

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146632.4 - phase: 0
(1038 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
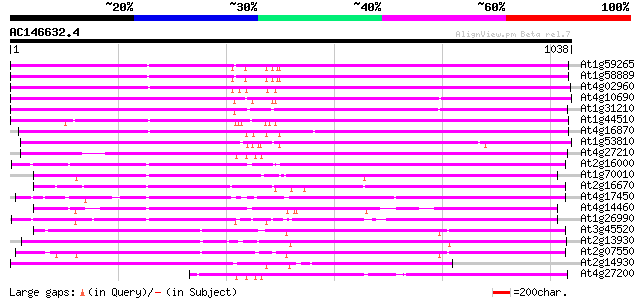
Score E
Sequences producing significant alignments: (bits) Value
At1g59265 polyprotein, putative 815 0.0
At1g58889 polyprotein, putative 813 0.0
At4g02960 putative polyprotein of LTR transposon 798 0.0
At4g10690 retrotransposon like protein 784 0.0
At1g31210 putative reverse transcriptase 783 0.0
At1g44510 polyprotein, putative 782 0.0
At4g16870 retrotransposon like protein 772 0.0
At1g53810 768 0.0
At4g27210 putative protein 717 0.0
At2g16000 putative retroelement pol polyprotein 652 0.0
At1g70010 hypothetical protein 642 0.0
At2g16670 putative retroelement pol polyprotein 608 e-174
At4g17450 retrotransposon like protein 580 e-165
At4g14460 retrovirus-related like polyprotein 579 e-165
At1g26990 polyprotein, putative 546 e-155
At3g45520 copia-like polyprotein 512 e-145
At2g13930 putative retroelement pol polyprotein 511 e-145
At2g07550 putative retroelement pol polyprotein 510 e-144
At2g14930 pseudogene 509 e-144
At4g27200 putative protein 501 e-142
>At1g59265 polyprotein, putative
Length = 1466
Score = 815 bits (2104), Expect = 0.0
Identities = 459/1130 (40%), Positives = 632/1130 (55%), Gaps = 101/1130 (8%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS+ NLS + + ++V +G +IPI T+LS + PL L N+L+ P + KNL
Sbjct: 341 ITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHNILYVPNIHKNL 400
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLY--PITTSQAIS--PSTFA 116
+SV + N VSVEF P F VKD TG+ L++ +++ +LY PI +SQ +S S +
Sbjct: 401 ISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASSQPVSLFASPSS 460
Query: 117 ALAPSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHI-CHSCSLGKHIKLPFVSSNSCT 175
S WHARLGHP +++S+ N + S + C C + K K+PF S +
Sbjct: 461 KATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDCLINKSNKVPFSQSTINS 520
Query: 176 IMPFDIIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRT 235
P + I+SD+W+SP+LS +RYYV+FVD ++ + W +PL +KSQV F++F+ +
Sbjct: 521 TRPLEYIYSDVWSSPILSHDNYRYYVIFVDHFTRYTWLYPLKQKSQVKETFITFKNLLEN 580
Query: 236 QFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRT 295
+F+ + DNG EF WE+ ++G++ S PHT NG +ERK R I T
Sbjct: 581 RFQTRIGTFYSDNGGEFVA--LWEYFSQHGISHLTSPPHTPEHNGLSERKHRHIVETGLT 638
Query: 296 LLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPL 355
LL HAS+P ++W +A +A YLIN LP L +SP + L+ P+Y LRVFGC CYP
Sbjct: 639 LLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNYDKLRVFGCACYPW 698
Query: 356 FPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKL---- 411
+KL +S CVFLGY +Y C L + ++ ISRHV FDE FPF+
Sbjct: 699 LRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDENCFPFSNYLATL 758
Query: 412 ----------------HNPQPYTYGFMDDGPSPYVIHHL--------------------- 434
H P T + PS HH
Sbjct: 759 SPVQEQRRESSCVWSPHTTLP-TRTPVLPAPSCSDPHHAATPPSSPSAPFRNSQVSSSNL 817
Query: 435 -----TSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPN--------------- 474
+S PS +P QPTTQPT + Q HS ++S N
Sbjct: 818 DSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQ-THSSQNTSQNNPTNESPSQLAQSLS 876
Query: 475 -PSPSTTATPSP-------PYQPTPISV----PKPV------------------TRSQHG 504
P+ S++++PSP PTP S+ P P+ TR++ G
Sbjct: 877 TPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPLNTHSMGTRAKAG 936
Query: 505 IFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPD-VN 563
I KP + +L S+ PR + AL+D W+ AM E NA I N TW+LVP PP V
Sbjct: 937 IIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDLVPPPPSHVT 996
Query: 564 VIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALS 623
++ WIFT K SDG RYKARLV G Q+ G+D ETFSPV+K +IR VL +A+
Sbjct: 997 IVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVD 1056
Query: 624 KAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKR 683
++W I QLDV NAFL G L + VYM QP GF D + PN+VC L+K+LYGLKQAPRAWY
Sbjct: 1057 RSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLKQAPRAWYVE 1116
Query: 684 FADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFA 743
+Y+ TIGF +S SD SLF+ ++G ++ Y+L+YVDDI++T + L + LS F+
Sbjct: 1117 LRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNTLDNLSQRFS 1176
Query: 744 MKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSS 803
+KD LHYFLGI GL LSQR+Y +++ R M K +TP+ PKLS S
Sbjct: 1177 VKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAPSPKLSLYSG 1236
Query: 804 APHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLD 863
DP+ YR + G+LQYL FTRPDI+YAV ++ FMH P EEH+ ALKRI+RY+ GT +
Sbjct: 1237 TKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRILRYLAGTPN 1296
Query: 864 HGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEA 923
HG+ L +T +L +Y+DADW G D ST+GY V+LG + ISWS+K+Q + RSS EA
Sbjct: 1297 HGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEA 1356
Query: 924 EYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIH 983
EYR VAN SE W+ +LL EL + + ++YCDNV A YL NPV H R KHI +D H
Sbjct: 1357 EYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDYH 1416
Query: 984 SVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVS 1033
+R +V G +RV+HV + Q+AD TK L F++F + + V + P S
Sbjct: 1417 FIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGVTRVPPS 1466
>At1g58889 polyprotein, putative
Length = 1466
Score = 813 bits (2100), Expect = 0.0
Identities = 458/1130 (40%), Positives = 631/1130 (55%), Gaps = 101/1130 (8%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS+ NLS + + ++V +G +IPI T+LS + PL L N+L+ P + KNL
Sbjct: 341 ITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHNILYVPNIHKNL 400
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLY--PITTSQAIS--PSTFA 116
+SV + N VSVEF P F VKD TG+ L++ +++ +LY PI +SQ +S S +
Sbjct: 401 ISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASSQPVSLFASPSS 460
Query: 117 ALAPSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHI-CHSCSLGKHIKLPFVSSNSCT 175
S WHARLGHP +++S+ N + S + C C + K K+PF S +
Sbjct: 461 KATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDCLINKSNKVPFSQSTINS 520
Query: 176 IMPFDIIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRT 235
P + I+SD+W+SP+LS +RYYV+FVD ++ + W +PL +KSQV F++F+ +
Sbjct: 521 TRPLEYIYSDVWSSPILSHDNYRYYVIFVDHFTRYTWLYPLKQKSQVKETFITFKNLLEN 580
Query: 236 QFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRT 295
+F+ + DNG EF WE+ ++G++ S PHT NG +ERK R I T
Sbjct: 581 RFQTRIGTFYSDNGGEFVA--LWEYFSQHGISHLTSPPHTPEHNGLSERKHRHIVETGLT 638
Query: 296 LLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPL 355
LL HAS+P ++W +A +A YLIN LP L +SP + L+ P+Y LRVFGC CYP
Sbjct: 639 LLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNYDKLRVFGCACYPW 698
Query: 356 FPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKL---- 411
+KL +S CVFLGY +Y C L + ++ ISRHV FDE FPF+
Sbjct: 699 LRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDENCFPFSNYLATL 758
Query: 412 ----------------HNPQPYTYGFMDDGPSPYVIHHL--------------------- 434
H P T + PS HH
Sbjct: 759 SPVQEQRRESSCVWSPHTTLP-TRTPVLPAPSCSDPHHAATPPSSPSAPFRNSQVSSSNL 817
Query: 435 -----TSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPN--------------- 474
+S PS +P QPTTQPT + Q HS ++S N
Sbjct: 818 DSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQ-THSSQNTSQNNPTNESPSQLAQSLS 876
Query: 475 -PSPSTTATPSP-------PYQPTPISV----PKPV------------------TRSQHG 504
P+ S++++PSP PTP S+ P P+ TR++ G
Sbjct: 877 TPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPLNTHSMGTRAKAG 936
Query: 505 IFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPD-VN 563
I KP + +L S+ PR + AL+D W+ AM E NA I N TW+LVP PP V
Sbjct: 937 IIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDLVPPPPSHVT 996
Query: 564 VIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALS 623
++ WIFT K SDG RYKAR V G Q+ G+D ETFSPV+K +IR VL +A+
Sbjct: 997 IVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVD 1056
Query: 624 KAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKR 683
++W I QLDV NAFL G L + VYM QP GF D + PN+VC L+K+LYGLKQAPRAWY
Sbjct: 1057 RSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLKQAPRAWYVE 1116
Query: 684 FADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFA 743
+Y+ TIGF +S SD SLF+ ++G ++ Y+L+YVDDI++T + L + LS F+
Sbjct: 1117 LRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNTLDNLSQRFS 1176
Query: 744 MKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSS 803
+KD LHYFLGI GL LSQR+Y +++ R M K +TP+ PKLS S
Sbjct: 1177 VKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAPSPKLSLYSG 1236
Query: 804 APHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLD 863
DP+ YR + G+LQYL FTRPDI+YAV ++ FMH P EEH+ ALKRI+RY+ GT +
Sbjct: 1237 TKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRILRYLAGTPN 1296
Query: 864 HGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEA 923
HG+ L +T +L +Y+DADW G D ST+GY V+LG + ISWS+K+Q + RSS EA
Sbjct: 1297 HGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEA 1356
Query: 924 EYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIH 983
EYR VAN SE W+ +LL EL + + ++YCDNV A YL NPV H R KHI +D H
Sbjct: 1357 EYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDYH 1416
