

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146632.7 + phase: 0
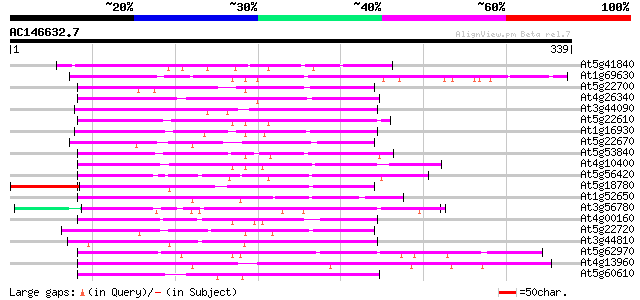
(339 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g41840 putative protein 82 4e-16
At1g69630 hypothetical protein 78 6e-15
At5g22700 unknown protein 77 1e-14
At4g26340 unknown protein 75 4e-14
At3g44090 putative protein 75 7e-14
At5g22610 unknown protein 72 6e-13
At1g16930 hypothetical protein 71 7e-13
At5g22670 unknown protein 71 1e-12
At5g53840 putative protein 70 1e-12
At4g10400 putative protein 70 1e-12
At5g56420 unknown protein 70 2e-12
At5g18780 putative protein 70 2e-12
At1g52650 F6D8.13 70 2e-12
At3g56780 putative protein 69 5e-12
At4g00160 unknown protein 68 8e-12
At5g22720 unknown protein 67 1e-11
At3g44810 hypothetical protein 67 1e-11
At5g62970 putative protein 67 2e-11
At4g13960 hypothetical protein 67 2e-11
At5g60610 unknown protein 66 3e-11
>At5g41840 putative protein
Length = 540
Score = 82.0 bits (201), Expect = 4e-16
Identities = 72/225 (32%), Positives = 111/225 (49%), Gaps = 27/225 (12%)
Query: 29 KRWRRVWLGMPNADRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDD 88
K ++V +G+ + DRIS LPD L+C ILS LP K+ T++L++RW+ L + +N DD
Sbjct: 2 KSRKKVVVGV-SGDRISGLPDALICHILSFLPTKEAASTTVLAKRWKPLLAFVPNLNFDD 60
Query: 89 TSYIHD---RDAYDRYYH-----VIALFLFEMKIHHPI----MKTVTILSASPFFNPLP- 135
+ Y H R+ Y + Y V ++ + K P+ +K ++ S +P
Sbjct: 61 SIYFHPRARRNKYSKSYESFMSFVDSVLALQAKTKTPLKRFHVKCEDVVDQSWVLEWIPK 120
Query: 136 -LWLGCLQVQHLDVTSSATLC-----LCVPYKVLNSTALVVLKLNALTIDYVH---RSST 186
L G L + L +TSS C +P K+ S LV LK+ D VH
Sbjct: 121 VLKRGVLDID-LHITSSRNYCENSSFYSLPSKIFVSKTLVRLKIQFQ--DGVHIDVEGGV 177
Query: 187 NLPSLKILHLTQVHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNT 231
+LP LK LHL ++ L K+LS LE+L+L +L D++
Sbjct: 178 SLPKLKTLHLDYFK-IETSMLNKLLSGCHALEELVLANLMWADSS 221
>At1g69630 hypothetical protein
Length = 451
Score = 78.2 bits (191), Expect = 6e-15
Identities = 94/370 (25%), Positives = 159/370 (42%), Gaps = 79/370 (21%)
Query: 37 GMPNADRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRD 96
G D IS LPD LLC +L LP K ++ TS+LSRRWR+L + +++D+T D
Sbjct: 13 GSDEVDWISKLPDCLLCEVLLNLPTKDVVKTSVLSRRWRNLWKHVPGLDLDNT----DFQ 68
Query: 97 AYDRYYHVIALFLFEMKIHHPIMKTVTILSASPFFNP----LPLWLGCL---QVQHLDV- 148
++ + + FL + + K + ++P + W+ + +VQH+DV
Sbjct: 69 EFNTFLSFVDSFL-DFNSESFLQKFILKYDCDDEYDPDIFLIGRWINTIVTRKVQHIDVL 127
Query: 149 -TSSATLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFL 207
S + + +P + +LV LKL LT+ +LPSLK++ L F L
Sbjct: 128 DDSYGSWEVQLPSSIYTCESLVSLKLCGLTL--ASPEFVSLPSLKVMDLIITKFADDMGL 185
Query: 208 IKILSMSPLLEDLLLKD--------LQVTDNTLAH------------DDAAALKPFPKLL 247
