

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146649.9 - phase: 0 /pseudo
(1684 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
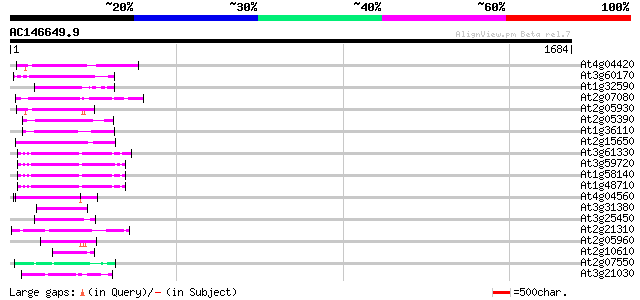
Score E
Sequences producing significant alignments: (bits) Value
At4g04420 putative transposon protein 114 4e-25
At3g60170 putative protein 110 9e-24
At1g32590 hypothetical protein, 5' partial 100 7e-21
At2g07080 putative gag-protease polyprotein 97 8e-20
At2g05930 copia-like retroelement pol polyprotein 92 2e-18
At2g05390 putative retroelement pol polyprotein 91 7e-18
At1g36110 hypothetical protein 90 1e-17
At2g15650 putative retroelement pol polyprotein 88 4e-17
At3g61330 copia-type polyprotein 85 3e-16
At3g59720 copia-type reverse transcriptase-like protein 85 3e-16
At1g58140 hypothetical protein 84 5e-16
At1g48710 hypothetical protein 84 5e-16
At4g04560 putative transposon protein 75 2e-13
At3g31380 hypothetical protein 75 3e-13
At3g25450 hypothetical protein 72 3e-12
At2g21310 putative retroelement pol polyprotein 67 1e-10
At2g05960 putative retroelement pol polyprotein 62 3e-09
At2g10610 pseudogene 60 1e-08
At2g07550 putative retroelement pol polyprotein 58 4e-08
At3g21030 hypothetical protein 53 2e-06
>At4g04420 putative transposon protein
Length = 1008
Score = 114 bits (286), Expect = 4e-25
Identities = 95/382 (24%), Positives = 168/382 (43%), Gaps = 46/382 (12%)
Query: 22 FSWWKTNMYSYIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRKIHTPAQK 70
+ WK M + I GL +E W EDG D L ++ A++
Sbjct: 21 YGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTKDQW--------NDAEE 72
Query: 71 KTYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQY 130
K + + +I + + ++ ++ + +AK + L YEG+ V+ ++ ML Q+
Sbjct: 73 AKAKANSRALSLIFNFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 132
Query: 131 ELFKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAK 190
E M++ E+IEE + + S L K Y LV K+LR LPSR+ K TA+ +
Sbjct: 133 ENLSMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 192
Query: 191 DLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDG 250
D +++ E++V L+ +E+ + + SK +AL + K ++ +E D
Sbjct: 193 DTDSIDFEEVVGMLQAYELEITSGKGG-YSKGLALAASAKKNEI----------QELKD- 240
Query: 251 DSDEDHSVKMAMLSNKLEYLARK-QKKFLSKRNGYKNWK----REDQKGCFNCKKPGHFI 305
M+M++ R+ +KK + G ++ + D+ C C+ GH
Sbjct: 241 --------TMSMMAKDFSRAMRRVEKKGFGRNQGTDRYRDRSSKRDEIQCHECQGYGHIK 292
Query: 306 ADCPDLQKEKSKSRPKKPSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMR 365
A+CP L K K + +KF KS +SES S+ +++D K +
Sbjct: 293 AECPSL-KRKDLKCSECNGLGHTKFDCVGSKSKPDKSCSSESESDSNDGDSEDYIKGFVS 351
Query: 366 LVATV-SSEAVSESESDSEDEN 386
V + + S+SE+D EDE+
Sbjct: 352 FVGIIEEKDESSDSEADGEDED 373
>At3g60170 putative protein
Length = 1339
Score = 110 bits (274), Expect = 9e-24
Identities = 77/301 (25%), Positives = 138/301 (45%), Gaps = 43/301 (14%)
Query: 13 PKFNGDPEEFSWWKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKT 72
P+F+G + +W M +++ ELW ++E+G+ + + G + A ++
Sbjct: 13 PRFDG---YYDFWSMTMENFLRS--RELWRLVEEGIPAIVV---GTTPVSEAQRSAVEEA 64
Query: 73 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYEL 132
K K++ + +I R + DKST+KA++ S+ Y+GS KV+ A+ L ++EL
Sbjct: 65 KLKDLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFEL 124