Query: 984 SVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVS 1033
+R +V G +RV+HV + Q+AD TK L F++F + + V + P S
Sbjct: 1417 FIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGVTRVPPS 1466
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 798 bits (2062), Expect = 0.0
Identities = 439/1136 (38%), Positives = 633/1136 (55%), Gaps = 101/1136 (8%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS+ NLS + + +++ +G +IPI +L + L L VL+ P + KNL
Sbjct: 320 ITSDFNNLSFHQPYTGGDDVMIADGSTIPITHTGSASLPTSSRSLDLNKVLYVPNIHKNL 379
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLY--PITTSQAIS--PSTFA 116
+SV + N VSVEF P F VKD TG+ L++ +++ +LY PI +SQA+S S +
Sbjct: 380 ISVYRLCNTNRVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASSQAVSMFASPCS 439
Query: 117 ALAPSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHI-CHSCSLGKHIKLPFVSSNSCT 175
S WH+RLGHP +++S+ N + S + C C + K K+PF +S +
Sbjct: 440 KATHSSWHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCFINKSHKVPFSNSTITS 499
Query: 176 IMPFDIIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRT 235
P + I+SD+W+SP+LS +RYYV+FVD ++ + W +PL +KSQV F+ F++ +
Sbjct: 500 SKPLEYIYSDVWSSPILSIDNYRYYVIFVDHFTRYTWLYPLKQKSQVKDTFIIFKSLVEN 559
Query: 236 QFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRT 295
+F+ + + DNG EF ++ ++G++ S PHT NG +ERK R I + T
Sbjct: 560 RFQTRIGTLYSDNGGEFVVLR--DYLSQHGISHFTSPPHTPEHNGLSERKHRHIVEMGLT 617
Query: 296 LLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPL 355
LL HAS+P ++W +A +A YLIN LP L QSP + L+ + P+Y L+VFGC CYP
Sbjct: 618 LLSHASVPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACYPW 677
Query: 356 FPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAK----- 410
+KL+ +S C F+GY +Y C + + ++ SRHV FDE FPF+
Sbjct: 678 LRPYNRHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNFGV 737
Query: 411 ------------------------LHNPQPYTYGFMDD-------GPSPYVIHHLTSQ-- 437
L P P G D PSP ++S
Sbjct: 738 STSQEQRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSPLCTTQVSSSNL 797
Query: 438 -------PSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPNP--------------- 475
PS +P QPT QP + +++SP ++PNP
Sbjct: 798 PSSSISSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPNPNSPSPNSPNQNSPLP 857
Query: 476 --------------------SPSTTATPSPPYQPT-------------PISVPKPVTRSQ 502
SPS+++T +PP P P++ TR++
Sbjct: 858 QSPISSPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQVNAQAPVNTHSMATRAK 917
Query: 503 HGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPP-D 561
GI KP ++ + TS+ + PR + A++D W+ AM E NA I N TW+LVP PP
Sbjct: 918 DGIRKPNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLVPPPPPS 977
Query: 562 VNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLA 621
V ++ WIFT K SDG RYKARLV G Q+ G+D ETFSPV+K +IR VL +A
Sbjct: 978 VTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVA 1037
Query: 622 LSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWY 681
+ ++W I QLDV NAFL G L + VYM QP GF D + P++VC L+K++YGLKQAPRAWY
Sbjct: 1038 VDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQAPRAWY 1097
Query: 682 KRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTE 741
Y+ T+GF +S SD SLF+ ++G ++ Y+L+YVDDI++T + L + LS
Sbjct: 1098 VELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTLDALSQR 1157
Query: 742 FAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSAN 801
F++K+ LHYFLGI GL LSQR+Y +++ R M K +TP+ T PKL+ +
Sbjct: 1158 FSVKEHEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMATSPKLTLH 1217
Query: 802 SSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGT 861
S DP+ YR + G+LQYL FTRPD++YAV ++ +MH P ++H +ALKR++RY+ GT
Sbjct: 1218 SGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVLRYLAGT 1277
Query: 862 LDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSA 921
DHG+ L +T +L +Y+DADW G D ST+GY V+LG + ISWS+K+Q + RSS
Sbjct: 1278 PDHGIFLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSST 1337
Query: 922 EAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMD 981
EAEYR VAN SE W+ +LL EL + ++YCDNV A YL NPV H R KHI +D
Sbjct: 1338 EAEYRSVANTSSELQWICSLLTELGIQLSHPPVIYCDNVGATYLCANPVFHSRMKHIALD 1397
Query: 982 IHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVSTAGV 1037
H +R +V G +RV+HV + Q+AD TK L V F++F + V + P S GV
Sbjct: 1398 YHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRVAFQNFSRKIGVIKVPPSCGGV 1453
>At4g10690 retrotransposon like protein
Length = 1515
Score = 784 bits (2025), Expect = 0.0
Identities = 436/1104 (39%), Positives = 624/1104 (56%), Gaps = 71/1104 (6%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+T+ L + S + +IVGNG +PI L+ L L++VL P + K+L
Sbjct: 334 ITNTTDGLQNSQTYSGDDSVIVGNGDFLPITHIGTIPLNISQGTLPLEDVLVCPGITKSL 393
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPI--TTSQAISPSTFAAL 118
+SV K T D S FD +KD +T L + LY + Q + +
Sbjct: 394 LSVSKLTDDYPCSFTFDSDSVVIKDKRTQQLLTQGNKHKGLYVLKDVPFQTYYSTRQQSS 453
Query: 119 APSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMP 178
+WH RLGHP V+ L K K I N+ S S++C +C +GK +LPFV+S + P
Sbjct: 454 DDEVWHQRLGHPNKEVLQHLIKTKAIVVNKTS-SNMCEACQMGKVCRLPFVASEFVSSRP 512
Query: 179 FDIIHSDIW-TSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQF 237
+ IH D+W +PV S+ G +YYV+F+D+YS F W +PL KS FS+F+ F+ + Q+
Sbjct: 513 LERIHCDLWGPAPVTSAQGFQYYVIFIDNYSRFTWFYPLKLKSDFFSVFVLFQQLVENQY 572
Query: 238 EREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLL 297
+ ++ QCD G EF + F G+ +SCPHT QNG AER+ R + + +L+
Sbjct: 573 QHKIAMFQCDGGGEFVSYKFVAHLASCGIKQLISCPHTPQQNGIAERRHRYLTELGLSLM 632
Query: 298 VHASLPPSFWHHALQMATYLINILPNKQLA-YQSPLKILYQKEPSYSHLRVFGCLCYPLF 356
H+ +P W A + +L N+LP+ L+ +SP ++L+ P Y+ LRVFG CYP
Sbjct: 633 FHSKVPHKLWVEAFFTSNFLSNLLPSSTLSDNKSPYEMLHGTPPVYTALRVFGSACYPYL 692
Query: 357 PSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHN--- 413
NK +S CVFLGY + ++ Y+C + K+ I RHV+FDE +FP++ +++
Sbjct: 693 RPYAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLFDERKFPYSDIYSQFQ 752
Query: 414 ----------------------------------PQPYTYGFMDDGPSPYVIHHLTSQPS 439
P + G +P + T+ P
Sbjct: 753 TISGSPLFTAWQKGFSSTALSRETPSTNVEDIIFPSATVSSSVPTGCAPNIAETATA-PD 811
Query: 440 LGQPAQHDL-----PNTQPTTQPTTPEEQHAHSPPSSSPNPSPSTTA-TPS--------- 484
+ A HD+ P T T+ PT PEE + S+ + + ++A TP
Sbjct: 812 VDVAAAHDMVVPPSPITS-TSLPTQPEESTSDQNHYSTDSETAISSAMTPQSINVSLFED 870
Query: 485 ---PPYQPT-------PISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDP 534
PP Q P + +TR++ GI KP + L + P P++ AL+D
Sbjct: 871 SDFPPLQSVISSTTAAPETSHPMITRAKSGITKPNPKYALFSVKSNYPEPKSVKEALKDE 930
Query: 535 NWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKT 594
W AM +E + + TW+LVP ++ W+F K SDG +R KARLV G
Sbjct: 931 GWTNAMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVFKTKLNSDGSLDRLKARLVARGYE 990
Query: 595 QQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGF 654
Q+ GVD ET+SPVV+ AT+R++L +A WS+ QLDVKNAFLH ELKETV+M QP GF
Sbjct: 991 QEEGVDYVETYSPVVRSATVRSILHVATINKWSLKQLDVKNAFLHDELKETVFMTQPPGF 1050
Query: 655 RDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYI 714
DP+ P++VC LKK++Y LKQAPRAW+ +F+ Y+ GF S SD SLF+Y KG + ++
Sbjct: 1051 EDPSRPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKYGFICSFSDPSLFVYLKGRDVMFL 1110
Query: 715 LLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAA 774
LLYVDD+ILT ++D L ++++LSTEF MKD+G+LHYFLGI +H GLFLSQ KY +
Sbjct: 1111 LLYVDDMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQAHYHNDGLFLSQEKYTS 1170
Query: 775 EIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQ 834
+++ AGM+ C S TP+ L ++ P +P+++R LAG LQYLT TRPDI +AV
Sbjct: 1171 DLLVNAGMSDCSSMPTPLQL--DLLQGNNKPFPEPTYFRRLAGKLQYLTLTRPDIQFAVN 1228
Query: 835 QVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRST 894
VC MH P H LKRI+ Y++GT+ G++L ++ S L Y+D+DW GC DTRRST
Sbjct: 1229 FVCQKMHAPTMSDFHLLKRILHYLKGTMTMGINLSSNTDSVLRCYSDSDWAGCKDTRRST 1288
Query: 895 SGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASL 954
G+C FLG N+ISWSAKR T+S+SS EAEYR ++ SE W+ LL E+ P ++
Sbjct: 1289 GGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRTLSFAASEVSWIGFLLQEIGLPQQQIPE 1348
Query: 955 VYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLP 1014
+YCDN+SA+YLS NP H R+KH ++D + VRE+VA G + V H+P+ Q+ADIFTK LP
Sbjct: 1349 MYCDNLSAVYLSANPALHSRSKHFQVDYYYVRERVALGALTVKHIPASQQLADIFTKSLP 1408
Query: 1015 LVLFEDFRNNLSVRQPPVSTAGVC 1038
F D R L V PP ++ C
Sbjct: 1409 QAPFCDLRFKLGVVLPPDTSLRGC 1432
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 783 bits (2021), Expect = 0.0
Identities = 426/1044 (40%), Positives = 593/1044 (55%), Gaps = 23/1044 (2%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS L S + ++VG+G +PI T + N + L VL P + K+L
Sbjct: 332 VTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSSNGKIPLNEVLVVPNIQKSL 391
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQ--AISPSTFAAL 118
+SV K D V FD + D QT + R LY + + A+ + A
Sbjct: 392 LSVSKLCDDYPCGVYFDANKVCIIDLQTQKVVTTGPRRNGLYVLENQEFVALYSNRQCAA 451
Query: 119 APSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMP 178
+WH RLGH + + L+ +K I+ N++ S +C C +GK +LPF+ S+S + P
Sbjct: 452 TEEVWHHRLGHANSKALQHLQNSKAIQINKSRTSPVCEPCQMGKSSRLPFLISDSRVLHP 511