+++ P+LE L ++ L+V +L +D PKL
Sbjct: 186 ETLITKCPVLESLTIERSFCDEIEVLRVRSQSLLRFTHVADSDEGVVEDLVVSIDAPKLE 245
Query: 248 RADVSESSISAFLL--PLKLF---------------YNVQFLRSQVLLQ----------- 279
+S+ +++F+L P KL +N L + +++
Sbjct: 246 YLRLSDHRVASFILNKPGKLVKADIDIVFNLSSVNKFNPDDLPKRTMIRNFLLGISTIKD 305
Query: 280 ------TLE--QDFSTTQ----FLNLTHMDLIFHDGYYWISLMKFICACPSLQTLTIHNI 327
TLE DFS + F NL+ + + F+ GY W L F+ +CP+L+TL + +
Sbjct: 306 MIIFSSTLEVIYDFSRCERLPLFRNLSVLCVEFY-GYMWEMLPIFLESCPNLKTLVVKS- 363
Query: 328 GGDYDDHNNN 337
Y + N
Sbjct: 364 -ASYQEKGEN 372
>At5g22700 unknown protein
Length = 452
Score = 77.4 bits (189), Expect = 1e-14
Identities = 63/190 (33%), Positives = 97/190 (50%), Gaps = 24/190 (12%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRS--LRPRMTEINI----DDTSYIHDR 95
DRIS LPDELLC+ILS LP K + TS+LS RWRS L + +I+I D T+++
Sbjct: 21 DRISSLPDELLCQILSNLPTKNAVTTSILSTRWRSIWLSTPVLDIDIDAFDDATTFVSFA 80
Query: 96 DAYDRYYHVIALFLFEMKIHHPIMKTVTILSASPFFNPLPLWLGCL---QVQHLDVTSSA 152
+ + L F++ + + TI+ W+ ++QHL+V
Sbjct: 81 SRFLEFSKDSCLHKFKLSVERDDVDMCTIMP----------WIQDAVNRRIQHLEVDCRF 130
Query: 153 TLCLCVPYKVLN-STALVVLKLNALTIDYVHR-SSTNLPSLKILHLTQVHFLKLKFLIKI 210
+ Y L S LV L+L+ +T+ HR +LP+LK++HL + + L+ L
Sbjct: 131 SFHFEAVYLTLYLSETLVSLRLHFVTL---HRYEFVSLPNLKVMHLEENIYYCLETLENF 187
Query: 211 LSMSPLLEDL 220
+S P+LEDL
Sbjct: 188 ISSCPVLEDL 197
>At4g26340 unknown protein
Length = 419
Score = 75.5 bits (184), Expect = 4e-14
Identities = 60/190 (31%), Positives = 98/190 (51%), Gaps = 15/190 (7%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
DRIS L D+LL +ILS +P K ++ TSLLS+RW+SL ++E+ DD+ + D ++ ++
Sbjct: 2 DRISQLSDDLLLQILSFIPGKDVVATSLLSKRWQSLWMLVSELEYDDSYHTGDYKSFSQF 61
Query: 102 YHVIALFLFEMKIHHPIMKTVTI-LSASPFFNPLPLWLGCLQVQHLDV------TSSATL 154
++ + + P++K + + L + LW+G + L TSS
Sbjct: 62 -----VYRSLLSNNAPVIKHLHLNLGPDCPAIDIGLWIGFALTRRLRQLKINIRTSSNDA 116
Query: 155 CLCVPYKVLNSTALVVLKL-NALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILSM 213
+P + S L L+L N + +D SS LPSLK+LHL V + L +L
Sbjct: 117 SFSLPSSLYTSDTLETLRLINFVLLDV--PSSVCLPSLKVLHLKTVDYEDDASLPSLLFG 174
Query: 214 SPLLEDLLLK 223
P LE+L ++
Sbjct: 175 CPNLEELFVE 184
>At3g44090 putative protein
Length = 449
Score = 74.7 bits (182), Expect = 7e-14
Identities = 57/195 (29%), Positives = 91/195 (46%), Gaps = 18/195 (9%)
Query: 40 NADRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYD 99
N + LPD+LL +IL LP K+ + TSLLS+RWR+L + +++DD + H D Y+
Sbjct: 22 NLASMDCLPDDLLVQILYFLPTKEAISTSLLSKRWRTLYSLVHNLDLDDYIFWHHEDGYN 81
Query: 100 RYYHVIALFLFEMKIHHPI-------MKTVTILSASPF-----FNPLPLWLGCLQVQHLD 147