Query: 133 FKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDL 192
MK+ E I+ R T+V+ ++ + S +VSKILRSL ++ V +IEE+ DL
Sbjct: 125 LAMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDL 184
Query: 193 NTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDGDS 252
+TLS+++L SL VHE LN H +++ + + + ++ S G S
Sbjct: 185 STLSIDELHGSLLVHEQRLNGHVQEEQALKVTHEERPSQGRGRGVFRGSRGRGRG-RGRS 243
Query: 253 DEDHSVKMAMLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQ 312
+ ++ C+ C GHF +CP+ +
Sbjct: 244 GTNRAI----------------------------------VECYKCHNLGHFQYECPEWE 269
Query: 313 K 313
K
Sbjct: 270 K 270
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 100 bits (249), Expect = 7e-21
Identities = 69/240 (28%), Positives = 114/240 (46%), Gaps = 32/240 (13%)
Query: 75 KHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFK 134
K HK++ + ASI +T + K T+K ++ S+ Y+G+ +V+ A+ L +E+ +
Sbjct: 24 KDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSFEVLE 83
Query: 135 MKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNT 194
MK E+I +SR + + ++ L + S +V KILR+L ++ V AIEE+ ++
Sbjct: 84 MKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEESNNIKE 143
Query: 195 LSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDGDSDE 254
L+V+ L SSL VHE +L+ H+ ++ KA + PDG
Sbjct: 144 LTVDGLQSSLMVHEQNLSRHDVEER-----------------VLKAET--QWRPDGGRGR 184
Query: 255 DHSVKMAMLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQKE 314
S R + + + GY N D CF C K GH+ A+CP +KE
Sbjct: 185 GGSPSRG----------RGRGGYQGRGRGYVN---RDTVECFKCHKMGHYKAECPSWEKE 231
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 97.1 bits (240), Expect = 8e-20
Identities = 94/391 (24%), Positives = 172/391 (43%), Gaps = 50/391 (12%)
Query: 18 DPEEFSWWKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDR-KIHTPAQKKTYKKH 76
D + + +WK M I G E+G I + K + A++K K
Sbjct: 17 DTKRYGYWKVCMTQIIRG------------------QEDGFKITKPKANWTAEEKLQSKF 58
Query: 77 H-KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKM 135
+ + I + E+ + +AK + +L ++EG+ V+ + + Q+E KM
Sbjct: 59 NARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSHEGTSSVKRTRLDHIATQFEYLKM 118
Query: 136 KDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNTL 195
+ DE I + S+ L + +++ K+Y LV K+LR LP ++ + A + + +
Sbjct: 119 EPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPKFAAHKAVMRVAGNTDKI 178
Query: 196 SVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDGDSDED 255
S DLV LK+ EM ++ + K SK+IA A + SE +E DG +
Sbjct: 179 SFVDLVGMLKLEEMKADQ-DKVKPSKNIAF----------NADQGSEQFQEIKDGMALLA 227
Query: 256 HSVKMAMLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQKEK 315
+ A+ +++ R + +F N K+E Q C+ C GH +CP + K K
Sbjct: 228 RNFGKALKRVEIDG-ERSRGRFSRSENDDLRKKKEIQ--CYECGGFGHIKPECP-ITKRK 283
Query: 316 SKSRPKKPSFSSSKF----RKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMRLVATVS 371
K +KF + ++K+ + ++ D E+DD+ + + VA ++
Sbjct: 284 EMKCLKCKGVGHTKFECPNKSKLKEKSLISFSD---------SESDDEGEELLNFVAFMA 334
Query: 372 SEAVSE--SESDSEDENEVYSKIPRQELIDS 400
S S+ S++DS+ + E+ K + L DS
Sbjct: 335 SSDSSKFMSDTDSDCDEELNPKDKYRVLYDS 365
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 92.4 bits (228), Expect = 2e-18