Query: 179 FDIIHSDIW-TSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQF 237
D IH D+W SPV+S+ G +YY +FVDDYS + W +PL KS+ S+F+SF+ + Q
Sbjct: 512 LDRIHCDLWGPSPVVSNQGLKYYAIFVDDYSRYSWFYPLHNKSEFLSVFISFQKLVENQL 571
Query: 238 EREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLL 297
++K Q D G EF + E+G+ R+SCP+T QNG AERK R + + ++L
Sbjct: 572 NTKIKVFQSDGGGEFVSNKLKTHLSEHGIHHRISCPYTPQQNGLAERKHRHLVELGLSML 631
Query: 298 VHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFP 357
H+ P FW + A Y+IN LP+ L SP + L+ ++P YS LRVFG CYP
Sbjct: 632 FHSHTPQKFWVESFFTANYIINRLPSSVLKNLSPYEALFGEKPDYSSLRVFGSACYPCLR 691
Query: 358 STTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNP--- 414
NK RS CVFLGY S ++ Y+C+ + K+ ISR+VIF+E++ PF + +
Sbjct: 692 PLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYISRNVIFNESELPFKEKYQSLVP 751
Query: 415 -------QPYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHS 467
Q + + + + P L S+P + +Q T Q T PE +
Sbjct: 752 QYSTPLLQAWQHNKISEISVPAAPVQLFSKPI----DLNTYAGSQVTEQLTDPEPTSNNE 807
Query: 468 PPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNP 527
NP A Q I+ TRS+ GI KP + L TS + P+
Sbjct: 808 GSDEEVNPVAEEIAAN----QEQVINSHAMTTRSKAGIQKPNTRYALITSRMNTAEPKTL 863
Query: 528 VSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKAR 587
SA++ P W A+ +E N + TW LVP D+N++ S W+F K DG ++ KAR
Sbjct: 864 ASAMKHPGWNEAVHEEINRVHMLHTWSLVPPTDDMNILSSKWVFKTKLHPDGSIDKLKAR 923
Query: 588 LVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVY 647
LV G Q+ GVD ETFSPVV+ ATIR VL ++ SK W I QLDV NAFLHGEL+E V+
Sbjct: 924 LVAKGFDQEEGVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVSNAFLHGELQEPVF 983
Query: 648 MHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRK 707
M+QP GF DP P HVC L K++YGLKQAPRAW+ F++++ GF S SD SLF+ +
Sbjct: 984 MYQPSGFIDPQKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFVCSKSDPSLFVCHQ 1043
Query: 708 GTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFL 767
+ Y+LLYVDDI+LT S +L ++ L F+MKDLG YFLGI + + GLFL
Sbjct: 1044 DGKILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFLGIQIEDYANGLFL 1103
Query: 768 SQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRP 827
Q YA +I+++AGM+ C TP+ +L +S A+P+++RSLAG LQYLT TRP
Sbjct: 1104 HQTAYATDILQQAGMSDCNPMPTPL--PQQLDNLNSELFAEPTYFRSLAGKLQYLTITRP 1161
Query: 828 DIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGC 887
DI +AV +C MH P LKRI+RYI+GT+ GL + +ST TL +Y+D+D GC
Sbjct: 1162 DIQFAVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMGLPIKRNSTLTLSAYSDSDHAGC 1221
Query: 888 PDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHC 947
+TRRST+G+C+ LG NLISWSAKRQ T+S SS EAEYR + E W+ LL +L
Sbjct: 1222 KNTRRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAAREITWISFLLRDLGI 1281
Query: 948 PIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIAD 1007
P + VYCDN+SA+YLS NP H R+KH + D H +RE+VA G + H+ + +Q+AD
Sbjct: 1282 PQYLPTQVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVALGLIETQHISATFQLAD 1341
Query: 1008 IFTKGLPLVLFEDFRNNLSVRQPP 1031
+FTK LP F D R+ L V P
Sbjct: 1342 VFTKSLPRRAFVDLRSKLGVSGSP 1365
>At1g44510 polyprotein, putative
Length = 1459
Score = 782 bits (2019), Expect = 0.0
Identities = 443/1118 (39%), Positives = 612/1118 (54%), Gaps = 90/1118 (8%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS+ LS + + +++ +G + I+ T L N L L VL+ P + KNL
Sbjct: 347 ITSDLNALSLHQPYNGGEYVMIADGTGLTIKQTGSTFLPSQNRDLALHKVLYVPDIRKNL 406
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLY------PITTSQAISPST 114
+SV + N VSVEF P F VKD TG L++ ++ DLY P T+ SPS
Sbjct: 407 ISVYRLCNTNQVSVEFFPASFQVKDLNTGTLLLQGRTKDDLYEWPVTNPPATALFTSPSP 466
Query: 115 FAALAPSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHI-CHSCSLGKHIKLPFVSSNS 173
L S WH+RLGHP A ++++L + + AS + C C + K KLPF +S+
Sbjct: 467 KTTL--SSWHSRLGHPSASILNTLLSKFSLPVSVASSNKTSCSDCLINKSHKLPFATSSI 524
Query: 174 CTIMPFDIIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFI 233
+ P + I +D+WTSP++S ++YY++ VD Y+ + W +PL +KSQV + F++F+ +
Sbjct: 525 HSSSPLEYIFTDVWTSPIISHDNYKYYLVLVDHYTRYTWLYPLQQKSQVKATFIAFKALV 584
Query: 234 RTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNII 293
+F+ +++ + DNG EF +F NG++ S PHT NG +ERK R I
Sbjct: 585 ENRFQAKIRTLYSDNGGEFIALR--DFLVSNGISHLTSPPHTPEHNGLSERKHRHIVETG 642
Query: 294 RTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCY 353
TLL AS+P +W +A A YLIN +P L QSP + L+ P+Y LRVFGCLC+
Sbjct: 643 LTLLTQASVPREYWTYAFATAVYLINRMPTPVLCLQSPFQKLFGSSPNYQRLRVFGCLCF 702
Query: 354 PLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHN 413
P T NKL+ RS CVFLGY +Y C D+ + ++ SRHV+FDE+ +PFA
Sbjct: 703 PWLRPYTRNKLEERSKRCVFLGYSLTQTAYLCLDVDNNRLYTSRHVMFDESTYPFAASIR 762
Query: 414 PQ----------------PYTYGF-----------------------MDDGP-------S 427
Q P GF +D P S
Sbjct: 763 EQSQSSLVTPPESSSSSSPANSGFPCSVLRLQSPPASSPETPSPPQQQNDSPVSPRQTGS 822
Query: 428 PYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSS-----PNPSPST--- 479
P HH + S P+ + N++PT E A S P+S PNP+P T
Sbjct: 823 PTPSHHSQVRDSTLSPSP-SVSNSEPTAPHENGPEPEAQSNPNSPFIGPLPNPNPETNPS 881
Query: 480 -----------TATPSPPYQPT-----------PISVPKPVTRSQHGIFKPKRQLNLNTS 517
T T PP Q T P + + TRS++ I KPK + +L +
Sbjct: 882 SSIEQRPVDKSTTTALPPNQTTIAATSNSRSQPPKNNHQMKTRSKNNITKPKTKTSLTVA 941
Query: 518 VPRSPL--PRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKE 575
+ + L P AL+D W+ AM DEF+A +N TW+LVP P +++ W+F K
Sbjct: 942 LTQPHLSEPNTVTQALKDKKWRFAMSDEFDAQQRNHTWDLVPPNPTQHLVGCRWVFKLKY 1001
Query: 576 KSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKN 635
+G+ ++YKARLV G QQ GVD ETFSPV+K TIR VL +A+ K W + QLDV N
Sbjct: 1002 LPNGLIDKYKARLVAKGFNQQYGVDYAETFSPVIKATTIRVVLDVAVKKNWPLKQLDVNN 1061
Query: 636 AFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSH 695
AFL G L E VYM QP GF D + P+HVC L+K++YGLKQAPRAWY ++ IGF +
Sbjct: 1062 AFLQGTLTEEVYMAQPPGFVDKDRPSHVCRLRKAIYGLKQAPRAWYMELKQHLLNIGFVN 1121
Query: 696 STSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLG 755
S +D SLFIY GT + Y+L+YVDDII+T S ++S L+ F++KD LHYFLG
Sbjct: 1122 SLADTSLFIYSHGTTLLYLLVYVDDIIVTGSDHKSVSAVLSSLAERFSIKDPTDLHYFLG 1181
Query: 756 IAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSL 815
I T GL L QRKY +++ + M K +TP+ T PKL+ + D S YRS+
Sbjct: 1182 IEATRTNTGLHLMQRKYMTDLLAKHNMLDAKPVATPLPTSPKLTLHGGTKLNDASEYRSV 1241
Query: 816 AGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTST 875
G+LQYL FTRPDIA+AV ++ FMH P +H A KR++RY+ GT HG+ L SS
Sbjct: 1242 VGSLQYLAFTRPDIAFAVNRLSQFMHQPTSDHWQAAKRVLRYLAGTTTHGIFLNSSSPIH 1301
Query: 876 LISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSES 935
L +++DADW G ST+ Y ++LG N ISWS+K+Q +SRSS E+EYR VAN SE
Sbjct: 1302 LHAFSDADWAGDSADYVSTNAYVIYLGRNPISWSSKKQRGVSRSSTESEYRAVANAASEI 1361
Query: 936 CWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVR 995
WL +LL ELH + ++CDN+ A Y+ NPV H R KHI +D H VR + +R
Sbjct: 1362 RWLCSLLTELHIRLPHGPTIFCDNIGATYICANPVFHSRMKHIALDYHFVRGMIQSRALR 1421
Query: 996 VLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVS 1033
V HV + Q+AD TK L F R+ + VRQ P S
Sbjct: 1422 VSHVSTNDQLADALTKSLSRPHFLSARSKIGVRQLPPS 1459
>At4g16870 retrotransposon like protein
Length = 1474
Score = 772 bits (1993), Expect = 0.0
Identities = 441/1121 (39%), Positives = 604/1121 (53%), Gaps = 106/1121 (9%)
Query: 16 NNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVE 75
N +++ +G S+ I T L LTL VL+ P + KNLVSV + N VSVE
Sbjct: 357 NGDDVMIADGTSLKITKTGSTFLPSNARDLTLNKVLYVPDIQKNLVSVYRLCNTNQVSVE 416
Query: 76 FDPFGFSVKDFQTGMRLMRCESRGDLY--PITTSQAISPSTFAALAPSL--WHARLGHPG 131
F P F VKD TG L++ ++ +LY P+T +A + T + +L WH+RLGHP
Sbjct: 417 FFPASFQVKDLNTGTLLLQGRTKDELYEWPVTNPKATALFTTPSPKTTLSSWHSRLGHPS 476
Query: 132 APVVDSLRKNKFIECN-RASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIWTSP 190
+ ++++L + + AS C C + K KLPF S+ + P + I SD+W SP
Sbjct: 477 SSILNTLISKFSLPVSVSASNKLACSDCFINKSHKLPFSISSIKSTSPLEYIFSDVWMSP 536
Query: 191 VLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGK 250
+LS ++YY++ VD ++ + W +PL +KSQV S F++F+ + +F+ +++ + DNG
Sbjct: 537 ILSPDNYKYYLVLVDHHTRYTWLYPLQQKSQVKSTFIAFKALVENRFQAKIRTLYSDNGG 596
Query: 251 EFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHA 310
EF EF NG++ S PHT NG +ERK R I TLL AS+P +W +A
Sbjct: 597 EFIALR--EFLVSNGISHLTSPPHTPEHNGLSERKHRHIVETGLTLLTQASVPREYWPYA 654
Query: 311 LQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTP 370
A YLIN +P L+ +SP + L+ +P+Y LRVFGCLC+P T NKL+ RS
Sbjct: 655 FAAAVYLINRMPTPVLSMESPFQKLFGSKPNYERLRVFGCLCFPWLRPYTHNKLEERSRR 714
Query: 371 CVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYGFMDDGPSPYV 430
CVFLGY +Y C+D+ +++ SRHV+FDE FPF+ L + + SP V
Sbjct: 715 CVFLGYSLTQTAYLCFDVEHKRLYTSRHVVFDEASFPFSNLTSQNSLPTVTFEQSSSPLV 774
Query: 431 IHHLTSQ--------------------------PSLGQPAQHDLP--------------- 449
L+S P QP P
Sbjct: 775 TPILSSSSVLPSCLSSPCTVLHQQQPPVTTPNSPHSSQPTTSPAPLSPHRSTTMDFQVPQ 834
Query: 450 --------------NTQPTTQPTTPEEQHAHSPPS---SSPN------PSPSTTATPSPP 486
N++PT E A SPP S+P P P+ P+
Sbjct: 835 VRSSSPLLSSSSSLNSEPTAPNENGPEPEAQSPPIGPLSNPTHEAFIGPLPNPNRNPTNE 894
Query: 487 YQPTPISVPKPV--------------------------------TRSQHGIFKPKRQLNL 514
+PTP PKPV TR+++ I KP + +L
Sbjct: 895 IEPTPAPHPKPVKPTTTTTTPNRTTVSDASHQPTAPQQNQHNMKTRAKNNIKKPNTKFSL 954
