIEGAILADEMGLGKTIQAITYLTLLSRLNN 261
Query: 355 VSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKT 414
P LV+ P + NW E W P V+QYHG A+A ++ + + L+K
Sbjct: 262 DPGPHLVVCPASVLENWERELRKWCPSFTVLQYHGAARAA--------YSRELNSLSKAG 313
Query: 415 EAYKFNVLLTSY-------EMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSIS 467
+ FNVLL Y E D + W +++DE H LK+ S + L S++
Sbjct: 314 KPPPFNVLLVCYSLFERHSEQQKDDRKVLKRWRWSCVLMDEAHALKDKNSYRWKNLMSVA 373
Query: 468 --FQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVS 525
R++LTGTPLQN+L E+++LL F+ P F + + ++ + E + +K ++
Sbjct: 374 RNANQRLMLTGTPLQNDLHELWSLLEFMLPDIFTTENVDLKKLLNAEDTELITRMKSILG 433
Query: 526 PHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYR-----------AMLTK-NYQILRN 573
P +LRRLK D MQ + PK +R+ V + Q + Y+ A L K + + L +
Sbjct: 434 PFILRRLKSDVMQQLVPKIQRVEYVLMERKQEDAYKEAIEEYRAASQARLVKLSSKSLNS 493
Query: 574 IGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEF--------------------- 612
+ K + ++ + N Q RK+ NHP LI D +
Sbjct: 494 LAKALPKRQISNYFTQFRKIANHPLLIRRIYSDEDVIRIARKLHPIGAFGFECSLDRVIE 553
Query: 613 -------------------------LHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQM 647
L + + SAK L +L + K GHRVLIFSQ
Sbjct: 554 EVKGFNDFRIHQLLFQYGVNDTKGTLSDKHVMLSAKCRTLAELLPSMKKSGHRVLIFSQW 613
Query: 648 TKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGIN 707
T +LDILE L++ TY R+DGS VTDRQT + FN DKS F LLSTR+ G G+N
Sbjct: 614 TSMLDILEWTLDVI--GVTYRRLDGSTQVTDRQTIVDTFNNDKSIFACLLSTRAGGQGLN 671
Query: 708 LATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLML 767
L ADTVII+D DFNP D QA +R HRIGQ+ + ++RLV +++V+E I ++AK+KL+L
Sbjct: 672 LTGADTVIIHDMDFNPQIDRQAEDRCHRIGQTKPVTIFRLVTKSTVDENIYEIAKRKLVL 731
Query: 768 D 768
D
Sbjct: 732 D 732
>At2g46020 putative SNF2 subfamily transcriptional activator
Length = 1245
Score = 280 bits (717), Expect = 2e-75
Identities = 209/634 (32%), Positives = 322/634 (49%), Gaps = 73/634 (11%)
Query: 290 NDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSL 349
N++V + QP L+ G+L +QL L W+ + N ILADEMGLGKT+ A I+ L
Sbjct: 19 NEVV--VRQPSMLQAGTLRDYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYL 76
Query: 350 YFEFKVSR-PCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPS 408
EFK + P L++VP + NW +E W P V+ + Y G R S
Sbjct: 77 -MEFKGNYGPHLIIVPNAVLVNWKSELHTWLPSVSCIYYVGTKDQR-------------S 122
Query: 409 GLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISF 468
L + + KFNVL+T+YE ++ D S V W+ +I+DE R+K+ ES L L+
Sbjct: 123 KLFSQVKFEKFNVLVTTYEFIMYDRSKLSKVDWKYIIIDEAQRMKDRESVLARDLDRYRC 182
Query: 469 QHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERF--------------NDLTSA 514
Q R+LLTGTPLQN+L E+++LLN L P F + AF + F +D
Sbjct: 183 QRRLLLTGTPLQNDLKELWSLLNLLLPDVFDNRKAFHDWFAQPFQKEGPAHNIEDDWLET 242