Identities = 69/265 (26%), Positives = 118/265 (44%), Gaps = 41/265 (15%)
Query: 22 FSWWKTNMYSYIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRKIHTPAQK 70
+ WK M + I GL +E W EDG D L +++ A++
Sbjct: 33 YGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTEDQW--------NDAEE 84
Query: 71 KTYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQY 130
+ + +I S+ + ++ ++ + +AK + L YEG+ V+ ++ ML Q+
Sbjct: 85 AKATANSRALSLIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 144
Query: 131 ELFKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAK 190
E M++ E+IEE + + S L K Y LV K+LR LPSR+ K TA+ +
Sbjct: 145 ENLTMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 204
Query: 191 DLNTLSVEDLVSSLKVHEMSLNEHES----------SKKSKS-----------IALPSKG 229
D N++ E++V + +E+ + + S K K I G
Sbjct: 205 DTNSIDFEEVVGMFQAYELEITSGKGGYGHIKAECPSLKRKDLKCSECKGLGHIKFDCVG 264
Query: 230 KSSKSSKAYKASESEEESPDGDSDE 254
SK ++ +SESE +S DGDS++
Sbjct: 265 SKSKPDRSC-SSESESDSNDGDSED 288
Score = 39.7 bits (91), Expect = 0.015
Identities = 66/299 (22%), Positives = 120/299 (40%), Gaps = 73/299 (24%)
Query: 113 EGSKKVREAKALML----VHQYELFKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLV 168
E +K ++AL L V+Q + ++++ ES +E + + G +K+S + D++
Sbjct: 83 EEAKATANSRALSLIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRI--DML 140
Query: 169 SKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSI--ALP 226
+ +L +EE +++ S + + + H + +++ K K + LP
Sbjct: 141 ASQFENL---------TMEETENIEEFSGKISAIASEAHNLG-KKYKDKKLVKKLLRCLP 190
Query: 227 SKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMAMLSNKLEYLARKQKKFLSKRNGY-- 284
S+ +S +++ S D +S + V + +LE S + GY
Sbjct: 191 SRFESKRTAMG--------TSLDTNSIDFEEVVGMFQAYELE--------ITSGKGGYGH 234
Query: 285 -----KNWKREDQKGCFNCKKPGHFIADCPDLQKEKSKSRPKKPSFSSSKFRKQIKKSLM 339
+ KR+D K C CK GH DC SKS+P + S
Sbjct: 235 IKAECPSLKRKDLK-CSECKGLGHIKFDCVG-----SKSKPDRSCSS------------- 275
Query: 340 ATWEDLDSESGSDKEEADDDAKAAMRLVATVS-------SEAVSESESDSEDENEVYSK 391
+SES S+ +++D K + V + SEA E E +S DE+ K
Sbjct: 276 ------ESESDSNDGDSEDYIKGFVSFVGIIEEKDESSDSEADGEDEDNSADEDSDIEK 328
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 90.5 bits (223), Expect = 7e-18
Identities = 61/274 (22%), Positives = 122/274 (44%), Gaps = 54/274 (19%)
Query: 39 ELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKTYKKHHKIRGIIVASIPRTEYMKMSDK 98
++W+ ++ G DD++ K+ R ++ S+P + +++
Sbjct: 9 KVWETIDPGSDDME----------------------KNDMARALLFQSVPESTILQVGKH 46
Query: 99 STAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEMYSRFQTLVSGLQIL 158
T+KAM+ ++ G+++V+EAK L+ +++ MKD+E+I+E R + + + L
Sbjct: 47 KTSKAMWEAIKTRNLGAERVKEAKLQTLMAEFDRLNMKDNETIDEFVGRISEISTKSESL 106
Query: 159 KKSYVTSDLVSKILRSLP-SRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHESS 217
+ S +V K L+SLP ++ + A+E+ DLNT ED+V +K +E + + + S
Sbjct: 107 GEEIEESKIVKKFLKSLPRKKYIHIIAALEQILDLNTTGFEDIVGRMKTYEDRVCDEDDS 166
Query: 218 KKSKSIALPSKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMAMLSNKLEYLARKQKKF 277
+ + + + +SS ++ + S G
Sbjct: 167 PEEQGKLMYANSESSYDTRGGRGRGRGRSSGRG--------------------------- 199
Query: 278 LSKRNGYKNWKREDQKG-CFNCKKPGHFIADCPD 310
R GY +R+ K C+ C K GH+ ++C D