Query: 515 NTSVP-RSPL-PRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFT 572
++P RSP P N AL+D W+ AM DEF+A +N TW+LVP + ++ W+F
Sbjct: 955 TATLPNRSPSEPTNVTQALKDKKWRFAMSDEFDAQQRNHTWDLVPHESQL-LVGCKWVFK 1013
Query: 573 HKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLD 632
K +G ++YKARLV G QQ GVD ETFSPV+K TIR VL +A+ K W I QLD
Sbjct: 1014 LKYLPNGAIDKYKARLVAKGFNQQYGVDYAETFSPVIKSTTIRLVLDVAVKKDWEIKQLD 1073
Query: 633 VKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIG 692
V NAFL G L E VYM QP GF D + P HVC L+K++YGLKQAPRAWY ++ IG
Sbjct: 1074 VNNAFLQGTLTEEVYMAQPPGFIDKDRPTHVCRLRKAIYGLKQAPRAWYMELKQHLFNIG 1133
Query: 693 FSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHY 752
F +S SD SLFIY GT Y+L+YVDDII+T S + +++ L+ F++KD LHY
Sbjct: 1134 FVNSLSDASLFIYCHGTTFVYVLVYVDDIIVTGSDKSSIDAVLTSLAERFSIKDPTDLHY 1193
Query: 753 FLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHY 812
FLGI T GL L QRKY +++ + MA K TP+ T PKL+ + D S Y
Sbjct: 1194 FLGIEATRTKQGLHLMQRKYIKDLLAKHNMADAKPVLTPLPTSPKLTLHGGTKLNDASEY 1253
Query: 813 RSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSS 872
RS+ G+LQYL FTRPDIAYAV ++ M P E+H A KR++RY+ GT HG+ L +S
Sbjct: 1254 RSVVGSLQYLAFTRPDIAYAVNRLSQLMPQPTEDHWQAAKRVLRYLAGTSTHGIFLDTTS 1313
Query: 873 TSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVV 932
L +++DADW G D ST+ Y ++LG N ISWS+K+Q ++RSS E+EYR VAN
Sbjct: 1314 PLNLHAFSDADWAGDSDDYVSTNAYVIYLGKNPISWSSKKQRGVARSSTESEYRAVANAA 1373
Query: 933 SESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARG 992
SE WL +LL +LH + ++CDN+ A YL NPV H R KHI +D H VR + G
Sbjct: 1374 SEVKWLCSLLSKLHIRLPIRPSIFCDNIGATYLCANPVFHSRMKHIAIDYHFVRNMIQSG 1433
Query: 993 EVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVS 1033
+RV HV +R Q+AD TK L F+ R + VRQ P S
Sbjct: 1434 ALRVSHVSTRDQLADALTKPLSRAHFQSARFKIGVRQLPPS 1474
>At1g53810
Length = 1522
Score = 768 bits (1983), Expect = 0.0
Identities = 428/1090 (39%), Positives = 608/1090 (55%), Gaps = 76/1090 (6%)
Query: 20 IIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPF 79
I+V +G+ +PI +++ + + LK VL P ++K+L+SV K T+D SVEFD
Sbjct: 355 IMVADGNFLPITHTGSGSIASSSGKIPLKEVLVCPDIVKSLLSVSKLTSDYPCSVEFDAD 414
Query: 80 GFSVKDFQTGMRLMRCESRGDLYPITTS--QAISPSTFAALAPSLWHARLGHPGAPVVDS 137
+ D T L+ +R LY + Q + + + + +WH RLGH A V+
Sbjct: 415 SVRINDKATKKLLVMGRNRDGLYSLEEPKLQVLYSTRQNSASSEVWHRRLGHANAEVLHQ 474
Query: 138 LRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIW-TSPVLSSSG 196
L +K I +C +C LGK +LPF+ S P + IH D+W SP S G
Sbjct: 475 LASSKSIIIINKVVKTVCEACHLGKSTRLPFMLSTFNASRPLERIHCDLWGPSPTSSVQG 534
Query: 197 HRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRH 256
RYYV+F+D YS F W +PL KS FS F+ F+ + Q ++K QCD G EF +
Sbjct: 535 FRYYVVFIDHYSRFTWFYPLKLKSDFFSTFVMFQKLVENQLGHKIKIFQCDGGGEFISSQ 594
Query: 257 FWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATY 316
F + +++G+ +SCP+T QNG AERK R I + +++ + LP +W + A +
Sbjct: 595 FLKHLQDHGIQQNMSCPYTPQQNGMAERKHRHIVELGLSMIFQSKLPLKYWLESFFTANF 654
Query: 317 LINILPNKQL-AYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLG 375
+IN+LP L +SP + LY K P YS LRVFGC CYP K RS CVFLG
Sbjct: 655 VINLLPTSSLDNNESPYQKLYGKAPEYSALRVFGCACYPTLRDYASTKFDPRSLKCVFLG 714
Query: 376 YPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLH---NPQPYTYGFMDDGPSPYVIH 432
Y ++ Y+C + +I ISRHV+FDE PF ++ +PQ T S H
Sbjct: 715 YNEKYKGYRCLYPPTGRIYISRHVVFDENTHPFESIYSHLHPQDKTPLLEAWFKS---FH 771
Query: 433 HLT-SQP--------SLGQPAQHDL-------------PNTQPTTQPT---------TPE 461
H+T +QP S+ QP DL PN T +PE
Sbjct: 772 HVTPTQPDQSRYPVSSIPQPETTDLSAAPASVAAETAGPNASDDTSQDNETISVVSGSPE 831
Query: 462 E----------QHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKP-------------V 498
HSP + S +PSP+ ++ S P Q +PI + V
Sbjct: 832 RTTGLDSASIGDSYHSPTADSSHPSPARSSPASSP-QGSPIQMAPAQQVQAPVTNEHAMV 890
Query: 499 TRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPR 558
TR + GI KP ++ L T P P+ AL+ P W AM +E + +TW LVP
Sbjct: 891 TRGKEGISKPNKRYVLLTHKVSIPEPKTVTEALKHPGWNNAMQEEMGNCKETETWTLVPY 950
Query: 559 PPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVL 618
P++NV+ SMW+F K +DG ++ KARLV G Q+ G+D ET+SPVV+ T+R +L
Sbjct: 951 SPNMNVLGSMWVFRTKLHADGSLDKLKARLVAKGFKQEEGIDYLETYSPVVRTPTVRLIL 1010
Query: 619 SLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPR 678
+A W + Q+DVKNAFLHG+L ETVYM QP GF D + P+HVCLL KSLYGLKQ+PR
Sbjct: 1011 HVATVLKWELKQMDVKNAFLHGDLTETVYMRQPAGFVDKSKPDHVCLLHKSLYGLKQSPR 1070
Query: 679 AWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLL 738
AW+ RF++++ GF S D SLF+Y + +LLYVDD+++T ++ +++ L
Sbjct: 1071 AWFDRFSNFLLEFGFICSLFDPSLFVYSSNNDVILLLLYVDDMVITGNNSQSLTHLLAAL 1130
Query: 739 STEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKL 798
+ EF MKD+G +HYFLGI + + GGLF+SQ+KYA +++ A MA C TP+ +
Sbjct: 1131 NKEFRMKDMGQVHYFLGIQIQTYDGGLFMSQQKYAEDLLITASMANCSPMPTPLPLQLDR 1190
Query: 799 SANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYI 858
+N +DP+++RSLAG LQYLT TRPDI +AV VC MH P + LKRI+RYI
Sbjct: 1191 VSNQDEVFSDPTYFRSLAGKLQYLTLTRPDIQFAVNFVCQKMHQPSVSDFNLLKRILRYI 1250
Query: 859 QGTLDHGLHLYPSSTSTLIS----------YTDADWGGCPDTRRSTSGYCVFLGDNLISW 908
+GT+ G+ Y S++S+++S Y+D+D+ C +TRRS GYC F+G N+ISW
Sbjct: 1251 KGTVSMGIQ-YNSNSSSVVSAYESDYDLSAYSDSDYANCKETRRSVGGYCTFMGQNIISW 1309
Query: 909 SAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGN 968
S+K+Q T+SRSS EAEYR ++ SE W+ ++L E+ + ++CDN+SA+YL+ N
Sbjct: 1310 SSKKQPTVSRSSTEAEYRSLSETASEIKWMSSILREIGVSLPDTPELFCDNLSAVYLTAN 1369
Query: 969 PVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVR 1028
P H RTKH ++D H +RE+VA + V H+P Q+ADIFTK LP F R L V
Sbjct: 1370 PAFHARTKHFDVDHHYIRERVALKTLVVKHIPGHLQLADIFTKSLPFEAFTRLRFKLGVD 1429
Query: 1029 QPPVSTAGVC 1038
PP + C
Sbjct: 1430 FPPTPSLRGC 1439
>At4g27210 putative protein
Length = 1318
Score = 717 bits (1852), Expect = 0.0
Identities = 403/1057 (38%), Positives = 570/1057 (53%), Gaps = 86/1057 (8%)
Query: 20 IIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPF 79
I+V +G+ +PI TNL+ + + L +VL P + K+L+S+ K T D +VEF+
Sbjct: 206 IMVDDGNYLPITHTGSTNLASSSGTVPLTDVLVCPSITKSLLSMSKLTQDFPCTVEFEYD 265
Query: 80 GFSVKDFQTGMRLMRCESRGDLYPITTS---QAISPSTFAALAPSLWHARLGHPGAPVVD 136
G V D T L+ +R LY + QA + + + +WH RLGHP +
Sbjct: 266 GVRVNDKATKKLLLMGSNRDGLYCLKDDKQFQAFFSTRQRSASDEVWHRRLGHPHPQI-- 323
Query: 137 SLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIWTSPVLSS-S 195
+ P + +H D+W ++S
Sbjct: 324 ---------------------------------------LQPLERVHCDLWGPTTITSVQ 344
Query: 196 GHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNR 255
G RYY +F+D YS F W +PL KS ++IFL+F + Q +++ QCD G EF +
Sbjct: 345 GFRYYAVFIDHYSRFSWIYPLKLKSDFYNIFLAFHKLVENQLSQKISVFQCDGGGEFVSH 404
Query: 256 HFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMAT 315
F + + +G+ +LSCPHT QNG AERK R + + ++L + +P FW A A
Sbjct: 405 KFLQHLQSHGIQQQLSCPHTPQQNGLAERKHRHLVELGLSMLFQSHVPHKFWVEAFFTAN 464
Query: 316 YLINILPNKQLAYQ-SPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFL 374
+LIN+LP L SP + LY K+P Y+ LR FG C+P NK S CVFL
Sbjct: 465 FLINLLPTSALKESISPYEKLYDKKPDYTSLRSFGSACFPTLRDYAENKFNPCSLKCVFL 524
Query: 375 GYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLH---NPQPYT-----YGFMDDGP 426
GY ++ Y+C + ++ ISRHVIFDE+ +PF+ + +PQP T + D P
Sbjct: 525 GYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPFSHTYKHLHPQPRTPLLAAWLRSSDSP 584
Query: 427 SPYVIHHLTSQ---------PSLGQPAQHDLPNTQP----------TTQPTTPEEQH--- 464
+P +S+ P L Q LP P TTQ + +
Sbjct: 585 APSTSTSPSSRSPLFTSADFPPLPQRKTPLLPTLVPISSVSHASNITTQQSPDFDSERTT 644
Query: 465 ----------AHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNL 514
+HS + S + A+ + +V VTR++ GI KP +
Sbjct: 645 DFDSASIGDSSHSSQAGSDSEETIQQASVNVHQTHASTNVHPMVTRAKVGISKPNPRYVF 704
Query: 515 NTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHK 574
+ P P+ +AL+ P W AM +E + +TW LVP D++V+ S W+F K
Sbjct: 705 LSHKVSYPEPKTVTAALKHPGWTGAMTEEIGNCSETQTWSLVPYKSDMHVLGSKWVFRTK 764
Query: 575 EKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVK 634
+DG + KAR+V G Q+ G+D ET+SPVV+ T+R VL LA + W I Q+DVK
Sbjct: 765 LHADGTLNKLKARIVAKGFLQEEGIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVK 824
Query: 635 NAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFS 694
NAFLHG+LKETVYM QP GF DP+ P+HVCLL KS+YGLKQ+PRAW+ +F+ ++ GF
Sbjct: 825 NAFLHGDLKETVYMTQPAGFVDPSKPDHVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFF 884
Query: 695 HSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFL 754
S SD SLFIY + +LLYVDD+++T +S ++++ L+ EF M D+G LHYFL
Sbjct: 885 CSKSDPSLFIYAHNNNLILLLLYVDDMVITGNSSQTLTSLLAALNKEFRMTDMGQLHYFL 944
Query: 755 GIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRS 814
GI V GLF+SQ+KYA +++ A M C TP+ + + +DP+++RS
Sbjct: 945 GIQVQRQQNGLFMSQQKYAEDLLIAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRS 1004
Query: 815 LAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTS 874
+AG LQYLT TRPDI +AV VC MH P H LKRI+RYI+GT+ G+ S +
Sbjct: 1005 IAGKLQYLTLTRPDIQFAVNFVCQKMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPT 1064
Query: 875 TLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSE 934
L +Y+D+DWG C TRRS G C F+G NL+SWS+K+ T+SRSS EAEY+ +++ SE