+Y+ FE + + +KT +++S + + + WL + +
Sbjct: 82 QYFSNDIQKSFEEFMERTLALLGGGHIKTFSLISDEIYRFDHVVDSIRPWL------YYN 135
Query: 148 VTSSATLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFL 207
+ + PYKV ST LV L L T+LP LK+L L + F +
Sbjct: 136 LQKDSLWQFGFPYKVFTSTKLVKLSLGTRLACPRIPQDTSLPVLKVLLLEYIWFEDNQLS 195
Query: 208 IKILSMSPLLEDLLL 222
L+ P LEDL +
Sbjct: 196 DVFLAACPALEDLTI 210
>At5g22610 unknown protein
Length = 502
Score = 71.6 bits (174), Expect = 6e-13
Identities = 57/199 (28%), Positives = 101/199 (50%), Gaps = 16/199 (8%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
D IS LP+ LL +ILS LP K I+ TS+LS+RW+S+ + +++D + + H YD +
Sbjct: 18 DLISKLPEVLLSQILSYLPTKDIVRTSVLSKRWKSVWLLIPGLDLDSSEFPH----YDTF 73
Query: 102 YHVIALFLFEMKIHHPIMKTVTILSASPFFNP--LPLWLGCL---QVQHLDVTSSATLC- 155
+ FLF + +P + + + +P + LW C+ ++QHLDV +
Sbjct: 74 VDFMNEFLFFSREENPCLHKLKLSIQKNENDPSCVTLWTDCVARGKLQHLDVEFGGRVME 133
Query: 156 ----LCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKIL 211
+P + L+ L+L + + +S +LPSLK + L + + L ++
Sbjct: 134 REFWEMMPLSLYICKTLLHLRLYRVLLGNFDQSVDSLPSLKSMCLEENVYSNEASLESLI 193
Query: 212 SMSPLLEDLLLKDLQVTDN 230
S +LEDL + +++ DN
Sbjct: 194 SSCRVLEDLTI--VKIDDN 210
>At1g16930 hypothetical protein
Length = 449
Score = 71.2 bits (173), Expect = 7e-13
Identities = 59/193 (30%), Positives = 98/193 (50%), Gaps = 20/193 (10%)
Query: 40 NADRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYD 99
N+DRIS LPD LLC+ILS L K+ + TS+LS+RWR+L + +++D ++ D D
Sbjct: 13 NSDRISNLPDSLLCQILSDLSTKESVCTSVLSKRWRNLWLHVPVLDLDSNNFPDD----D 68
Query: 100 RYYHVIALFLFEMKIHH----PIMKTVTILSASPFFNPLPLWLGCL---QVQHLDVTSSA 152
+ + FL H ++ V AS F + W+ + +V H +V +
Sbjct: 69 VFVSFVNRFLGSENEQHLERFKLIYEVNEHDASRFKS----WINAVIKRRVCHFNVHNEV 124
Query: 153 ---TLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIK 209
+ +P + + LV L+L + +D H S +LP +KI+HL V + L
Sbjct: 125 DDDDELVKMPLSLYSCERLVNLQLYRVALD--HPESVSLPCVKIMHLDMVKYDADSTLEI 182
Query: 210 ILSMSPLLEDLLL 222
++S P+LE+L +
Sbjct: 183 LISGCPVLEELTI 195
>At5g22670 unknown protein
Length = 443
Score = 70.9 bits (172), Expect = 1e-12
Identities = 64/202 (31%), Positives = 96/202 (46%), Gaps = 38/202 (18%)
Query: 37 GMPNADRISILPDELLCRILSLLPAKQIMVTSLLSRRWR--SLRPRMTEINIDDTSYIHD 94
G+ + D IS+LPD+LLCRILS LP K + TS+LS+RW+ SL + E N+ +
Sbjct: 6 GVTSQDSISLLPDDLLCRILSNLPTKVAVRTSVLSKRWKRFSLSVPLLEFNVSEF----- 60
Query: 95 RDAYDRYYHVIALFL--------------FEMKIHHPIMKTVTILSASPFFNPLPLWLGC 140
Y + V+ FL FE K H T I +A
Sbjct: 61 -HGYYEFARVVHGFLDTSRETCIHKLKLAFEKKQHDRSYLTQWIHNAVK----------- 108
Query: 141 LQVQHLDVTSSATLCL-CVPYKVLNSTALVVLKLNALTI-DYVHRSSTNLPSLKILHLTQ 198