Sbjct: 200 ---RGGYGYQQRDKSKVICYRCDKTGHYASECLD 230
>At1g36110 hypothetical protein
Length = 745
Score = 90.1 bits (222), Expect = 1e-17
Identities = 72/283 (25%), Positives = 120/283 (41%), Gaps = 66/283 (23%)
Query: 39 ELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKTYKKHHKIRGIIVASIPRTEYMKMSDK 98
++W+ +E G+DD K+ RG+I SIP + +++
Sbjct: 42 KMWETIEPGIDD----------------------GYKNTMARGLIFQSIPESLTLQVGTL 79
Query: 99 STAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEMYSRFQTLVSGLQIL 158
+TAK ++ S+ Y G+ +V+EA+ L+ ++E KMK+ E I+ R L + L
Sbjct: 80 ATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMKMKESEKIDVFAGRLAELATRSDAL 139
Query: 159 KKSYVTSDLVSKILRSLPSR-WRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHESS 217
+ TS LV K L +LP R + + ++E+ DLN S ED+V +KV+
Sbjct: 140 GSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLNNTSFEDIVGRIKVY---------- 189
Query: 218 KKSKSIALPSKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMAMLSNKLE---YLARKQ 274
EE DG+ ED K+ + + Y +R +
Sbjct: 190 --------------------------EERVWDGEEQEDDQGKLMYANTDTQDSWYASRGR 223
Query: 275 KK---FLSKRNGYKNWKREDQK-GCFNCKKPGHFIADCPDLQK 313
+ F + G R+ K C+ C K GH+ ++CPD K
Sbjct: 224 GQGGGFNGRGRGRGRGSRDTSKVTCYRCDKLGHYASNCPDSVK 266
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 88.2 bits (217), Expect = 4e-17
Identities = 68/299 (22%), Positives = 135/299 (44%), Gaps = 16/299 (5%)
Query: 18 DPEEFSWWKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKTYKKHH 77
D E++ +W M + +LW ++E+GV + E + T ++
Sbjct: 13 DGEKYDFWSIKMATIFR--TRKLWSVVEEGVPVEPVQAEETPETARAKTLREEAVTNDTM 70
Query: 78 KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKD 137
++ I+ ++ + +++ S++K + L Y+GS +VR K L +YE KM D
Sbjct: 71 ALQ-ILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENLKMYD 129
Query: 138 DESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNTLSV 197
+++I+ + L L + + L+ KIL SLP+++ V+ +E+ +DL+ L++
Sbjct: 130 NDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLDALTM 189
Query: 198 EDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDGDSDEDHS 257
+L+ LK E + E S K + + SKG+ S + D ++ +
Sbjct: 190 SELLGILKAQEARVTAREESTKEGAFYVRSKGRESGFKQ------------DNTNNRVNQ 237
Query: 258 VKMAMLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQKEKS 316
K +K ++ + K + + KR + K C+ C K GH+ +C KE++
Sbjct: 238 DKKWCGFHKSSKHTEEECREKPKNDDHGKNKRSNIK-CYKCGKIGHYANECRSKNKERA 295
>At3g61330 copia-type polyprotein
Length = 1352
Score = 85.1 bits (209), Expect = 3e-16
Identities = 75/343 (21%), Positives = 156/343 (44%), Gaps = 21/343 (6%)
Query: 25 WKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKTYKKHHKIRGIIV 84
W M + I+G ++W+I+E G ++ + EG+ + + + K+ K +I
Sbjct: 21 WSLRMKA-ILGA-HDVWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEM 144
+ + K+ + ++AK + L +Y+G+ +V++ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + ++ K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHESSKKSKSIA---LPSKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMA 261
+ +E E KK + IA L + ++ ++Y+ + G +
Sbjct: 195 QAYE------EKKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 262 MLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQKEKSKSRPK 321
+ + K + + + K C+NC K GH+ ++C + +K +