Query: 515 EK----VDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYY-------RAM 563
EK + L +++ P MLRR +D ++P K ++ +S+IQ+ Y
Sbjct: 243 EKKVIVIHRLHQILEPFMLRRRVEDVEGSLPAKVSVVLRCRMSAIQSAVYDWIKATGTLR 302
Query: 564 LTKNYQILRNIGKGIAQ----QSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIK 619
+ + + LR I Q +++ N M+LRK CNHP L P +F + ++
Sbjct: 303 VDPDDEKLRAQKNPIYQAKIYRTLNNRCMELRKACNHPLL---NYPYFN--DFSKDFLVR 357
Query: 620 ASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDR 679
+ KL +L +L L + GHRVL+FS MTKLLDILE+YL ++ Y R+DG+ S+ DR
Sbjct: 358 SCGKLWILDRILIKLQRTGHRVLLFSTMTKLLDILEEYL--QWRRLVYRRIDGTTSLEDR 415
Query: 680 QTAIARFNQ-DKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQ 738
++AI FN D F+FLLS R+ G G+NL TADTV+IYD D NP + QA+ RAHRIGQ
Sbjct: 416 ESAIVDFNDPDTDCFIFLLSIRAAGRGLNLQTADTVVIYDPDPNPKNEEQAVARAHRIGQ 475
Query: 739 SNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKEVEDILKWGTEELFNDSCA 798
+ + +++ +V E++ K+ D+L +SG ++ED + G +
Sbjct: 476 TREV---KVIYMEAVVEKLSSHQKE----DEL---RSGGSVDLEDDMA-GKDRYIGSIEG 524
Query: 799 LNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDR 858
L + + DE + R +E++ + + +E + D
Sbjct: 525 LIRNNIQQYKIDMADEVINAGRFDQR-------TTHEERRMTLETLLHDEERYQETVHDV 577
Query: 859 SNLQDASTDIAEGDSENDMLGSM-KALEWNDEPT 891
+L + + IA + E ++ M + +W +E T
Sbjct: 578 PSLHEVNRMIARSEEEVELFDQMDEEFDWTEEMT 611
>At4g31900 putative protein
Length = 1067
Score = 254 bits (648), Expect = 2e-67
Identities = 161/388 (41%), Positives = 233/388 (59%), Gaps = 58/388 (14%)
Query: 530 RRLKKDAMQN-IPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVM 588
+RLKKD +++ +PPK E ++ V++SS Q E Y+A++T NYQ+L K A+ S N++M
Sbjct: 325 KRLKKDVLKDKVPPKKELILRVDMSSQQKEVYKAVITNNYQVLTK--KRDAKIS--NVLM 380
Query: 589 QLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMT 648
+LR+VC+HPYL+P EP ++AS KL LL M+ L ++GHRVLI++Q
Sbjct: 381 KLRQVCSHPYLLPDFEPRFEDANEAFTKLLEASGKLQLLDKMMVKLKEQGHRVLIYTQFQ 440
Query: 649 KLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKS-RFVFLLSTRSCGLGIN 707
L +LEDY F YER+DG +S +RQ I RFN + S RF FLLSTR+ G+GIN
Sbjct: 441 HTLYLLEDYFT--FKNWNYERIDGKISGPERQVRIDRFNAENSNRFCFLLSTRAGGIGIN 498
Query: 708 LATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLML 767
LATADTVIIYDSD+NPHAD+QAM R HR+GQ+N++++YRL+ + +VEER++++ K K++L
Sbjct: 499 LATADTVIIYDSDWNPHADLQAMARVHRLGQTNKVMIYRLIHKGTVEERMMEITKNKMLL 558
Query: 768 DQLFKGKSG-SQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKHRKR 826
+ L GK Q E++DI+K+G++ELF SE N DEA
Sbjct: 559 EHLVVGKQHLCQDELDDIIKYGSKELF-----------SEEN----DEA----------- 592
Query: 827 TGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMK--AL 884
S KI +D+ AI +LLDR+++ DA + + E D L + K +
Sbjct: 593 -------------GRSGKIHYDDAAIEQLLDRNHV-DAVEVSLDDEEETDFLKNFKVASF 638
Query: 885 EWNDEPTE-------EHVEGESPPHGAD 905