+VQHLD+ + L +P+ + LV L+L+ +++ D+ H +LP LK +HL
Sbjct: 109 RKVQHLDIGRWSYLGQELIPHSLYTCETLVSLRLHNVSLPDFDH---VSLPRLKTMHLID 165
Query: 199 VHFLKLKFLIKILSMSPLLEDL 220
+ L ++S P+LEDL
Sbjct: 166 NIYPNDALLENLISSCPVLEDL 187
>At5g53840 putative protein
Length = 444
Score = 70.5 bits (171), Expect = 1e-12
Identities = 63/205 (30%), Positives = 99/205 (47%), Gaps = 23/205 (11%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
+R+S LPD L+C ILS L K + TS+LS RWR+L + ++ D + ++ +
Sbjct: 18 ERLSQLPDHLICVILSHLSTKDAVRTSILSTRWRNLWQLVPVLDFDS----RELRSFSEF 73
Query: 102 YHVIALFLFEMKIHHPIMKTVTILSASPFFNPLPLWLGCL---QVQHLDVTSSATLCL-- 156
F + K + V I + + L W+ + ++QH+D+ S C
Sbjct: 74 VSFAGSFFYLHKDSYIQKLRVCIYDLAGNYY-LTSWIDLVTRHRIQHIDI--SVFTCSGF 130
Query: 157 -CVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILSMSP 215
+P + LV LKL+ +T+ V+ +LP LKIL L V+F L KI+S SP
Sbjct: 131 GVIPLSLYTCDTLVHLKLSRVTM--VNVEFVSLPCLKILDLDFVNFTNETTLDKIISCSP 188
Query: 216 LLEDLLL--------KDLQVTDNTL 232
+LE+L + K +QV TL
Sbjct: 189 VLEELTIVKSSEDNVKIIQVRSQTL 213
>At4g10400 putative protein
Length = 326
Score = 70.5 bits (171), Expect = 1e-12
Identities = 65/229 (28%), Positives = 108/229 (46%), Gaps = 22/229 (9%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
DRIS LPDE+L +ILS +P K + TS+LS+RW L +T++ Y + +
Sbjct: 2 DRISGLPDEVLVKILSFVPTKVAVSTSILSKRWEFLWMWLTKLKFGSKRY-----SESEF 56
Query: 102 YHVIALFLFEMKIHH-PIMKTVTILSASPFFNP--LPLWLGCL---QVQHLDVTSS--AT 153
+ + +H P++++ ++ + F P + +W+ ++ L + SS
Sbjct: 57 KRLQCFLDRNLPLHRAPVIESFRLVLSDSHFKPEDIRMWVVVAVSRYIRELKIYSSHYGE 116
Query: 154 LCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILSM 213
+P + +LV+LKL+ + V R LPSLK L L V + K L ++L
Sbjct: 117 KQNILPSSLYTCKSLVILKLDGGVLLDVPR-MVCLPSLKTLELKGVRYFKQGSLQRLLCN 175
Query: 214 SPLLEDLLLKDLQVTDNTLAHDDAAALKPF-PKLLRADVSESSISAFLL 261
P+LEDL++ N HD+ L P L R +S S F++
Sbjct: 176 CPVLEDLVV-------NLSHHDNMGKLTVIVPSLQRLSLSTPSSREFVI 217
>At5g56420 unknown protein
Length = 422
Score = 69.7 bits (169), Expect = 2e-12
Identities = 69/240 (28%), Positives = 107/240 (43%), Gaps = 35/240 (14%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
DR+S LPD+ L +ILS LP K ++VTSLLS+RWR L + +N D +HD + R+
Sbjct: 6 DRLSQLPDDFLLQILSWLPTKDVLVTSLLSKRWRFLWTLVPRLNYD--LRLHD-NTCPRF 62
Query: 102 YHVIALFLFEMKIHHPIMKTVTILSASPFF---NPLPLWLGCLQVQHLDVTSSATLC--- 155
+ L K P ++++ I S F + +W+ + V S + C
Sbjct: 63 SQFVDRSLLLHKA--PTLESLNIKIGSICFTAEKDVGVWVR-IGVDRFVRELSVSYCSGE 119
Query: 156 --LCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILSM 213
+ +P + + L VLKL +T++ SLK LHL V +L + L +I+S
Sbjct: 120 EPIRLPKCLFTCSTLAVLKLENITLEDA-SCYVCFQSLKTLHLLDVKYLDDQSLPRIISS 178