Sbjct: 249 PHEDNTNQRGENSSRGRGKGHPKSRYDKSSVK-CYNCGKFGHYASEC---KAPSNKKFEE 304
Query: 322 KPSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAM 364
K + K +++ LMA+++ D + + K D A M
Sbjct: 305 KAHYVEEKIQEE-DMLLMASYKK-DEQKENHKWYLDSGASNHM 345
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 85.1 bits (209), Expect = 3e-16
Identities = 71/327 (21%), Positives = 151/327 (45%), Gaps = 20/327 (6%)
Query: 25 WKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKTYKKHHKIRGIIV 84
W M + I+G ++W+I+E G ++ + EG+ + + + K+ K +I
Sbjct: 21 WSLRMKA-ILGA-HDVWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEM 144
+ + K+ + ++AK + L +Y+G+ +V++ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + ++ K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHESSKKSKSIA---LPSKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMA 261
+ +E E KK + I L + ++ ++Y+ + G +
Sbjct: 195 QAYE------EKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 262 MLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQKEKSKSRPK 321
+ + K + + + K C+NC K GH+ ++C +K K +
Sbjct: 249 PHEDNTNQRGENSSRGRGKGHPKSRYDKSSVK-CYNCGKFGHYASECKAPSNKKFK---E 304
Query: 322 KPSFSSSKFRKQIKKSLMATWEDLDSE 348
K ++ K +++ LMA+++ + E
Sbjct: 305 KANYVEEKIQEE-DMLLMASYKKDEQE 330
>At1g58140 hypothetical protein
Length = 1320
Score = 84.3 bits (207), Expect = 5e-16
Identities = 70/327 (21%), Positives = 151/327 (45%), Gaps = 20/327 (6%)
Query: 25 WKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKTYKKHHKIRGIIV 84
W M + I+G ++W+I+E G ++ + EG+ + + + K+ K +I
Sbjct: 21 WSLRMKA-ILGA-HDVWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEM 144
+ + K+ + ++AK + L +Y+G+ +V++ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + ++ K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHESSKKSKSIA---LPSKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMA 261
+ +E E KK + I L + ++ ++Y+ + G +
Sbjct: 195 QAYE------EKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 262 MLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQKEKSKSRPK 321
+ + K + + + K C+NC K GH+ ++C + +K +
Sbjct: 249 PHEDNTNQRGENSSRGRGKGHPKSRYDKSSVK-CYNCGKFGHYASEC---KAPSNKKFEE 304
Query: 322 KPSFSSSKFRKQIKKSLMATWEDLDSE 348
K ++ K +++ LMA+++ + E
Sbjct: 305 KANYVEEKIQEE-DMLLMASYKKDEQE 330
>At1g48710 hypothetical protein
Length = 1352
Score = 84.3 bits (207), Expect = 5e-16
Identities = 70/327 (21%), Positives = 151/327 (45%), Gaps = 20/327 (6%)
Query: 25 WKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKKTYKKHHKIRGIIV 84
W M + I+G ++W+I+E G ++ + EG+ + + + K+ K +I
Sbjct: 21 WSLRMKA-ILGA-HDVWEIVEKGF--IEPENEGSL--SQTQKDGLRDSRKRDKKALCLIY 74
Query: 85 ASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEM 144
+ + K+ + ++AK + L +Y+G+ +V++ + L ++E +MK+ E + +
Sbjct: 75 QGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY 134
Query: 145 YSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 204
+SR T+ + L+ + ++ K+LRSL ++ VT IEE KDL +++E L+ SL
Sbjct: 135 FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSL 194
Query: 205 KVHEMSLNEHESSKKSKSI---ALPSKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMA 261
+ +E E KK + I L + ++ ++Y+ + G +
Sbjct: 195 QAYE------EKKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWR 248