Sbjct: 1065 LLQAYSDSDWGNCKQTRRSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASE 1124
Query: 935 SCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEV 994
WL LL EL P+ ++CDN+SA+YL+ NP H RTKH ++D H VRE+VA +
Sbjct: 1125 ILWLSTLLRELRIPLPDTPELFCDNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKAL 1184
Query: 995 RVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPP 1031
V H+P QIADIFTK LP F R L V P
Sbjct: 1185 VVKHIPGSEQIADIFTKSLPYEAFIHLRGKLGVTLSP 1221
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 652 bits (1683), Expect = 0.0
Identities = 383/1031 (37%), Positives = 554/1031 (53%), Gaps = 32/1031 (3%)
Query: 3 SEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVS 62
S +L S + S + + G ++ I G L N + LKNVL P+ NL+S
Sbjct: 442 SHDRSLFSSLDTSVLSAVNLPTGPTVKISGVGTLKL---NDDILLKNVLFIPEFRLNLIS 498
Query: 63 VRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAPSL 122
+ T D V FD ++D G L + +LY + S S A + S+
Sbjct: 499 ISSLTDDIGSRVIFDKNSCEIQDLIKGRMLGQGRRVANLYLLDVGDQ-SISVNAVVDISM 557
Query: 123 WHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDII 182
WH RLGH +D++ + ++ GS CH C L K KL F +SN FD++
Sbjct: 558 WHRRLGHASLQRLDAISDSLGTTRHKNKGSDFCHVCHLAKQRKLSFPTSNKVCKEIFDLL 617
Query: 183 HSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREV 241
H D+W V + G++Y++ VDD+S W + L KS+V ++F +F + Q++ +V
Sbjct: 618 HIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLLKTKSEVLTVFPAFIQQVENQYKVKV 677
Query: 242 KNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHAS 301
K ++ DN E F F E G+ SCP T QN ERK + I N+ R L+ +
Sbjct: 678 KAVRSDNAPELK---FTSFYAEKGIVSFHSCPETPEQNSVVERKHQHILNVARALMFQSQ 734
Query: 302 LPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTI 361
+P S W + A +LIN P++ L ++P +IL P Y LR FGCLCY
Sbjct: 735 VPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILTGTAPVYEQLRTFGCLCYSSTSPKQR 794
Query: 362 NKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYGF 421
+K Q RS C+FLGYPS ++ YK DL S + ISR+V F E FP AK + F
Sbjct: 795 HKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQFHEEVFPLAKNPGSESSLKLF 854
Query: 422 MDDGP-SPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPNPSPSTT 480
P S +I T PS LP+ Q + P Q PP+ N T
Sbjct: 855 TPMVPVSSGIISDTTHSPS-------SLPS-QISDLPPQISSQRVRKPPAHL-NDYHCNT 905
Query: 481 ATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAM 540
Y PIS + + P +N ++ + P+P N A W A+
Sbjct: 906 MQSDHKY---PIS-----STISYSKISPSHMCYIN-NITKIPIPTNYAEAQDTKEWCEAV 956
Query: 541 DDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVD 600
D E A+ K TWE+ P + W+FT K +DG ERYKARLV G TQ+ G+D
Sbjct: 957 DAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLD 1016
Query: 601 CGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPN-- 658
+TFSPV K TI+ +L ++ SK W + QLDV NAFL+GEL+E ++M P G+ +
Sbjct: 1017 YTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGI 1076
Query: 659 -LPNHVCL-LKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILL 716
LP++V L LK+S+YGLKQA R W+K+F+ + ++GF + DH+LF+ +L+
Sbjct: 1077 VLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLV 1136
Query: 717 YVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEI 776
YVDDI++ ++S+A + L F ++DLG L YFLG+ V T G+ + QRKYA E+
Sbjct: 1137 YVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQRKYALEL 1196
Query: 777 IERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQV 836
++ GM ACK S P+ K+ + D YR + G L YLT TRPDI +AV ++
Sbjct: 1197 LQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKL 1256
Query: 837 CLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSG 896
C F PR H+ A R+++YI+GT+ GL SS TL + D+DW C D+RRST+
Sbjct: 1257 CQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQDSRRSTTS 1316
Query: 897 YCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVY 956
+ +F+GD+LISW +K+Q T+SRSSAEAEYR +A E WL LL+ L ++Y
Sbjct: 1317 FTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQAS-PPVPILY 1375
Query: 957 CDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLV 1016
D+ +AIY++ NPV H+RTKHI++D H+VRE++ GE+++LHV + Q+ADI TK L
Sbjct: 1376 SDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADILTKPLFPY 1435
Query: 1017 LFEDFRNNLSV 1027
FE ++ +S+
Sbjct: 1436 QFEHLKSKMSI 1446
>At1g70010 hypothetical protein
Length = 1315
Score = 642 bits (1656), Expect = 0.0
Identities = 365/982 (37%), Positives = 522/982 (52%), Gaps = 23/982 (2%)
Query: 45 LTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPI 104
L L +VL PQ NL+SV T + FD ++D + + + +LY +
Sbjct: 323 LILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMGKQVANLYIV 382
Query: 105 TTSQAISPSTFAALAPS------LWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSC 158
P T +++ + LWH RLGHP + + + + C C
Sbjct: 383 DLDSLSHPGTDSSITVASVTSHDLWHKRLGHPSVQKLQPMSSLLSFPKQKNNTDFHCRVC 442
Query: 159 SLGKHIKLPFVSSNSCTIMPFDIIHSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLS 217
+ K LPFVS N+ + PFD+IH D W V + G+RY++ VDDYS W + L
Sbjct: 443 HISKQKHLPFVSHNNKSSRPFDLIHIDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLR 502
Query: 218 KKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSS 277
KS V ++ +F T + QFE +K ++ DN E + F +F G+ SCP T
Sbjct: 503 NKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPELN---FTQFYHSKGIVPYHSCPETPQ 559
Query: 278 QNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQ 337
QN ERK + I N+ R+L + +P S+W + A YLIN LP L + P ++L +
Sbjct: 560 QNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAPILEDKCPFEVLTK 619
Query: 338 KEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISR 397
P+Y H++VFGCLCY +K R+ C F+GYPS + YK DL + II+SR
Sbjct: 620 TVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSR 679
Query: 398 HVIFDETQFPFAKLHNPQPYTYGFMDDGPSPYVIHHLTS--QPSLGQPAQHDLPNTQPTT 455
HV+F E FPF Q F D P+P + + PS + LP+ PT
Sbjct: 680 HVVFHEELFPFLGSDLSQEEQNFFPDLNPTPPMQRQSSDHVNPSDSSSSVEILPSANPTN 739
Query: 456 QPTTPEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLN 515
P Q +H + P+ TP + K + S I P L
Sbjct: 740 NVPEPSVQTSH---RKAKKPAYLQDYYCHSVVSSTPHEIRKFL--SYDRINDP--YLTFL 792
Query: 516 TSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKE 575
+ ++ P N A + W+ AM EF+ L TWE+ P D I WIF K
Sbjct: 793 ACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKY 852
Query: 576 KSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKN 635
SDG ERYKARLV G TQ+ G+D ETFSPV K +++ +L +A S+ QLD+ N
Sbjct: 853 NSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISN 912
Query: 636 AFLHGELKETVYMHQPMGFR----DPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTI 691
AFL+G+L E +YM P G+ D PN VC LKKSLYGLKQA R WY +F+ + +
Sbjct: 913 AFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGL 972
Query: 692 GFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLH 751
GF S DH+ F+ +L+Y+DDII+ +++DA + S + + F ++DLG L
Sbjct: 973 GFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELK 1032
Query: 752 YFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSH 811
YFLG+ + G+ +SQRKYA ++++ G CK SS P+D + +S +
Sbjct: 1033 YFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGP 1092
Query: 812 YRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPS 871
YR L G L YL TRPDI +AV ++ F PR+ H+ A+ +I++YI+GT+ GL +
Sbjct: 1093 YRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFYSAT 1152
Query: 872 STSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANV 931
S L Y +AD+ C D+RRSTSGYC+FLGD+LI W +++Q +S+SSAEAEYR ++
Sbjct: 1153 SELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVA 1212
Query: 932 VSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVAR 991
E WL N L EL P+ K +L++CDN +AI+++ N V H+RTKHIE D HSVRE++ +
Sbjct: 1213 TDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRERLLK 1272
Query: 992 GEVRVLHVPSRYQIADIFTKGL 1013
G + H+ + QIAD FTK L
Sbjct: 1273 GLFELYHINTELQIADPFTKPL 1294
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 608 bits (1567), Expect = e-174
Identities = 369/1022 (36%), Positives = 545/1022 (53%), Gaps = 45/1022 (4%)
Query: 45 LTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESR--GDLY 102
L + +V + + +L+S+ + +N ++ V+D +T +M R G +
Sbjct: 313 LVMISVFYVEEFQSDLISIGQLMDENRCVLQMSDRFLVVQD-RTSRMVMGAGRRVGGTFH 371
Query: 103 PITTSQAISPSTFAALAPSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGK 162
+T A S + LWH+R+GHP A VV SL + + + C C K
Sbjct: 372 FRSTEIAASVTVKEEKNYELWHSRMGHPAARVV-SLIPESSVSVSSTHLNKACDVCHRAK 430
Query: 163 HIKLPFVSSNSCTIMPFDIIHSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQ 221
+ F S + T+ F++I+ D+W S +G RY++ +DDYS +W + L+ KS+
Sbjct: 431 QTRNSFPLSINKTLRIFELIYCDLWGPYRTPSHTGARYFLTIIDDYSRGVWLYLLNDKSE 490
Query: 222 VFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGK 281
+F QF ++K ++ DNG EF +F +E GV SC T +N +
Sbjct: 491 APCHLKNFFAMTDRQFNVKIKTVRSDNGTEF--LCLTKFFQEQGVIHERSCVATPERNDR 548
Query: 282 AERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPS 341
ERK R + N+ R L A+LP FW + A YLIN P+ L +P + L++K+P
Sbjct: 549 VERKHRHLLNVARALRFQANLPIQFWGECVLTAAYLINRTPSSVLNDSTPYERLHKKQPR 608
Query: 342 YSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIF 401
+ HLRVFG LCY + +K RS CVF+GYP + ++ +DL + +SR V+F
Sbjct: 609 FDHLRVFGSLCYAHNRNRGGDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVF 668