Query: 214 SPLLEDLLLK--------------------DLQVTDNTLAHDDAAALKPFPKLLRADVSE 253
LEDL+++ L + DD L PKL R D+ +
Sbjct: 179 CSSLEDLVVQRCPGDNVKVVTVTAPSLKTLSLHKSSQAFEGDDDGFLIDTPKLKRVDIED 238
>At5g18780 putative protein
Length = 441
Score = 69.7 bits (169), Expect = 2e-12
Identities = 55/186 (29%), Positives = 90/186 (47%), Gaps = 16/186 (8%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDR------ 95
DRISILP+ LLC ILS L K + TS+LS RWR L + +++D + + D
Sbjct: 11 DRISILPEPLLCHILSFLRTKDSVRTSVLSSRWRDLWLWVPRLDLDKSDFSDDNPSASFV 70
Query: 96 DAYDRYYHVIALFLFEMKIHHPIMKTVTILSASPFFNPLPLWLGCLQVQHLDVTSSATLC 155
D + + L F++ H + T+ + + L ++QH ++ + C
Sbjct: 71 DKFLNFRGESYLRGFKLNTDHDVYDISTL-------DACLMRLDKCKIQHFEIENCFGFC 123
Query: 156 -LCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILSMS 214
L +P + LV LKL+ + + S +LP L+I+H +V F K +++ S
Sbjct: 124 ILLMPLIIPMCHTLVSLKLSFVILSKF--GSLSLPCLEIMHFEKVIFPSDKSAEVLIACS 181
Query: 215 PLLEDL 220
P+L DL
Sbjct: 182 PVLRDL 187
Score = 48.1 bits (113), Expect = 7e-06
Identities = 22/42 (52%), Positives = 30/42 (71%)
Query: 1 MSGLPDELLCEILSFLPSEEFPSTSLLSKRWRRVWLGMPNAD 42
+S LP+ LLC ILSFL +++ TS+LS RWR +WL +P D
Sbjct: 13 ISILPEPLLCHILSFLRTKDSVRTSVLSSRWRDLWLWVPRLD 54
>At1g52650 F6D8.13
Length = 465
Score = 69.7 bits (169), Expect = 2e-12
Identities = 58/209 (27%), Positives = 95/209 (44%), Gaps = 17/209 (8%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
D +S LP+ +LC I S L K+ +TS+L +RWR+L + + IDD+ ++H + +
Sbjct: 2 DHVSSLPEGVLCNIFSFLTTKEAALTSILCKRWRNLLAFVPNLVIDDSVFLHPEEGKEER 61
Query: 102 YHVIALFL------FEMKIHHPIMKTVTILSASPFFNPLPLWL------GCLQVQHLDVT 149
Y + F+ ++ + PI K + + W+ G ++ L +
Sbjct: 62 YEIQQSFMEFVDRVLALQGNSPIKKFSLKFRTDFASHRVNAWISNVLARGVSELDVLVIL 121
Query: 150 SSATLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIK 209
A P K S LV LK+N+L ID++ LP LK L L V KF +
Sbjct: 122 YGAEFLPLSP-KCFKSRNLVTLKINSLGIDWL-AGDIFLPMLKTLVLHSVKLCVDKFFFR 179
Query: 210 ILSMSPLLEDLLLKDLQVTDNTLAHDDAA 238
L P LE+L+L + D + +A+
Sbjct: 180 AL---PALEELVLFAVSWRDRDVTVSNAS 205
>At3g56780 putative protein
Length = 431
Score = 68.6 bits (166), Expect = 5e-12
Identities = 65/231 (28%), Positives = 111/231 (47%), Gaps = 22/231 (9%)
Query: 44 ISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRYYH 103
I+ LPD+L+ +ILS + K ++VTSLLS++W+SL R+ + D + D ++R+
Sbjct: 9 INELPDDLILKILSFVSTKHVVVTSLLSKKWKSLWTRVPILKYD----VRDHTRFERF-- 62
Query: 104 VIALFLFEMKIHHPIMKTVTI-LSASPFFNPLPLWLGCLQVQHLDVTSSATLCLCVPYKV 162
+ LF + H +++++ + LS + + + W+ H + C+ + V
Sbjct: 63 -LDKSLFSHQSH--VLESLHVELSVTLWNKDIGPWIRTALHHHHCHLRELEIDACIVHTV 119
Query: 163 L-----NSTALVVLKLNALTIDY-VHRSSTNLPSLKILHLTQVHFLKLKFLIKILSMSPL 216