Query: 262 MLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDLQKEKSKSRPK 321
+ + K + + + K C+NC K GH+ ++C + +K +
Sbjct: 249 PHEDNTNQRGENSSRGRGKGHPKSRYDKSSVK-CYNCGKFGHYASEC---KAPSNKKFEE 304
Query: 322 KPSFSSSKFRKQIKKSLMATWEDLDSE 348
K ++ K +++ LMA+++ + E
Sbjct: 305 KANYVEEKIQEE-DMLLMASYKKDEQE 330
>At4g04560 putative transposon protein
Length = 590
Score = 75.5 bits (184), Expect = 2e-13
Identities = 46/199 (23%), Positives = 97/199 (48%), Gaps = 3/199 (1%)
Query: 18 DPEEFSWWKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTP--AQKKTYKK 75
D E + +WK + I +D + W +EDG + I K T A +KT
Sbjct: 349 DAEHYGYWKVLIKRSIQSIDMDAWFAVEDGWMPPTTKDAKRDIVSKSRTEWIADEKTAAN 408
Query: 76 HH-KIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFK 134
H+ + +I S+ R ++ ++ +AK ++ L ++E + V+ + ML ++E
Sbjct: 409 HNSQALSVIFGSLLRNKFTQVQGCLSAKEVWEILQVSFECTNNVKRTRLDMLASEFENLT 468
Query: 135 MKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNT 194
M+ +ES+++ + ++ +L K+Y +V K LRSLP +++ +AI+ + + +
Sbjct: 469 MEAEESVDDFNGKLSSITQEAVVLGKTYKDKKMVKKFLRSLPDKFQSHKSAIDVSLNSDQ 528
Query: 195 LSVEDLVSSLKVHEMSLNE 213
L + +V ++ ++ E
Sbjct: 529 LKFDQVVGMMQAYDTDKEE 547
Score = 47.4 bits (111), Expect = 7e-05
Identities = 57/264 (21%), Positives = 109/264 (40%), Gaps = 43/264 (16%)
Query: 13 PKFNGDPEEFSWWKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTP--AQK 70
P FN E + +W+ M + ++LW+I+++GV + + H+P Q+
Sbjct: 8 PIFN--KENYGFWRIKMKTIFQ--TKKLWEIVDEGVPKPPAEGD--------HSPEAVQQ 55
Query: 71 KTYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQY 130
KT + ++ + I +T +SD + AS AL +Y
Sbjct: 56 KTRCEAASLKDLTALQILQTA---VSDSIFPRIAPAS--------------SALGKPWEY 98
Query: 131 ELFKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAK 190
E KMK+ ++I ++ + + L++ + +V KIL SLP R+ V +++ K
Sbjct: 99 ENLKMKESDNINTFMTKLIEMGNQLRVHGEEKSDYQIVQKILISLPKRFDIIVAMMKQTK 158
Query: 191 DLNTLSVEDL--VSSLKVHEMS----------LNEHESSKKSKSIALPSKGKSSKSSKAY 238
DL +LS V K H S L++ + + + G +K+ +
Sbjct: 159 DLTSLSAGKWCDVCERKNHNESDCWMKKNKGVLSQQVGNNERRCFVCNKPGHLAKNCRLR 218
Query: 239 KASESEEESPDGDSDEDHSVKMAM 262
+ + + + DEDH + A+
Sbjct: 219 RTERVDLSLEETNDDEDHMLFSAV 242
>At3g31380 hypothetical protein
Length = 262
Score = 75.1 bits (183), Expect = 3e-13
Identities = 46/156 (29%), Positives = 80/156 (50%), Gaps = 1/156 (0%)
Query: 80 RGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDE 139
RG++V SIP +++ + TAK ++ S+ + G+++V+EA+ L+ +E KMK+ E
Sbjct: 3 RGLLVQSIPEAFTLQVGNLQTAKEVWDSIKTRHVGAERVKEARVQTLMADFEKMKMKEAE 62
Query: 140 SIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLP-SRWRPKVTAIEEAKDLNTLSVE 198
I++ R L + L + LV K L SLP ++ + ++E+ DLN + E
Sbjct: 63 KIDDFAGRLSELSTKSAPLGVNIEVPKLVKKFLNSLPRKKYIHIIASLEQVLDLNNTTFE 122
Query: 199 DLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKS 234
D+V +KV+E + + E L SKS
Sbjct: 123 DIVGCMKVNEERVYDPEEETNEDQNKLMYTSSDSKS 158
>At3g25450 hypothetical protein
Length = 1343
Score = 72.0 bits (175), Expect = 3e-12
Identities = 45/186 (24%), Positives = 94/186 (50%), Gaps = 16/186 (8%)
Query: 74 KKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELF 133
+K+ R ++ SIP + +++ + T+ A++ ++ + G+++V+EA+ L+ +++