Query: 402 DETQFPFAKLHNPQPYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLP---NTQPTTQPT 458
E +FPF ++ + Q + +P V + + LGQ A P ++ + T
Sbjct: 669 SELEFPF-RISHEQNVIEEEEEALWAPIVDGLIEEEVHLGQNAGPTPPICVSSPISPSAT 727
Query: 459 TPEEQHAHSPPSSS---PNPSPSTTATPSPPYQPT-----PISVPKPVTR---SQHGIFK 507
+ +H+ S P + P P+ STT+ SP PT P+S KP T + +
Sbjct: 728 SSRSEHSTSSPLDTEVVPTPATSTTSASSPS-SPTNLQFLPLSRAKPTTAQAVAPPAVPP 786
Query: 508 PKRQLNLNTSVP--------RSPLPRNPVSALRDPNWKMAMDDE-------------FNA 546
P+RQ N + P + + + S L +++ D+ +A
Sbjct: 787 PRRQSTRNKAPPVTLKDFVVNTTVCQESPSKLNSILYQLQKRDDTRRFSASHTTYVAIDA 846
Query: 547 LIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFS 606
+N TW + PP I S W++ K SDG ERYKARLV G Q+ G D GETF+
Sbjct: 847 QEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKEGEDYGETFA 906
Query: 607 PVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLL 666
PV K AT+R L +A+ + W IHQ+DV NAFLHG+L+E VYM P GF + + PN VC L
Sbjct: 907 PVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGF-EASHPNKVCRL 965
Query: 667 KKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTAS 726
+K+LYGLKQAPR W+++ + GF S +D+SLF KG+ IL+YVDD+I+T +
Sbjct: 966 RKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYVDDLIITGN 1025
Query: 727 SDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACK 786
S L++ F MKDLG L YFLGI V T G+++ QRKYA +II G+ K
Sbjct: 1026 SQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIYICQRKYALDIISETGLLGVK 1085
Query: 787 SSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREE 846
++ P++ KL ++S DP YR L G L YL TR D+A++V + FM +PRE+
Sbjct: 1086 PANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSVHILARFMQEPRED 1145
Query: 847 HMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLI 906
H A R+VRY++ G+ L S + + D+DW G P +RRS +GY V GD+ I
Sbjct: 1146 HWAAALRVVRYLKADPGQGVFLRRSGDFQITGWCDSDWAGDPMSRRSVTGYFVQFGDSPI 1205
Query: 907 SWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLS 966
SW K+Q T+S+SSAEAEYR ++ + SE WL+ LL L + ++ CD+ SAIY++
Sbjct: 1206 SWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCDSKSAIYIA 1265
Query: 967 GNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLS 1026
NPV H+RTKHIE+D H VR++ +G + HV + Q+ADIFTK L F FR L
Sbjct: 1266 TNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFTKPLGRDCFSAFRIKLG 1325
Query: 1027 VR 1028
+R
Sbjct: 1326 IR 1327
>At4g17450 retrotransposon like protein
Length = 1433
Score = 580 bits (1495), Expect = e-165
Identities = 351/1030 (34%), Positives = 508/1030 (49%), Gaps = 71/1030 (6%)
Query: 12 FNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNS 71
F N + + N ++ I G LS ++L NVL+ P+ NL+S + T +
Sbjct: 452 FRSLENTFVRLPNDCTVKIAGIGFIQLS---DAISLHNVLYIPEFKFNLIS--ELTKELM 506
Query: 72 VSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAPSLWHARLGHPG 131
+ V DF + + + P S + + WH RLGHP
Sbjct: 507 IGRGSQVGNLYVLDFNENNHTVSLKGTTSMCP-----EFSVCSSVVVDSVTWHKRLGHPA 561
Query: 132 APVVDSLR-----KNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDI 186
+D L K K I + H+CH C L K L F S + FD++H D
Sbjct: 562 YSKIDLLSDVLNLKVKKINKEHSPVCHVCHVCHLSKQKHLSFQSRQNMCSAAFDLVHIDT 621
Query: 187 WTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQC 246
W F ++ W + L KS V +F +F + TQ++ ++K+++
Sbjct: 622 WGP-------------FSVPTNDATWIYLLKNKSDVLHVFPAFINMVHTQYQTKLKSVRS 668
Query: 247 DNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSF 306
DN E F + +G+ SCP T QN ERK + I N+ R LL +++P F
Sbjct: 669 DNAHELK---FTDLFAAHGIVAYHSCPETPEQNSVVERKHQHILNVARALLFQSNIPLEF 725
Query: 307 WHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQA 366
W + A +LIN LP L +SP + L P+Y L+ FGCLCY +K +
Sbjct: 726 WGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAYESLKTFGCLCYSSTSPKQRHKFEP 785
Query: 367 RSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYGFMDDGP 426
R+ CVFLGYP ++ YK D+ + + ISRHVIF E FPF + DD
Sbjct: 786 RARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFHEDIFPFI--------SSTIKDDIK 837
Query: 427 SPYVIHHLTSQPSLGQPAQ-HDLPNTQPTTQPTTPEEQHAHSPPSSSPNP---SPSTTAT 482
+ P L PA+ DLP Q T+ + H H SSS P +
Sbjct: 838 DFF--------PLLQFPARTDDLPLEQ-----TSIIDTHPHQDVSSSKALVPFDPLSKRQ 884
Query: 483 PSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAMDD 542
PP T H ++ + +P+ A W AM +
Sbjct: 885 KKPPKHLQDFHCYNNTTEPFHAFIN---------NITNAVIPQRYSEAKDFKAWCDAMKE 935
Query: 543 EFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCG 602
E A+++ TW +V PP+ I W+FT K +DG ERYKARLV G TQ+ G+D
Sbjct: 936 EIGAMVRTNTWSVVSLPPNKKAIGCKWVFTIKHNADGSIERYKARLVAKGYTQEEGLDYE 995
Query: 603 ETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRD---PNL 659
ETFSPV K ++R +L LA WS+HQLD+ NAFL+G+L E +YM P G+ D L
Sbjct: 996 ETFSPVAKLTSVRMMLLLAAKMKWSVHQLDISNAFLNGDLDEEIYMKIPPGYADLVGEAL 1055
Query: 660 PNH-VCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFI-YRKGTAMAYILLY 717
P H +C L KS+YGLKQA R WY + ++ + +GF S +DH+LFI Y G M +L+Y
Sbjct: 1056 PPHAICRLHKSIYGLKQASRQWYLKLSNTLKGMGFQKSNADHTLFIKYANGVLMG-VLVY 1114
Query: 718 VDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEII 777
VDDI++ ++SD + L + F ++DLG+ YFLGI + G+ + QRKY E++
Sbjct: 1115 VDDIMIVSNSDDAVAQFTAELKSYFKLRDLGAAKYFLGIEIARSEKGISICQRKYILELL 1174
Query: 778 ERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVC 837
G K SS P+D KL+ P D + YR L G L YL TRPDIAYAV +C
Sbjct: 1175 STTGFLGSKPSSIPLDPSVKLNKEDGVPLTDSTSYRKLVGKLMYLQITRPDIAYAVNTLC 1234
Query: 838 LFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGY 897
F H P H+ A+ +++RY++GT+ GL L YTD+D+G C D+RR + Y
Sbjct: 1235 QFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYSADDKFDLRGYTDSDFGSCTDSRRCVAAY 1294
Query: 898 CVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYC 957
C+F+GD L+SW +K+Q T+S S+AEAE+R ++ E WL L + P + +YC
Sbjct: 1295 CMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMSQGTKEMIWLSRLFDDFKVPFIPPAYLYC 1354
Query: 958 DNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVL 1017
DN +A+++ N V H+RTK +E+D + RE V G ++ + V + Q+AD TK +
Sbjct: 1355 DNTAALHIVNNSVFHERTKFVELDCYKTREAVESGFLKTMFVETGEQVADPLTKAIHPAQ 1414
Query: 1018 FEDFRNNLSV 1027
F + V
Sbjct: 1415 FHKLIGKMGV 1424
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 579 bits (1492), Expect = e-165
Identities = 360/1037 (34%), Positives = 505/1037 (47%), Gaps = 156/1037 (15%)
Query: 45 LTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPI 104
L L NVLH P NL+SV S S F +++ G+ + R +LY +
Sbjct: 515 LILHNVLHVPDFKFNLMSVSSLVKTISCSAHFYVDCCLIQELSQGLMIGRGRLYHNLYIL 574
Query: 105 TTSQAISPSTFAALA----------PSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHI 154
T SPST A LWH RLGHP + V+ L++
Sbjct: 575 ETENT-SPSTSTPAACLFTGSVLNDGHLWHQRLGHPSSVVLQKLKR-------------- 619
Query: 155 CHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWT 213
L ++S N+ PFD++H DIW + S G RY++ VDD + W
Sbjct: 620 -----------LAYISHNNLASNPFDLVHLDIWGPFSIESIEGFRYFLTVVDDCTRTTWV 668
Query: 214 FPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCP 273
+ L K V S+F F + TQF ++K I+ DN E F E KE+G+ SC
Sbjct: 669 YMLRNKKDVSSVFPEFIKLVSTQFNAKIKAIRSDNAPELG---FTEIVKEHGMLHHFSCA 725
Query: 274 HTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLK 333
+T QN ERK + I N+ R LL +++P +W + A +LIN LP+ L +SP +
Sbjct: 726 YTPQQNSVVERKHQHILNVARALLFQSNIPMQYWSDCVTTAVFLINRLPSPLLNNKSPYE 785
Query: 334 ILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKI 393
++ K+P YS L+ FGCLC+ + K R+ CVFLGYPS ++ YK DL S +
Sbjct: 786 LILNKQPDYSLLKNFGCLCFVSTNAHERTKFTPRARACVFLGYPSGYKGYKVLDLESHSV 845
Query: 394 IISRHVIFDETQFPFAKLHNPQPYTYGFMDDG-PSPYVIHHLTSQPSLGQPAQHDLPNTQ 452
+SR+V+F E FPF F + P P +H + + P + D +
Sbjct: 846 TVSRNVVFKEHVFPFKTSELLNKAVDMFPNSILPLPAPLHFVETMPLI------DEDSLI 899
Query: 453 PTTQPTTPEEQHAHSPPSSSPN--PSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKR 510
PTT + + HA S S+ P+ P S T T PI+ K TR+ + +
Sbjct: 900 PTTTDSRTADNHASSSSSALPSIIPPSSNTETQDIDSNAVPITRSKRTTRAPSYLSEYHC 959
Query: 511 QL--------NLNTSVPRSPLPR------------NPVS--------------------- 529
L ++S+P PLP P+S
Sbjct: 960 SLVPSISTLPPTDSSIPIHPLPEIFTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAYNT 1019
Query: 530 ---------ALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGV 580
A++ W +E A+ NKTW + PPD NV+ W+FT K DG
Sbjct: 1020 ETEPKTFSQAMKSEKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDGT 1079
Query: 581 FERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHG 640
ERYKARLV G TQQ G+D +TFSPV K + + +L LA W++ Q+DV +AFLHG
Sbjct: 1080 VERYKARLVAQGFTQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLHG 1139
Query: 641 ELKETVYMHQPMGFRDPN----LPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHS 696
+L E ++M P G+ P PN VC L KS+YGLKQA R WYKRF
Sbjct: 1140 DLDEEIFMSLPQGYTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKRFVA---------- 1189
Query: 697 TSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGI 756
L+Y+DDI++ +++DA + +LL +EF +KDLG +FLG+
Sbjct: 1190 -----------------ALVYIDDIMIASNNDAEVENLKALLRSEFKIKDLGPARFFLGL 1232
Query: 757 AVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLA 816
CK SS P+D L + P +P+ YR L
Sbjct: 1233 L--------------------------GCKPSSIPMDPTLHLVRDMGTPLPNPTAYRKLI 1266
Query: 817 GALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTL 876