L LVVLKL + ID ++ +LPSLK LH+ L +LS
Sbjct: 120 LPPELFTCKTLVVLKLKGIVIDVEAPLTTVSLPSLKTLHIDHSSLFDFGSLQMLLSNCNF 179
Query: 217 LEDLLLKDLQVTDNTLAHDDAAALKPFPKL----LRADVSESSISAFLLPL 263
+ DL++ ++ + A D + K L L+ +S SS SA LPL
Sbjct: 180 ITDLMV--IRESRFFFAEYDVSWCKTLMALKLEGLKDVISISSSSAVCLPL 228
Score = 45.4 bits (106), Expect = 4e-05
Identities = 72/299 (24%), Positives = 118/299 (39%), Gaps = 54/299 (18%)
Query: 4 LPDELLCEILSFLPSEEFPSTSLLSKRWRRVWLGMPNADRISILPDELLCRILSLLPAKQ 63
LPD+L+ +ILSF+ ++ TSLLSK+W+ +W R+ IL + + ++
Sbjct: 12 LPDDLILKILSFVSTKHVVVTSLLSKKWKSLWT------RVPILKYD----VRDHTRFER 61
Query: 64 IMVTSLLSRRWRSLRPRMTEINID---------DTSYIHDRDAYDRYYHVIALF------ 108
+ SL S + L E+++ + +H + R + A
Sbjct: 62 FLDKSLFSHQSHVLESLHVELSVTLWNKDIGPWIRTALHHHHCHLRELEIDACIVHTVLP 121
Query: 109 --LFEMK--------------------IHHPIMKTVTILSASPF-FNPLPLWLG-CLQVQ 144
LF K + P +KT+ I +S F F L + L C +
Sbjct: 122 PELFTCKTLVVLKLKGIVIDVEAPLTTVSLPSLKTLHIDHSSLFDFGSLQMLLSNCNFIT 181
Query: 145 HLDVTSSATLCLCVPYKVLNSTALVVLKLNAL--TIDYVHRSSTNLPSLKILHLTQVHFL 202
L V + Y V L+ LKL L I S+ LP LK LH+ ++
Sbjct: 182 DLMVIRESRFFFA-EYDVSWCKTLMALKLEGLKDVISISSSSAVCLPLLKTLHVARMEDF 240
Query: 203 KLKFLIKILSMSPLLEDLLLKDLQVTDNTLAHD-DAAALKPFPKLLRADVSESSISAFL 260
++LS P+L DL L++ + +D L D D L+ + R D I + +
Sbjct: 241 NNDSFCRLLSNCPVLSDLTLEE-KTSDVLLNLDIDMPYLQRLSIITRVDADSKHIFSLM 298
>At4g00160 unknown protein
Length = 453
Score = 67.8 bits (164), Expect = 8e-12
Identities = 55/197 (27%), Positives = 93/197 (46%), Gaps = 29/197 (14%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
DRIS LPD LL +ILS LP K ++ TS+ S++WR L + + D Y D++ Y
Sbjct: 16 DRISELPDALLIKILSFLPTKIVVATSVFSKQWRPLWKLVPNLEFDSEDY-DDKEQYTFS 74
Query: 102 YHVIALFLFEMKIHHPIMKTVTILSASPFFNP--LPLWLGCLQVQHL--------DVTSS 151
V FL P++++ + S +P + LW+G +HL D +
Sbjct: 75 EIVCKSFLSHKA---PVLESFRLEFESEKVDPVDIGLWVGIAFSRHLRELVLVAADTGTG 131
Query: 152 ------ATLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLK 205
++LC C + L L+++ +++ + + SL+ LHL V +
Sbjct: 132 TAFKFPSSLCTCNTLETLRLVLLILVDISSPVV---------MKSLRTLHLELVSYKDES 182
Query: 206 FLIKILSMSPLLEDLLL 222
+ +LS P+LE+LL+
Sbjct: 183 SIRNLLSGCPILEELLV 199
>At5g22720 unknown protein
Length = 422
Score = 67.4 bits (163), Expect = 1e-11
Identities = 60/198 (30%), Positives = 96/198 (48%), Gaps = 16/198 (8%)
Query: 32 RRVWLGMPNADRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSL---RPRMTEINIDD 88
R V D IS LPD L+ +IL L K+ + TS+LS+RWRSL P + I+ D
Sbjct: 13 RTVCSSKAKEDLISQLPDSLITQILFYLQTKKAVTTSVLSKRWRSLWLSTPGLVLISNDF 72
Query: 89 TSYIHDRDAYDRYYHVIALFLFEMKIHHPIMKTVTILSASPFFNPLPLWLGCL---QVQH 145