Sbjct: 58 EKNDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMAEFDKL 117
Query: 134 KMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLP-SRWRPKVTAIEEAKDL 192
KMKD E+I++ R + + L + S +V K L+SLP ++ V A+E+ DL
Sbjct: 118 KMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALEQVLDL 177
Query: 193 NTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDGDS 252
T + ED+ +K +E + + + S + + + +E EEE D
Sbjct: 178 KTTTFEDIAGRIKTYEDRVWDDDDSHEDQGKLM---------------TEVEEEVVDDLE 222
Query: 253 DEDHSV 258
+E+ V
Sbjct: 223 EEEEEV 228
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 66.6 bits (161), Expect = 1e-10
Identities = 79/361 (21%), Positives = 148/361 (40%), Gaps = 67/361 (18%)
Query: 7 SDSGKAPKFNGDPEEFSWWKTNMYSYIMGLDEELWDILEDGVDDLDLDEEGAAIDR-KIH 65
S + K +G+ + + WK + ++I EL +LE +D ++EE + + +
Sbjct: 3 SGHSEVEKLDGEGD-YVLWKEKLLAHI-----ELLGLLEGLEEDEAIEEEESTAETDSLL 56
Query: 66 TPAQKKTYK-KHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKAL 124
T + K K K K R ++ S+ K+ + TA M L K+ AK+L
Sbjct: 57 TKTEDKVLKEKRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVL-------DKLFMAKSL 109
Query: 125 ----MLVHQYELFKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWR 180
L + +KM D +IEE + F L+S L+ +K S D +L SLP ++
Sbjct: 110 PNRIYLKQRLYGYKMSDSMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFD 169
Query: 181 PKVTAIEEAKDLNTLSVEDLVSSL--KVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAY 238
++ K TL+++++ ++ KV E+ + S ++ + +G+S K K+
Sbjct: 170 QLKDTLKYGK--TTLALDEITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSS 227
Query: 239 KASESEEESPDGDSDEDHSVKMAMLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNC 298
+ ++S+ S K ++K C+ C
Sbjct: 228 ERNKSQSRS----------------------------------------KSREKKVCWVC 247
Query: 299 KKPGHFIADCPDLQKEKSKSRPKKPSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADD 358
K GHF C +++ K + SS+ + + +A E ES +D +E D+
Sbjct: 248 GKEGHFKKQCYVWKEKNKKGNNSEKGESSNVIGQAADAAALAVRE----ESNADNQEVDN 303
Query: 359 D 359
+
Sbjct: 304 E 304
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 62.0 bits (149), Expect = 3e-09
Identities = 49/199 (24%), Positives = 93/199 (46%), Gaps = 31/199 (15%)
Query: 93 MKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEMYSRFQTLV 152
+++ T+KA++ + + G+++V+EAK L+ +++ KMKD+E+I+E R +
Sbjct: 35 LQVGKLKTSKAVWDKIQSRNLGAERVKEAKLKTLMAEFDKLKMKDNETIDECAGRLSEIS 94
Query: 153 SGLQILKKSYVTSDLVSKILRSLPS-RWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSL 211
+ L + + +V K L+SLP+ ++ V A+E+ DL + +D+V +K +E +
Sbjct: 95 TKSTSLGEDIEETKVVKKFLKSLPTKKYIHIVAALEQVLDLKNTTFKDIVGRIKTYEDKI 154
Query: 212 --------NEHESSKK-------------SKSIALP---------SKGKSSKSSKAYKAS 241
E E +K S+ I L + + K K
Sbjct: 155 WVLITCLKKEAEEEEKSVVGVEAEEELVISREITLRLLVIAVINYASNCPDRLLKLIKLQ 214
Query: 242 ESEEESPDGDSDEDHSVKM 260
E ++E+ D D DE S+ M
Sbjct: 215 ERQQEAEDDDDDEVESLMM 233
>At2g10610 pseudogene
Length = 531
Score = 60.1 bits (144), Expect = 1e-08
Identities = 40/128 (31%), Positives = 64/128 (49%), Gaps = 5/128 (3%)
Query: 129 QYELFKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLP-SRWRPKVTAIE 187
++ KMKD I++ + L S LV K L+S+P R+ V A+E
Sbjct: 3 EFHRLKMKDSHKIDDFAGMLSEISVKSAALGVIIEESRLVKKFLKSVPRKRYIHIVAALE 62