E+ D+ E + +E S AD
Sbjct: 639 EYVDDENEAAALEEAQAIENNSSVRNAD 666
Score = 140 bits (354), Expect = 3e-33
Identities = 94/257 (36%), Positives = 137/257 (52%), Gaps = 27/257 (10%)
Query: 165 EFLVKWVGKSHIHNSWISES----------HLKVIAK-----RKLENYKAKYGTATINIC 209
++LVKW G S++H SW+ E HLK+ + ++ + A+ G
Sbjct: 78 QYLVKWKGLSYLHCSWVPEQEFEKAYKSHPHLKLKLRVTRFNAAMDVFIAENGAHEFIAI 137
Query: 210 EEQWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMFET 269
+WK +R++A R G E VK+ Y WES E + + + I RF +
Sbjct: 138 RPEWKTVDRIIACREGDDG-EEYLVKYKELSYRNSYWES--ESDISDFQNEIQRFKDINS 194
Query: 270 LTLEREASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVI 329
+ R+ EN + +Q D+ E G+L +QLE LN+LR W K NVI
Sbjct: 195 SS-RRDKYVENERNREEFKQFDLT------PEFLTGTLHTYQLEGLNFLRYSWSKKTNVI 247
Query: 330 LADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHG 389
LADEMGLGKTI + AF++SL FE +S P LV+ PL T+ NW EFA WAP +NVV Y G
Sbjct: 248 LADEMGLGKTIQSIAFLASL-FEENLS-PHLVVAPLSTIRNWEREFATWAPHMNVVMYTG 305
Query: 390 CAKARAIIRQYEWHASD 406
++AR +I ++E++ S+
Sbjct: 306 DSEARDVIWEHEFYFSE 322
>At2g18760 putative SNF2/RAD54 family DNA repair and recombination
protein
Length = 1187
Score = 246 bits (629), Expect = 3e-65
Identities = 177/602 (29%), Positives = 291/602 (47%), Gaps = 68/602 (11%)
Query: 307 LFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLV 366
LF +Q + WL + + I+ DEMGLGKTI +F+ SL+F K+ +P +++ P+
Sbjct: 385 LFDYQRVGVQWLWELHCQRAGGIIGDEMGLGKTIQVLSFLGSLHFS-KMYKPSIIICPVT 443
Query: 367 TMGNWLAEFALWAPDVNVV--------QYHGCAKARAIIRQYEWHAS-----DPSGLNKK 413
+ W E W PD +V HG + +A Y+ +S +P N K
Sbjct: 444 LLRQWRREAQKWYPDFHVEILHDSAQDSGHGKGQGKASESDYDSESSVDSDHEPKSKNTK 503
Query: 414 TEAYKFN--------VLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNS 465
N +L+T+YE + + W ++DEGHR++N S + +
Sbjct: 504 KWDSLLNRVLNSESGLLITTYEQLRLQGEKLLNIEWGYAVLDEGHRIRNPNSDITLVCKQ 563
Query: 466 ISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFND--------------- 510
+ HR+++TG P+QN L E+++L +F+ P L FE F+
Sbjct: 564 LQTVHRIIMTGAPIQNKLTELWSLFDFVFPGKLGVLPVFEAEFSVPITVGGYANASPLQV 623
Query: 511 LTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQI 570
T+ L+ L+ P++LRR+K D ++ KTE ++ L+ Q YRA L +
Sbjct: 624 STAYRCAVVLRDLIMPYLLRRMKADVNAHLTKKTEHVLFCSLTVEQRSTYRAFLASSE-- 681
Query: 571 LRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTE----PDSGSVEFLHEMRIKASAKLTL 626
+ I G ++ L + +RK+CNHP L+ PD G+ E S K+ +
Sbjct: 682 VEQIFDG--NRNSLYGIDVMRKICNHPDLLEREHSHQNPDYGNPE--------RSGKMKV 731
Query: 627 LHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARF 686
+ +LK+ ++GHRVL+FSQ ++LDILE +L +Y R+DG V R I F
Sbjct: 732 VAEVLKVWKQQGHRVLLFSQTQQMLDILESFLVAN--EYSYRRMDGLTPVKQRMALIDEF 789
Query: 687 NQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYR 746