G L YLT TRPDI YAV Q+ F+ P + H+ A +++RYI+ GL L
Sbjct: 1267 GRLLYLTITRPDITYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMYSADYEICL 1326
Query: 877 ISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESC 936
++DADW C DTRRS SG+C++LG +LISW +K+QA SRSS E+EYR +A E
Sbjct: 1327 NGFSDADWAACKDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSMAQATCEII 1386
Query: 937 WLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRV 996
WL+ LL +LH P+ + ++CDN SA++ S NPV H+RTKHIE+D H+VR+++ G ++
Sbjct: 1387 WLQQLLKDLHIPLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQIKAGNLKA 1446
Query: 997 LHVPSRYQIADIFTKGL 1013
LHVP+ Q ADI TK L
Sbjct: 1447 LHVPTENQHADILTKAL 1463
>At1g26990 polyprotein, putative
Length = 1436
Score = 546 bits (1406), Expect = e-155
Identities = 337/1035 (32%), Positives = 519/1035 (49%), Gaps = 76/1035 (7%)
Query: 3 SEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVS 62
+ + NL + + + + NGH++ I G L+ L+L NVL P+ NL+S
Sbjct: 432 THERNLYHTYKALDRTFVRLPNGHTVKIEGTGFIQLT---DALSLHNVLFIPEFKFNLLS 488
Query: 63 VRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQA-ISPSTFAALA-- 119
V T V F ++ + L + G+LY + ++ + S+F +
Sbjct: 489 VSVLTKTLQSKVSFTSDECMIQALTKELMLGKGSQVGNLYILNLDKSLVDVSSFPGKSVC 548
Query: 120 ------PSLWHARLGHPGAPVVDSLRKNKFIECNRAS--GSHICHSCSLGKHIKLPFVSS 171
+WH RLGHP +D+L + + + SH CH C L K LPF S
Sbjct: 549 SSVKNESEMWHKRLGHPSFAKIDTLSDVLMLPKQKINKDSSH-CHVCHLSKQKHLPFKSV 607
Query: 172 NSCTIMPFDIIHSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFR 230
N F+++H D W V + +RY++ VDD+S W + L +KS V ++F SF
Sbjct: 608 NHIREKAFELVHIDTWGPFSVPTVDSYRYFLTIVDDFSRATWIYLLKQKSDVLTVFPSFL 667
Query: 231 TFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTIN 290
+ TQ+ +V +++ DN E F E + G+ CP T QN ERK + +
Sbjct: 668 KMVETQYHTKVCSVRSDNAHELK---FNELFAKEGIKADHPCPETPEQNFVVERKHQHLL 724
Query: 291 NIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGC 350
N+ R L+ + +P +W + A +LIN L + + ++P + L + +P YS L+ FGC
Sbjct: 725 NVARALMFQSGIPLEYWGDCVLTAVFLINRLLSPVINNETPYERLTKGKPDYSSLKAFGC 784
Query: 351 LCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAK 410
LCY + K R+ C+FLGYP ++ YK D+ + + ISRHVIF E FPFA
Sbjct: 785 LCYCSTSPKSRTKFDPRAKACIFLGYPMGYKGYKLLDIETYSVSISRHVIFYEDIFPFAS 844
Query: 411 LHNPQ------PYTYGFMDDGPSPYVIHHLTS-QPSLGQPAQHDLPNTQPTTQPTTPEEQ 463
+ P+ Y P+P HL Q S P HD ++ P+ P+
Sbjct: 845 SNITDAAKDFFPHIYL-----PAPNNDEHLPLVQSSSDAPHNHD-ESSSMIFVPSEPKST 898
Query: 464 HAHSPPSSSP-----NPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSV 518
PS N +P+TT T PY P + G F +N+ T+
Sbjct: 899 RQRKLPSHLQDFHCYNNTPTTTKTS--PY-PLTNYISYSYLSEPFGAF-----INIITA- 949
Query: 519 PRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSD 578
+ LP+ A D W AM E +A ++ TW + P + WI T K +D
Sbjct: 950 --TKLPQKYSEARLDKVWNDAMGKEISAFVRTGTWSICDLPAGKVAVGCKWIITIKFLAD 1007
Query: 579 GVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFL 638
G ER+KARLV G TQQ G+D TFSPV K T++ +LSLA W +HQLD+ NA L
Sbjct: 1008 GSIERHKARLVAKGYTQQEGIDFFNTFSPVAKMVTVKVLLSLAPKMKWYLHQLDISNALL 1067
Query: 639 HGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTS 698
+G+L+E +YM P G+ + + G + +P A
Sbjct: 1068 NGDLEEEIYMKLPPGYSE-------------IQGQEVSPNA---------------KCHG 1099
Query: 699 DHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAV 758
DH+LF+ + +L+YVDDI++ ++++A + S LS+ F ++DLG +FLGI +
Sbjct: 1100 DHTLFVKAQDGFFLVVLVYVDDILIASTTEAASAELTSQLSSFFQLRDLGEPKFFLGIEI 1159
Query: 759 THHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGA 818
+ G+ L QRKY +++ + + CK SS P++ KLS ++ D YR + G
Sbjct: 1160 ARNADGISLCQRKYVLDLLASSDFSDCKPSSIPMEPNQKLSKDTGTLLEDGKQYRRILGK 1219
Query: 819 LQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLIS 878
LQYL TRPDI +AV ++ + P + H+ AL +I+RY++GT+ GL + L
Sbjct: 1220 LQYLCLTRPDINFAVSKLAQYSSAPTDIHLQALHKILRYLKGTIGQGLFYGADTNFDLRG 1279
Query: 879 YTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWL 938
++D+DW CPDTRR +G+ +F+G++L+SW +K+Q +S SSAEAEYR ++ E WL
Sbjct: 1280 FSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSKKQDVVSMSSAEAEYRAMSVATKELIWL 1339
Query: 939 RNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLH 998
+L P + +YCDN +A++++ N V H+RTKHIE D H VRE + G ++ +
Sbjct: 1340 GYILTAFKIPFTHPAYLYCDNEAALHIANNSVFHERTKHIENDCHKVRECIEAGILKTIF 1399
Query: 999 VPSRYQIADIFTKGL 1013
V + Q+AD TK L
Sbjct: 1400 VRTDNQLADTLTKPL 1414
>At3g45520 copia-like polyprotein
Length = 1363
Score = 512 bits (1318), Expect = e-145
Identities = 339/1013 (33%), Positives = 500/1013 (48%), Gaps = 50/1013 (4%)
Query: 45 LTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPI 104
+TL+NV + P + +NL+S+ F +F+ ++ L+ LY +
Sbjct: 370 VTLQNVRYIPDMDRNLLSLGTF---EKAGHKFESENGMLRIKSGNQVLLEGRRYDTLYIL 426
Query: 105 TTSQAISPSTFAALAPS---LWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLG 161
A S A A LWH RL H + L K F++ + S C C G
Sbjct: 427 HGKPATDESLAVARANDDTVLWHRRLCHMSQKNMSLLIKKGFLDKKKVSMLDTCEDCIYG 486
Query: 162 KHIKLPFVSSNSCTIMPFDIIHSDIWTSPV--LSSSGHRYYVLFVDDYSNFLWTFPLSKK 219
+ K+ F + T + +HSD+W +P +S +Y++ F+DDY+ +W + L K
Sbjct: 487 RAKKIGFNLAQHDTKKKLEYVHSDLWGAPTVPMSLGNCQYFISFIDDYTRKVWVYFLKTK 546
Query: 220 SQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQN 279
+ F F+S+ + + Q VK ++ DNG EF NR F FC+E G +C +T QN
Sbjct: 547 DEAFEKFVSWISLVENQSGERVKTLRTDNGLEFCNRMFDGFCEEKGFQRHRTCAYTPQQN 606
Query: 280 GKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKE 339
G ER RTI +R++L + LP FW A A LIN P + ++ P K K
Sbjct: 607 GVVERMNRTIMEKVRSMLCDSGLPKRFWAEATHTAVLLINKTPCSAINFEFPDKRWSGKA 666
Query: 340 PSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHV 399
P YS+LR +GC+ F T KL R+ V +GYPS + YK + + +K ++SR+V
Sbjct: 667 PIYSYLRRYGCVT---FVHTDGGKLNLRAKKGVLIGYPSGVKGYKVWLIEEKKCVVSRNV 723
Query: 400 IFDETQFPFAKLHNPQPYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTT 459
F E + L + DD Y+ L + + P T+
Sbjct: 724 SFQENAV-YKDLMQRKEQVSCEEDDHAGSYIDLDLEADKDNSSGGEQSQAQVTPATRGAV 782
Query: 460 PEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKR--------Q 511
+S P T +P+S R + I P+R +
Sbjct: 783 -----------TSTPPRYETDDIEETDVHQSPLSYHLVRDRERREIRAPRRFDDEDYYAE 831
Query: 512 LNLNTSVPRSPLPRNPVSALRDPNW---KMAMDDEFNALIKNKTWELVPRPPDVNVIRSM 568
T + P + A+RD NW ++AM++E + +KN TW V RP +I S
Sbjct: 832 ALYTTEDGDAVEPADYKEAVRDENWDKWRLAMNEEIESQLKNDTWTTVTRPEKQRIIGSR 891
Query: 569 WIFTHKEKSDGVFE-RYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWS 627
WI+ +K+ GV E R+KARLV G Q+ GVD E F+PVVK +IR +LS+ +
Sbjct: 892 WIYKYKQGIPGVEEPRFKARLVAKGYAQREGVDYHEIFAPVVKHVSIRILLSIVAQENLE 951
Query: 628 IHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADY 687
+ QLDVK AFLHGELKE +YM P G N VCLL KSLYGLKQAPR W ++F Y
Sbjct: 952 LEQLDVKTAFLHGELKEKIYMMPPEGCESLFKENEVCLLNKSLYGLKQAPRQWNEKFNHY 1011
Query: 688 VSTIGFSHSTSDHSLFIYR-KGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKD 746
++ IGF S D + + + Y+L YVDD+++ A++ + LS +F MKD
Sbjct: 1012 MTEIGFKRSDYDSCAYTKKLSDDSTMYLLFYVDDMLVAANNMQAIDALKKELSIKFEMKD 1071
Query: 747 LGSLHYFLG--IAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVD--------TKP 796
LG+ LG I + G L+LSQ Y ++++ M K + TP+ T+
Sbjct: 1072 LGAAKKILGIEIIIDREAGVLWLSQESYLNKVLKTFNMLESKPALTPLGAHLKMKSATEE 1131
Query: 797 KLSANSSAPHADPSHYRSLAGALQY-LTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIV 855
KLS ++ P Y S G++ Y + TRPD+AY V V FM P +EH +K ++
Sbjct: 1132 KLSTEEEYMNSVP--YSSAVGSIMYAMIGTRPDLAYPVGVVSRFMSQPAKEHWLGVKWVL 1189
Query: 856 RYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQAT 915
RYI+GT+D L +S ++ Y DAD+ D RRS +G LG N ISW + Q
Sbjct: 1190 RYIKGTVDTRLCYKRNSDFSICGYCDADYAADLDKRRSITGLVFTLGGNTISWKSGLQRV 1249
Query: 916 LSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRT 975
+++SS E EY + V E+ WL+ LL + +K ++CD+ SAI LS N V H+RT
Sbjct: 1250 VAQSSTECEYMSLTEAVKEAIWLKGLLKDFGYE-QKNVEIFCDSQSAIALSKNNVHHERT 1308
Query: 976 KHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVR 1028
KHI++ H +RE +A G+V V + + ADIFTK LP+ F+ + L V+
Sbjct: 1309 KHIDVKFHFIREIIADGKVEVSKISTEKNPADIFTKVLPVNKFQTALDFLRVK 1361
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 511 bits (1316), Expect = e-145
Identities = 339/1034 (32%), Positives = 509/1034 (48%), Gaps = 40/1034 (3%)
Query: 22 VGNGHSIPIRGYSHTNL-SFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFG 80
+GN P++G + + + L +V + P + +NL+S+ D
Sbjct: 315 MGNDTYSPVKGIGSIKIRNSDGSQVILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGIL 374
Query: 81 FSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAPSLWHARLGHPGAPVVDSLRK 140
VK T ++ + ++ L +T S +LWH+RLGH ++ L K
Sbjct: 375 KIVKGCSTILKGQKRDTLYILDGVTEEGESHSSAEVKDETALWHSRLGHMSQKGMEILVK 434
Query: 141 NKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIWTSP--VLSSSGHR 198
+ C C GK ++ F + T +HSD+W SP S +
Sbjct: 435 KGCLRREVIKELEFCEDCVYGKQHRVSFAPAQHVTKEKLAYVHSDLWGSPHNPASLGNSQ 494
Query: 199 YYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFW 258
Y++ FVDDYS +W + L KK + F F+ ++ + Q +R+VK ++ DNG E+ N +F
Sbjct: 495 YFISFVDDYSRKVWIYFLRKKDEAFEKFVEWKKMVENQSDRKVKKLRTDNGLEYCNHYFE 554
Query: 259 EFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLI 318
+FCKE G+ +C +T QNG AER RTI + +R++L + + FW A A YLI