Query: 188 EAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEES 247
+ DLN ++ +D+V LKV+E + + + ++ +S L S SSK ++ EEE
Sbjct: 63 QVLDLNAITFDDIVGRLKVYEERVYDEDDPEEDQSKLLYSSMDSSKR----ESEGEEEED 118
Query: 248 PDGDSDED 255
P G+ DED
Sbjct: 119 PIGEEDED 126
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 58.2 bits (139), Expect = 4e-08
Identities = 64/306 (20%), Positives = 120/306 (38%), Gaps = 47/306 (15%)
Query: 14 KFNGDPEEFSWWKTNMYSY--IMGLDEELWDILEDGVDDLDLDEEGAAIDRKIHTPAQKK 71
KF+G + ++ WK + ++ I+GL+ L + G LDE + K+ +
Sbjct: 10 KFDGRGD-YTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEALE 68
Query: 72 TYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYE 131
KK K R IV S+ K+ +STA AM +L Y ++
Sbjct: 69 EKKK--KARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYS-- 124
Query: 132 LFKMKDDESIEEMYSRFQTLVSGLQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKD 191
FKM ++ S+E F +++ L+ + D +L +LP + ++ +
Sbjct: 125 -FKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSG 183
Query: 192 LNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDGD 251
+ L+++++ +++ E+ L + S K ++ L K K+ K + +
Sbjct: 184 KSILTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENKGKGEQKGKG-------- 235
Query: 252 SDEDHSVKMAMLSNKLEYLARKQKKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADCPDL 311
K KK SK+ + GC+ C + GHF + CP+
Sbjct: 236 ---------------------KGKKGKSKK----------KPGCWTCGEEGHFRSSCPNQ 264
Query: 312 QKEKSK 317
K + K
Sbjct: 265 NKPQFK 270
>At3g21030 hypothetical protein
Length = 411
Score = 52.8 bits (125), Expect = 2e-06
Identities = 56/274 (20%), Positives = 113/274 (40%), Gaps = 36/274 (13%)
Query: 36 LDEELWDILEDGVD-DLDLDEEGAAIDRKIHTPAQKKTYKKHHKIRGIIVASIPRTEYMK 94
++ +WD++++GV + + E AA + + + K +K I+ +S+P + + K
Sbjct: 27 VENGVWDVVQNGVSPNPTTNPELAATIKAVDLAQWRNLVVKDNKALKIMQSSLPDSVFRK 86
Query: 95 MSDKSTAKAMFASLCANYEGSKKVREAKALMLVHQYELFKMKDDESIEEMYSRFQTLVSG 154
++AK ++ L + +EAK L Q+E M + E ++ R + ++
Sbjct: 87 TISIASAKELWDLL----KKGNDTKEAKLRRLEKQFENLMMYEGERMKLYLKRLEKIIEE 142
Query: 155 LQILKKSYVTSDLVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEH 214
L +L +++K+L SLP + V ++E L L+ DL LK EM ++
Sbjct: 143 LFVLGNPISDDKVIAKLLTSLPRSYDDSVPVLKEFMTLPELTHRDL---LKAFEMFGSDP 199
Query: 215 ESSKKSKSIALPSKGKSSKSSKAYKASESEEESPDGDSDEDHSVKMAMLSNKLEYLARKQ 274
++ + + K + + SE +H+ + + ++ YL +
Sbjct: 200 KTMPQ----------ELMKFIMILRRAWSERMWCSACETYNHNQEDCYYNPRVVYLGGGR 249
Query: 275 KKFLSKRNGYKNWKREDQKGCFNCKKPGHFIADC 308
+ CF C + GHF DC
Sbjct: 250 GQ------------------CFQCGERGHFARDC 265
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.361 0.158 0.581
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,796,746
Number of Sequences: 26719
Number of extensions: 1180885
Number of successful extensions: 11855
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 150
Number of HSP's that attempted gapping in prelim test: 11102
Number of HSP's gapped (non-prelim): 660
length of query: 1684
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1571
effective length of database: 8,299,349
effective search space: 13038277279
effective search space used: 13038277279
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146649.9