N + FVF+L+T+ GLG NL A+ VII+D D+NP D+QA RA RIGQ + VYR
Sbjct: 790 NNSEDMFVFVLTTKVGGLGTNLTGANRVIIFDPDWNPSNDMQARERAWRIGQKKDVTVYR 849
Query: 747 LVVRASVEERIL-QLAKKKLMLDQLFKG-------KSGSQKEVEDILKWGTEELFNDSCA 798
L+ R ++EE++ + K + +++ K K+ K++ + G ++
Sbjct: 850 LITRGTIEEKVYHRQIYKHFLTNKILKNPQQRRFFKARDMKDLFILKDDGDSNASTETSN 909
Query: 799 LNGKDTSENN----NSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILK 854
+ + E N S+K + ++ + K G + + + TD + + M +E ILK
Sbjct: 910 IFSQLAEEINIVGVQSDK-KPESDTQLALHKTAEGSSEQTDVEMTDKTGEAMDEETNILK 968
Query: 855 LL 856
L
Sbjct: 969 SL 970
>At5g63950 unknown protein
Length = 1090
Score = 226 bits (577), Expect = 4e-59
Identities = 170/574 (29%), Positives = 281/574 (48%), Gaps = 87/574 (15%)
Query: 307 LFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLV 366
L+PHQ E LNWL + + IL D+MGLGKT+ C+F++ L F K+ + LV+ P
Sbjct: 377 LYPHQREGLNWLWSLHTQGKGGILGDDMGLGKTMQICSFLAGL-FHSKLIKRALVVAPKT 435
Query: 367 TMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSY 426
+ +W+ E A +Y+G + R+Y+ H L K +LLT+Y
Sbjct: 436 LLPHWMKELATVGLSQMTREYYGTSTKA---REYDLHHI----LQGK------GILLTTY 482
Query: 427 EMV------LADYSHFR------GVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
++V L H+ G W+ +I+DEGH +KN ++ L I HR+++
Sbjct: 483 DIVRNNTKALQGDDHYTDEDDEDGNKWDYMILDEGHLIKNPNTQRAKSLLEIPSSHRIII 542
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERF---------NDLTSAEK------VDE 519
+GTP+QNNL E++ L NF P + F++ + + T E+
Sbjct: 543 SGTPIQNNLKELWALFNFSCPGLLGDKNWFKQNYEHYILRGTDKNATDREQRIGSTVAKN 602
Query: 520 LKKLVSPHMLRRLKKD------AMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRN 573
L++ + P LRRLK + A + K E +V + L++ Q + Y A L N +I+ +
Sbjct: 603 LREHIQPFFLRRLKSEVFGDDGATSKLSKKDEIVVWLRLTACQRQLYEAFL--NSEIVLS 660
Query: 574 IGKGIAQQSMLNIVMQLRKVCNHPYLIPG--------------TEPDSGSVEFL------ 613
G S L + L+K+C+HP L+ T+ ++G E L
Sbjct: 661 AFDG----SPLAALTILKKICDHPLLLTKRAAEDVLEGMDSTLTQEEAGVAERLAMHIAD 716
Query: 614 -------HEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKT 666
S KL+ + S+L+ L EGHRVLIFSQ K+L++++D ++ +
Sbjct: 717 NVDTDDFQTKNDSISCKLSFIMSLLENLIPEGHRVLIFSQTRKMLNLIQD--SLTSNGYS 774
Query: 667 YERVDGSVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHAD 726
+ R+DG+ DR + F + +FLL+++ GLG+ L AD VI+ D +NP D
Sbjct: 775 FLRIDGTTKAPDRLKTVEEFQEGHVAPIFLLTSQVGGLGLTLTKADRVIVVDPAWNPSTD 834
Query: 727 IQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKEVEDILK 786
Q+++RA+RIGQ+ ++VYRL+ A+VEE+I +K++ LFK + ++++ +
Sbjct: 835 NQSVDRAYRIGQTKDVIVYRLMTSATVEEKIY---RKQVYKGGLFKTATEHKEQIRYFSQ 891
Query: 787 WGTEELFNDSCALNGKDTSENNNSNKDEAVAEVE 820
ELF S G D S +E +++
Sbjct: 892 QDLRELF--SLPKGGFDVSPTQQQLYEEHYNQIK 923
>At3g54280 TATA box binding protein (TBP) associated factor (TAF)
-like protein
Length = 2049
Score = 213 bits (543), Expect = 3e-55