T Y +A+ + F E K+ +K ++I + + W+ + +++H
Sbjct: 73 TDY----NAFVSFVDKFLGFSREQKLCLHKLK-LSIRKGENDQDCVTRWIDFVATPKLKH 127
Query: 146 LDVTSSATLCLC---VPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFL 202
LDV T C C +P + + +L+ L+LN + + S +LP LK + L Q +
Sbjct: 128 LDVEIGPTRCECFEVIPLSLYSCESLLYLRLNHVCLGKF--ESVSLPCLKTMSLEQNIYA 185
Query: 203 KLKFLIKILSMSPLLEDL 220
L ++S P+LEDL
Sbjct: 186 NEADLESLISTCPVLEDL 203
>At3g44810 hypothetical protein
Length = 452
Score = 67.4 bits (163), Expect = 1e-11
Identities = 64/202 (31%), Positives = 88/202 (42%), Gaps = 17/202 (8%)
Query: 36 LGMPNADRISI--LPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIH 93
LGM + S+ LPDELL +LS L KQ TS+LS+RWR+L ++ DD+ H
Sbjct: 3 LGMSSVSTASLNCLPDELLVHVLSSLETKQAASTSVLSKRWRTLFAVRRNLDFDDSIISH 62
Query: 94 DR-------DAYDRYYHVIALFL-FEMKIHHPIMKTVTILSASPFFNPLPLWLGC----- 140
D + + + L F+ + PI K I + W+
Sbjct: 63 PEVGEQNMDDVQESFRDFVDKRLAFQGSV--PINKFSLIYGDKHDDVRVDRWINTALEHG 120
Query: 141 LQVQHLDVTSSATLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVH 200
+ HL +TS P V ST LV L L S T LP LKIL L +
Sbjct: 121 VSELHLCLTSVTRRLHRFPSNVFRSTTLVKLTLGTNLFIVYFPSDTCLPVLKILVLDSIW 180
Query: 201 FLKLKFLIKILSMSPLLEDLLL 222
F ++KF +L+ P LEDL +
Sbjct: 181 FDRIKFSNVLLAGCPALEDLTI 202
>At5g62970 putative protein
Length = 449
Score = 66.6 bits (161), Expect = 2e-11
Identities = 92/356 (25%), Positives = 150/356 (41%), Gaps = 82/356 (23%)
Query: 42 DRIS-ILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDR 100
D+IS DELL +ILS LP K + TS+LS++W+ L R+ ++ D+ S ++ + R
Sbjct: 2 DKISGFSDDELLVKILSFLPFKFAITTSVLSKQWKFLWMRVPKLEYDEDS-MYSFEYSFR 60
Query: 101 YY-------------------HVIALFLFE-MKIH-HPIMKTVTILSASPFFNP--LPLW 137
Y+ H + F+ + + +H P+++++ + + F P + LW
Sbjct: 61 YFLPKAKEVDSETYSIVSESGHRMRSFIEKNLPLHSSPVIESLRLKFFTEVFQPEDIKLW 120
Query: 138 L-----GCLQVQHLDVTSSATLCLCVPYKVLNSTALVVLKL-NALTIDYVHRSSTNLPSL 191
+ C Q +D +P + +LV LKL N + +D H S LPSL
Sbjct: 121 VEIAVSRCAQELSVDFFPKEKHNALLPRNLYTCKSLVTLKLRNNILVDVPHVFS--LPSL 178
Query: 192 KILHLTQVHFLKLKFLIKILSMSPLLEDLLLKDLQVTDNTLAHD---------------- 235
KILHL +V + + L ++LS +LEDL++ +L DN D
Sbjct: 179 KILHLERVTYGDGESLQRLLSNCSVLEDLVV-ELDTGDNVRKLDVIIPSLLSLSFGMSRY 237
Query: 236 --------DAAALKPF----------------PKLLRADVSESSISAFLLPL-----KLF 266
D +LK F PKL A+++ LL L +L
Sbjct: 238 CAYEGYRIDTPSLKYFKLTDLSKTFSGLIENMPKLEEANITARHNFKKLLELVTSVKRLS 297
Query: 267 YNVQFLRSQVLLQTLEQDFSTTQFLNLTHMDLIFHDGYYWISLMKFICACPSLQTL 322
N++ ++ L D F L H++ H+ Y+ L + A P LQ L
Sbjct: 298 LNIENNDAEALTAIYGDDI---VFNELEHLNFHIHNAYWSELLYWLLKASPKLQNL 350
>At4g13960 hypothetical protein
Length = 434
Score = 66.6 bits (161), Expect = 2e-11