Identities = 157/531 (29%), Positives = 260/531 (48%), Gaps = 90/531 (16%)
Query: 310 HQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSR------PCLVLV 363
+Q E +NWL + IL D+MGLGKT+ A A ++S E + S P +++
Sbjct: 1447 YQQEGINWLGFLKRFKLHGILCDDMGLGKTLQASAIVASDAAERRGSTDELDVFPSIIVC 1506
Query: 364 PLVTMGNWLAEFALWAPD--VNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNV 421
P +G+W E + ++V+QY G A+ R +R+ + NV
Sbjct: 1507 PSTLVGHWAFEIEKYIDLSLLSVLQYVGSAQDRVSLRE---------------QFNNHNV 1551
Query: 422 LLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQN 481
++TSY++V D + W I+DEGH +KN++SK+ + + + QHR++L+GTP+QN
Sbjct: 1552 IITSYDVVRKDVDYLTQFSWNYCILDEGHIIKNAKSKITAAVKQLKAQHRLILSGTPIQN 1611
Query: 482 NLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEK---------------VDELKKLVSP 526
N+ E+++L +FL P + F+ + A + ++ L K V P
Sbjct: 1612 NIMELWSLFDFLMPGFLGTERQFQASYGKPLLAARDPKCSAKDAEAGVLAMEALHKQVMP 1671
Query: 527 HMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQ-----ILR--------- 572
+LRR K++ + ++P K + +LS +Q + Y + + I++
Sbjct: 1672 FLLRRTKEEVLSDLPEKIIQDRYCDLSPVQLKLYEQFSGSSAKQEISSIIKVDGSADSGN 1731
Query: 573 -NIGKGIAQQSMLNIVMQLRKVCNHPYLIPG---TEP--------DSGSVEFLHEM-RIK 619
++ A + + L K+C+HP L+ G TEP +G + + E+ +++
Sbjct: 1732 ADVAPTKASTHVFQALQYLLKLCSHPLLVLGDKVTEPVASDLAAMINGCSDIITELHKVQ 1791
Query: 620 ASAKLTLLHSMLK-------------ILYKEGHRVLIFSQMTKLLDILE-DYLNIEFGPK 665
S KL L +L+ L HRVLIF+Q LLDI+E D
Sbjct: 1792 HSPKLVALQEILEECGIGSDASSSDGTLSVGQHRVLIFAQHKALLDIIEKDLFQAHMKSV 1851
Query: 666 TYERVDGSVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHA 725
TY R+DGSV R + FN D + V LL+T GLG+NL +ADT++ + D+NP
Sbjct: 1852 TYMRLDGSVVPEKRFEIVKAFNSDPTIDVLLLTTHVGGLGLNLTSADTLVFMEHDWNPMR 1911
Query: 726 D-----------IQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKL 765
D QAM+RAHR+GQ + V+RL++R ++EE+++ L K K+
Sbjct: 1912 DHQFANIELNKLWQAMDRAHRLGQKRVVNVHRLIMRGTLEEKVMSLQKFKV 1962
>At1g03750 hypothetical protein
Length = 874
Score = 204 bits (519), Expect = 2e-52
Identities = 176/632 (27%), Positives = 289/632 (44%), Gaps = 124/632 (19%)
Query: 307 LFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEF----------KVS 356
L HQ E + ++ + + IL D+MGLGKTI AF++++Y +
Sbjct: 151 LLEHQREGVKFMYNLYKNNHGGILGDDMGLGKTIQTIAFLAAVYGKDGDAGESCLLESDK 210
Query: 357 RPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEA 416
P L++ P + NW +EF+ WA V YHG S+ + +K +A
Sbjct: 211 GPVLIICPSSIIHNWESEFSRWASFFKVSVYHG---------------SNRDMILEKLKA 255
Query: 417 YKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTG 476
VL+TS++ G+ WE++I DE HRLKN +SKL+ I + R+ LTG
Sbjct: 256 RGVEVLVTSFDTFRIQGPVLSGINWEIVIADEAHRLKNEKSKLYEACLEIKTKKRIGLTG 315
Query: 477 TPLQNNLGEMYNLLNFLQPASFPSLSAFE----------------ERFNDLTSAEKVDEL 520
T +QN + E++NL ++ P S + F ERF + K L
Sbjct: 316 TVMQNKISELFNLFEWVAPGSLGTREHFRDFYDEPLKLGQRATAPERFVQIADKRK-QHL 374
Query: 521 KKLVSPHMLRRLKKDAMQNI-PPKTERMVPVELSSIQAEYYRAML-TKNYQILRN----- 573
L+ +MLRR K++ + ++ K + +V ++S +Q Y+ M+ Q L N
Sbjct: 375 GSLLRKYMLRRTKEETIGHLMMGKEDNVVFCQMSQLQRRVYQRMIQLPEIQCLVNKDNPC 434
Query: 574 -IGKGIAQQS------------------------------MLNIVMQLRKVCNHPYLI-- 600
G + Q +L +M+L+++ NH LI
Sbjct: 435 ACGSPLKQSECCRRIVPDGTIWSYLHRDNHDGCDSCPFCLVLPCLMKLQQISNHLELIKP 494
Query: 601 -PGTEPD-----------------------SGSVEFLHEMRIKASAKLTLLHSMLKILYK 636
P EP+ S S F+ +K K+ L ++
Sbjct: 495 NPKDEPEKQKKDAEFVSTVFGTDIDLLGGISASKSFMDLSDVKHCGKMRALEKLMASWIS 554
Query: 637 EGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFL 696
+G ++L+FS ++LDILE +L I G ++ R+DGS RQ+ + FN S+ VFL
Sbjct: 555 KGDKILLFSYSVRMLDILEKFL-IRKG-YSFARLDGSTPTNLRQSLVDDFNASPSKQVFL 612
Query: 697 LSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEER 756
+ST++ GLG+NL +A+ V+I+D ++NP D+QA +R+ R GQ ++V+RL+ S+EE
Sbjct: 613 ISTKAGGLGLNLVSANRVVIFDPNWNPSHDLQAQDRSFRYGQKRHVVVFRLLSAGSLEEL 672
Query: 757 IL--QLAKKKL--------MLDQLFKGKSGSQKEVEDILKWGTEELFNDSCALNGK-DTS 805
+ Q+ K++L M + F+G KE + L +G LF D L+ K TS
Sbjct: 673 VYTRQVYKQQLSNIAVAGKMETRYFEGVQDC-KEFQGEL-FGISNLFRD---LSDKLFTS 727
Query: 806 ENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDK 837
+ ++D + E + + TG D E++
Sbjct: 728 DIVELHRDSNIDENKKRSLLETGVSEDEKEEE 759
>At3g57300 helicase-like protein
Length = 1507
Score = 197 bits (501), Expect = 2e-50
Identities = 116/325 (35%), Positives = 185/325 (56%), Gaps = 21/325 (6%)
Query: 298 QPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSR 357
Q EL G+L +Q++ L WL C+ + N ILADEMGLGKTI A AF++ L E +
Sbjct: 577 QTPELFKGTLKEYQMKGLQWLVNCYEQGLNGILADEMGLGKTIQAMAFLAHLAEEKNIWG 636
Query: 358 PCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAY 417
P LV+ P + NW E + + PD+ + Y G + R I+R+ +P + ++
Sbjct: 637 PFLVVAPASVLNNWADEISRFCPDLKTLPYWGGLQERTILRKN----INPKRMYRRDAG- 691
Query: 418 KFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGT 477
F++L+TSY++++ D +FR V W+ +++DE +K+S S + L S + ++R+LLTGT
Sbjct: 692 -FHILITSYQLLVTDEKYFRRVKWQYMVLDEAQAIKSSSSIRWKTLLSFNCRNRLLLTGT 750
Query: 478 PLQNNLGEMYNLLNFLQPASFPSLSAFEERFN----------DLTSAEKVDELKKLVSPH 527
P+QNN+ E++ LL+F+ P F + F E F+ + +++ L ++ P
Sbjct: 751 PIQNNMAELWALLHFIMPMLFDNHDQFNEWFSKGIENHAEHGGTLNEHQLNRLHAILKPF 810
Query: 528 MLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTK-NYQILRNIGKGIAQQ----S 582
MLRR+KKD + + KTE V +LSS Q +Y+A+ K + L + +G +
Sbjct: 811 MLRRVKKDVVSELTTKTEVTVHCKLSSRQQAFYQAIKNKISLAELFDSNRGQFTDKKVLN 870
Query: 583 MLNIVMQLRKVCNHPYLIPGTEPDS 607
++NIV+QLRKVCNHP L E S
Sbjct: 871 LMNIVIQLRKVCNHPELFERNEGSS 895
Score = 147 bits (371), Expect = 3e-35
Identities = 81/165 (49%), Positives = 110/165 (66%), Gaps = 4/165 (2%)