Identities = 72/333 (21%), Positives = 139/333 (41%), Gaps = 58/333 (17%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTSYIHDRDAYDRY 101
D +S LPDE+L ILS L K+ +TS+LS+RWR+L + ++IDD+ ++H ++ +
Sbjct: 2 DHVSSLPDEVLYHILSFLTTKEAALTSILSKRWRNLFTFVPNLDIDDSVFLHPQEGKEDR 61
Query: 102 YHVIALFL------FEMKIHHPIMKTVTILSASPFFNPLPLWLGCLQVQHLDVTSSATLC 155
Y + F+ ++ + PI K L + + W+ S+A
Sbjct: 62 YEIQKSFMKFVDRVLALQGNSPIKKLSLKLRTGFDSHRVDGWI-----------SNALAR 110
Query: 156 LCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLK--ILHLTQVHFLKLKFLIKILSM 213
+L LV LKLN++ +D++ LP LK +LH ++ + + K +++
Sbjct: 111 GVTELDLLIILNLVTLKLNSVRVDWLAAGDIFLPMLKTLVLHSVRLCVDNVNWGGKDVNV 170
Query: 214 SPLLEDLLLKDLQVTDNTLAHDDAAALK---------PFPKLLRADVSESSISAFLLPLK 264
S +L + + T + D + + +P ++ ++ E+ IS + +
Sbjct: 171 SNASLKILTVNYNICLGTFSVDTPSLVYFCFTEYVAIDYPVVIMQNLFEARISLLVTQDQ 230
Query: 265 L----FYNVQFLRSQVLLQTLEQD--------------------------FSTTQFLNLT 294
+ N+ ++ V+L+ + + S F NL
Sbjct: 231 IERARAPNIDWVEDDVVLRLVNMEKLIKGIRNVQYMYLSPNTLEVLSLCCESMPVFNNLK 290
Query: 295 HMDLIFHDGYYWISLMKFICACPSLQTLTIHNI 327
+ L H W ++ + CP L+TL + I
Sbjct: 291 SLTLKSHKSRGWQAMPVLLRNCPHLETLVLEGI 323
>At5g60610 unknown protein
Length = 388
Score = 65.9 bits (159), Expect = 3e-11
Identities = 61/191 (31%), Positives = 92/191 (47%), Gaps = 21/191 (10%)
Query: 42 DRISILPDELLCRILSLLPAKQIMVTSLLSRRWRSLRPRMTEINIDDTS-YIHDRDAYDR 100
DRIS LPDELL +I+S +P K + TS+LS+RW SL + ++ D T + D
Sbjct: 2 DRISGLPDELLVKIISFVPTKVAVSTSILSKRWESLWKWVPKLECDCTEPALRD------ 55
Query: 101 YYHVIALFLFEMKIHHPIMKTVTI--LSASPFFNPLPLWLG-----CLQVQHLDVTSS-A 152
L + + I++++ + S F + LW G CL+ +D S A
Sbjct: 56 ------FILKNLPLQARIIESLYLRFRRESFLFQDIKLWGGIAISHCLRELRIDFFSHYA 109
Query: 153 TLCLCVPYKVLNSTALVVLKLNALTIDYVHRSSTNLPSLKILHLTQVHFLKLKFLIKILS 212
+ +P + +LV LKL L I LPSLK L L V + + L +LS
Sbjct: 110 NPYVILPRSLYTCKSLVTLKLLGLGIRVDVPRDVCLPSLKTLLLQCVAYSEEDPLRLLLS 169
Query: 213 MSPLLEDLLLK 223
P+LEDL+++
Sbjct: 170 CCPVLEDLVIE 180
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,458,201
Number of Sequences: 26719
Number of extensions: 293211
Number of successful extensions: 1951
Number of sequences better than 10.0: 254
Number of HSP's better than 10.0 without gapping: 227
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 1274
Number of HSP's gapped (non-prelim): 627
length of query: 339
length of database: 11,318,596
effective HSP length: 100
effective length of query: 239
effective length of database: 8,646,696
effective search space: 2066560344
effective search space used: 2066560344
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146632.7