Query: 621 SAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQ 680
S KL L +LK L HRVL+F+QMTK+L+ILEDY+N + Y R+DGS ++ DR+
Sbjct: 1205 SGKLQTLDILLKRLRAGNHRVLLFAQMTKMLNILEDYMN--YRKYKYLRLDGSSTIMDRR 1262
Query: 681 TAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSN 740
+ F FVFLLSTR+ GLGINL ADTVI Y+SD+NP D+QAM+RAHR+GQ+
Sbjct: 1263 DMVRDFQHRSDIFVFLLSTRAGGLGINLTAADTVIFYESDWNPTLDLQAMDRAHRLGQTK 1322
Query: 741 RLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKEVEDIL 785
+ VYRL+ + +VEE+IL A +K + QL +G + +D L
Sbjct: 1323 DVTVYRLICKETVEEKILHRASQKNTVQQLV--MTGGHVQGDDFL 1365
>At3g19210 DNA repair protein, putative
Length = 688
Score = 185 bits (469), Expect = 1e-46
Identities = 149/520 (28%), Positives = 242/520 (45%), Gaps = 69/520 (13%)
Query: 307 LFPHQLEALNWLRKC---WYKSRNV---ILADEMGLGKTISACAFISSLYFEFKVSRP-- 358
L PHQ E + ++ C + S N+ ILAD+MGLGKT+ + + +L + P
Sbjct: 76 LRPHQREGVQFMFDCVSGLHGSANINGCILADDMGLGKTLQSITLLYTLLCQGFDGTPMV 135
Query: 359 --CLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEA 416
+++ P + NW AE W D + C R + SG++ T
Sbjct: 136 KKAIIVTPTSLVSNWEAEIKKWVGD-RIQLIALCESTRDDVL---------SGIDSFTRP 185
Query: 417 YK-FNVLLTSYEMVLADYSHF-RGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
VL+ SYE S F + ++LI DE HRLKN ++ L S++ + RVLL
Sbjct: 186 RSALQVLIISYETFRMHSSKFCQSESCDLLICDEAHRLKNDQTLTNRALASLTCKRRVLL 245
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAF-----------------EERFNDLTSAEKV 517
+GTP+QN+L E + ++NF P S + F EE N +A++
Sbjct: 246 SGTPMQNDLEEFFAMVNFTNPGSLGDAAHFRHYYEAPIICGREPTATEEEKN--LAADRS 303
Query: 518 DELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKG 577
EL V+ +LRR ++PPK +V +++++Q+ Y ++ + R +
Sbjct: 304 AELSSKVNQFILRRTNALLSNHLPPKIIEVVCCKMTTLQSTLYNHFISSK-NLKRALADN 362
Query: 578 IAQQSMLNIVMQLRKVCNHPYLI--------PGTEPDSGSVEFLHEMR------------ 617
Q +L + L+K+CNHP LI PGT +EF
Sbjct: 363 AKQTKVLAYITALKKLCNHPKLIYDTIKSGNPGTVGFENCLEFFPAEMFSGRSGAWTGGD 422
Query: 618 ---IKASAKLTLLHSMLKILY-KEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGS 673
++ S K+ +L +L L K R+++ S T+ LD+ P + R+DGS
Sbjct: 423 GAWVELSGKMHVLSRLLANLRRKTDDRIVLVSNYTQTLDLFAQLCRERRYP--FLRLDGS 480
Query: 674 VSVTDRQTAIARFNQD-KSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNR 732
+++ RQ + R N K F FLLS+++ G G+NL A+ ++++D D+NP D QA R
Sbjct: 481 TTISKRQKLVNRLNDPTKDEFAFLLSSKAGGCGLNLIGANRLVLFDPDWNPANDKQAAAR 540
Query: 733 AHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFK 772
R GQ R+ VYR + ++EE++ Q K L ++ +
Sbjct: 541 VWRDGQKKRVYVYRFLSTGTIEEKVYQRQMSKEGLQKVIQ 580
>At3g12810 Snf2-related CBP activator protein, putative
Length = 1048
Score = 180 bits (457), Expect = 3e-45
Identities = 110/307 (35%), Positives = 165/307 (52%), Gaps = 24/307 (7%)
Query: 306 SLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPL 365
SL +Q L+WL + K N ILADEMGLGKTI A ++ L + + P L++VP
Sbjct: 120 SLREYQHIGLDWLVTMYEKKLNGILADEMGLGKTIMTIALLAHLACDKGIWGPHLIVVPT 179
Query: 366 VTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTS 425
M NW EF W P ++ Y G AK R + RQ W + F+V +T+
Sbjct: 180 SVMLNWETEFLKWCPAFKILTYFGSAKERKLKRQ-GW-----------MKLNSFHVCITT 227
Query: 426 YEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGE 485
Y +V+ D F+ W+ LI+DE H +KN +S+ + L + + + R+LLTGTPLQN+L E
Sbjct: 228 YRLVIQDSKMFKRKKWKYLILDEAHLIKNWKSQRWQTLLNFNSKRRILLTGTPLQNDLME 287
Query: 486 MYNLLNFLQPASFPSLSAFEERF----------NDLTSAEKVDELKKLVSPHMLRRLKKD 535
+++L++FL P F S F++ F + + E +D L ++ P +LRRLK+D
Sbjct: 288 LWSLMHFLMPHVFQSHQEFKDWFCNPIAGMVEGQEKINKEVIDRLHNVLRPFLLRRLKRD 347
Query: 536 AMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCN 595
+ +P K E ++ LS Q Y + + + G + M++I+MQLRKVCN
Sbjct: 348 VEKQLPSKHEHVIFCRLSKRQRNLYEDFIAST-ETQATLTSG-SFFGMISIIMQLRKVCN 405
Query: 596 HPYLIPG 602
HP L G
Sbjct: 406 HPDLFEG 412
Score = 153 bits (387), Expect = 4e-37
Identities = 112/313 (35%), Positives = 169/313 (53%), Gaps = 32/313 (10%)
Query: 623 KLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTA 682
KL L +L+ L GHR LIF+QMTK+LD+LE ++N+ +G TY R+DGS +RQT
Sbjct: 661 KLQELAMLLRKLKFGGHRALIFTQMTKMLDVLEAFINL-YG-YTYMRLDGSTPPEERQTL 718
Query: 683 IARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRL 742
+ RFN + F+F+LSTRS G+GINL ADTVI YDSD+NP D QA +R HRIGQ+ +
Sbjct: 719 MQRFNTNPKIFLFILSTRSGGVGINLVGADTVIFYDSDWNPAMDQQAQDRCHRIGQTREV 778
Query: 743 LVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKEVEDILKWGTEELFNDSCALNGK 802
+YRL+ +++EE IL+ A +K +LD L + E K ELF+ AL K
Sbjct: 779 HIYRLISESTIEENILKKANQKRVLDNLV--IQNGEYNTEFFKKLDPMELFSGHKALTTK 836
Query: 803 D---TSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRS 859
D TS++ ++ + A+VE ++ ED+ + K + E A+ D
Sbjct: 837 DEKETSKHCGADIPLSNADVEAALKQA--------EDEADYMALKRVEQEEAV----DNQ 884
Query: 860 NLQDASTDIAEGD---SENDMLGSMKALEWNDEPTEEHVEGESPPHGADDMCTQNSEKKE 916
+ + E D +E+D+ DEP ++ + P ++M +S+ ++
Sbjct: 885 EFTEEPVERPEDDELVNEDDIKA--------DEPADQGLVAAGP--AKEEMSLLHSDIRD 934
Query: 917 DNAVIGGEENEWD 929
+ AVI E D
Sbjct: 935 ERAVITTSSQEDD 947
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,380,744
Number of Sequences: 26719
Number of extensions: 956793
Number of successful extensions: 2937
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 2611
Number of HSP's gapped (non-prelim): 219
length of query: 935
length of database: 11,318,596
effective HSP length: 109
effective length of query: 826
effective length of database: 8,406,225
effective search space: 6943541850
effective search space used: 6943541850
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146651.1