Sbjct: 555 KFCKEEGIVRHKTCAYTPQQNGIAERLNRTIMDKVRSMLSRSGMEKKFWAEAASTAVYLI 614
Query: 319 NILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPS 378
N P+ + + P + P S LR FGCL Y KL RS +F YP
Sbjct: 615 NRSPSTAINFDLPEEKWTGALPDLSSLRKFGCLAY---IHADQGKLNPRSKKGIFTSYPE 671
Query: 379 NHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYGFMDDGPSPYVIHHLTSQP 438
+ YK + L +K +ISR+VIF E Q F L T D + L P
Sbjct: 672 GVKGYKVWVLEDKKCVISRNVIFRE-QVMFKDLKGDSQNTISESD-------LEDLRVNP 723
Query: 439 SLGQPAQHDLPN-TQPTTQPTTPEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKP 497
+ D TQ + P+ E +H+P +SP S + +S +
Sbjct: 724 DMNDQEFTDQGGATQDNSNPS--EATTSHNPVLNSPTHSQDEESEEEDSDAVEDLSTYQL 781
Query: 498 VTRSQHGIFKPKRQLNLNTSV--------PRSPLPRNPVSALRDPNWK---MAMDDEFNA 546
V K + N + V P P++ AL DP+W+ AM +E +
Sbjct: 782 VRDRVRRTIKANPKYNESNMVGFAYYSEDDGKPEPKSYQEALLDPDWEKWNAAMKEEMVS 841
Query: 547 LIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGV-FERYKARLVGDGKTQQVGVDCGETF 605
+ KN TW+LV +P V +I W+FT K GV R+ ARLV G TQ+ GVD E F
Sbjct: 842 MSKNHTWDLVTKPEKVKLIGCRWVFTRKAGIPGVEAPRFIARLVAKGFTQKEGVDYNEIF 901
Query: 606 SPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCL 665
SPVVK +IR +LS+ + + Q+DVK AFLHG L+E +YM QP GF N VCL
Sbjct: 902 SPVVKHVSIRYLLSMVVHYNMELQQMDVKTAFLHGFLEEEIYMAQPEGFEIKRGSNKVCL 961
Query: 666 LKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYR-KGTAMAYILLYVDDIILT 724
LK+SLYGLKQ+PR W RF +++ I ++ S D ++ + G Y+LLYVDD+++
Sbjct: 962 LKRSLYGLKQSPRQWNLRFDEFMRGIKYTRSAYDSCVYFKKCNGDTYIYLLLYVDDMLIA 1021
Query: 725 ASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVT--HHTGGLFLSQRKYAAEIIERAGM 782
+++ + + LLS EF MKDLG LG+ ++ G L LSQ Y +++ M
Sbjct: 1022 SANKSEVNELKQLLSREFEMKDLGDAKKILGMEISRDRDAGLLTLSQEGYVKKVLRSFQM 1081
Query: 783 AACKSSSTPVDTKPKLSANSSAPHADPSH------YRSLAGALQY-LTFTRPDIAYAVQQ 835
K STP+ KL A + + + Y + G++ Y + TRPD+AY++
Sbjct: 1082 DNAKPVSTPLGIHFKLKAATDKEYEEQFERMKIVPYANTIGSIMYSMIGTRPDLAYSLGV 1141
Query: 836 VCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTS 895
+ FM P ++H A+K ++RY++GT L L Y D+D+G DTRRS +
Sbjct: 1142 ISRFMSKPLKDHWQAVKWVLRYMRGTEKKKLCFRKQEDFLLRGYCDSDYGSNFDTRRSIT 1201
Query: 896 GYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLV 955
GY +G N ISW +K Q ++ SS EAEY + V E+ WL+ EL + V
Sbjct: 1202 GYVFTVGGNTISWKSKLQKVVAISSTEAEYMALTEAVKEALWLKGFAAELG-HSQDYVEV 1260
Query: 956 YCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPL 1015
+ D+ SAI L+ N V H+RTKHI++ +H +R+ + G ++V+ + + A+IFTK +PL
Sbjct: 1261 HSDSQSAITLAKNSVHHERTKHIDIRLHFIRDIICAGLIKVVKIATECNPANIFTKTVPL 1320
Query: 1016 VLFEDFRNNLSVRQ 1029
FE N L V +
Sbjct: 1321 AKFEGALNMLRVTE 1334
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 510 bits (1313), Expect = e-144
Identities = 342/1053 (32%), Positives = 524/1053 (49%), Gaps = 63/1053 (5%)
Query: 12 FNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNP-PLTLKNVLHSPQLIKNLVSVRKFTTDN 70
FN + +GN +RG + + + L NV + P + +NL+S+ F
Sbjct: 329 FNEDAGGSVRMGNKTVSRVRGVGTIRVKNSDGLTIVLTNVRYIPDMDRNLLSLGTF---- 384
Query: 71 SVSVEFDPFGFSVKD----FQTGMRLMRCESRGDLYPITTSQAISPSTFAALAPS----L 122
E + F +D + G +++ R D + + ++ + A + + L
Sbjct: 385 ----EKAGYKFESEDGILRIKAGNQVLLTGRRYDTLYLLNWKPVASESLAVVKRADDTVL 440
Query: 123 WHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDII 182
WH RL H ++ L + F++ + S +C C GK + F ++ T + I
Sbjct: 441 WHQRLCHMSQKNMEILVRKGFLDKKKVSSLDVCEDCIYGKAKRKSFSLAHHDTKEKLEYI 500
Query: 183 HSDIWTSPV--LSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFERE 240
HSD+W +P LS +Y++ +DD++ +W + + K + F F+ + + Q +R
Sbjct: 501 HSDLWGAPFVPLSLGKCQYFMSIIDDFTRKVWVYFMKTKDEAFEKFVEWVNLVENQTDRR 560
Query: 241 VKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHA 300
VK ++ DNG EF N+ F FC+ G+ +C +T QNG AER RTI +R++L +
Sbjct: 561 VKTLRTDNGLEFCNKLFDGFCESIGIHRHRTCAYTPQQNGVAERMNRTIMEKVRSMLSDS 620
Query: 301 SLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTT 360
LP FW A LIN P+ L ++ P K P YS+LR +GC+ F T
Sbjct: 621 GLPKRFWAEATHTTVLLINKTPSSALNFEIPDKKWSGNPPVYSYLRRYGCVA---FVHTD 677
Query: 361 INKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYG 420
KL+ R+ V +GYP + YK + L RK ++SR++IF E + L Q
Sbjct: 678 DGKLEPRAKKGVLIGYPVGVKGYKVWILDERKCVVSRNIIFQENAV-YKDLMQRQENVST 736
Query: 421 FMDDGPSPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPNPSPSTT 480
DD Y+ L ++ + ++ NT P PE +P + N +
Sbjct: 737 EEDDQTGSYLEFDLEAERDVISGGDQEMVNTIPA-----PESPVVSTPTTQDTNDDEDSD 791
Query: 481 ATPSPPYQPTPISVPKPVTRSQHGIFKPKR--------QLNLNTSVPRSPLPRNPVSALR 532
SP +S R + I P+R + T + P N A
Sbjct: 792 VNQSP------LSYHLVRDRDKREIRAPRRFDDEDYYAEALYTTEDGEAVEPENYRKAKL 845
Query: 533 DPN---WKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFE-RYKARL 588
D N WK+AMD+E ++ KN TW +V RP + +I WIF +K GV E R+KARL
Sbjct: 846 DANFDKWKLAMDEEIDSQEKNNTWTIVTRPENQRIIGCRWIFKYKLGILGVEEPRFKARL 905
Query: 589 VGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYM 648
V G Q+ G+D E F+PVVK +IR +LS+ + + QLDVK AFLHGELKE +YM
Sbjct: 906 VAKGYAQKEGIDYHEIFAPVVKHVSIRVLLSIVAQEDLELEQLDVKTAFLHGELKEKIYM 965
Query: 649 HQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLF--IYR 706
P G+ N VCLL K+LYGLKQAP+ W ++F +++ I F S D + +
Sbjct: 966 SPPEGYESMFKANEVCLLNKALYGLKQAPKQWNEKFDNFMKEICFVKSAYDSCAYTKVLP 1025
Query: 707 KGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHH--TGG 764
G+ M Y+L+YVDDI++ + + + + L F MKDLG+ LG+ + G
Sbjct: 1026 DGSVM-YLLIYVDDILVASKNKEAITALKANLGMRFEMKDLGAAKKILGMEIIRDRTLGV 1084
Query: 765 LFLSQRKYAAEIIERAGMAACKSSSTPVD--------TKPKLSANSSAPHADPSHYRSLA 816
L+LSQ Y +I+E MA K + TP+ T+ KL + + P Y S
Sbjct: 1085 LWLSQEGYLNKILETYNMAEAKPAMTPLGAHFKFQAATEQKLIRDEDFMKSVP--YSSAV 1142
Query: 817 GALQY-LTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTST 875
G++ Y + TRPD+AY V + FM P +EH +K ++RYI+GTL L SS+ +
Sbjct: 1143 GSIMYAMLGTRPDLAYPVGIISRFMSQPIKEHWLGVKWVLRYIKGTLKTRLCYKKSSSFS 1202
Query: 876 LISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSES 935
++ Y DAD+ D RRS +G LG N ISW + Q +++S+ E+EY + V E+
Sbjct: 1203 IVGYCDADYAADLDKRRSITGLVFTLGGNTISWKSGLQRVVAQSTTESEYMSLTEAVKEA 1262
Query: 936 CWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVR 995
WL+ LL + +K+ ++CD+ SAI LS N V H+RTKHI++ H +RE ++ G V
Sbjct: 1263 IWLKGLLKDFGYE-QKSVEIFCDSQSAIALSKNNVHHERTKHIDVKYHFIREIISDGTVE 1321
Query: 996 VLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVR 1028
VL + + ADIFTK L + F+ N L V+
Sbjct: 1322 VLKISTEKNPADIFTKVLAVSKFQAALNLLRVK 1354
>At2g14930 pseudogene
Length = 1149
Score = 509 bits (1312), Expect = e-144
Identities = 298/841 (35%), Positives = 441/841 (52%), Gaps = 37/841 (4%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+T+ L N ++ +G+ +PI NL + L LK+VL P + K+L
Sbjct: 324 ITNNSSRLQQMQPYLGNDTVMASDGNFLPITHIGSANLPSTSGNLPLKDVLVCPNIAKSL 383
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGD-LYPITTS--QAISPSTFAA 117
+SV K T D S FD G VKD T L + S + LY + Q +
Sbjct: 384 LSVSKLTKDYPCSFTFDADGVLVKDKATCKVLTKGSSTSEGLYKLENPKFQMFYSTRQVK 443
Query: 118 LAPSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIM 177
+WH RLGHP V+ L K I+ N+++ S +C SC LGK +LPF++S+
Sbjct: 444 ATDEVWHMRLGHPNPQVLQLLANKKAIQINKST-SKMCESCRLGKSSRLPFIASDFIASR 502
Query: 178 PFDIIHSDIW-TSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQ 236
P + +H D+W +PV S G +YYV+F+D+ S F W +PL KS S+F+ F++F+
Sbjct: 503 PLERVHCDLWGPAPVSSIQGFQYYVIFIDNRSRFCWFYPLKHKSDFCSLFMKFQSFVENL 562
Query: 237 FEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTL 296
+ ++ Q D G EF + F + +E+G+ +SCPHT QNG AERK R + TL
Sbjct: 563 LQTKIGTFQSDGGGEFTSNRFLQHLQESGIQHYISCPHTPQQNGLAERKHRQLTERGLTL 622
Query: 297 LVHASLPPSFWHHALQMATYLINILPNKQL-AYQSPLKILYQKEPSYSHLRVFGCLCYPL 355
+ + P FW A A +L N+LP L + +P ++L+ K P YS LR FGC C+P
Sbjct: 623 MFQSKAPQRFWVEAFFTANFLSNLLPTSALDSSTTPYQVLFGKAPDYSALRTFGCACFPT 682
Query: 356 FPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQ 415
+ NK RS C+FLGY ++ Y+C+ + ++ +SRHV+FDE+ FPF +
Sbjct: 683 LRAYARNKFDPRSLKCIFLGYTEKYKGYRCFFPPTNRVYLSRHVLFDESSFPFIDTYT-- 740
Query: 416 PYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLP---NTQPTTQPTTPEEQHAHSPPSSS 472
P+P L S PS P ++D N+ + T ++
Sbjct: 741 ----SLQHPSPTPMFDAWLKSFPSSSSPLENDQTAGFNSGASVPVITAQQTQPILSLKDG 796
Query: 473 PN----PSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNT--------SVPR 520
PN T ++ + + PI V T S K L++ + S
Sbjct: 797 PNILLPEGEITVSSNNQDIEDEPICVTPLQTLSSEDNAKSSETLSMGSEECSECTASFDL 856
Query: 521 SPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEK--SD 578
P+ N +S+ D+ + I E P + + + + K D
Sbjct: 857 DPIGNNALSS-------SPRHDQLTSSIPRAATEST-HPMTTRLKKGIIKLNQRVKLNVD 908
Query: 579 GVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFL 638
G ++YKARLV G Q+ G+D ET+SPVV+ AT+R VL L+ W + Q+DVKN FL
Sbjct: 909 GSLDKYKARLVAQGFKQEEGIDYLETYSPVVRSATVRAVLHLSTIMNWELKQMDVKNGFL 968
Query: 639 HGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTS 698
HG+L ETVYM QP GF D P+HVCLL K+LYGLKQAPRAW+ +F+ ++ + GF S S
Sbjct: 969 HGDLTETVYMKQPAGFIDKAHPDHVCLLHKALYGLKQAPRAWFDKFSKFLLSFGFVCSMS 1028
Query: 699 DHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAV 758
D SLF+ K + +LLYVDD+++T +S L +++S L+ +F MKDLG L YFLGI
Sbjct: 1029 DPSLFVCVKNKDVIMLLLYVDDMVITGNSSKLLSSLLSELNKQFKMKDLGRLSYFLGIQA 1088
Query: 759 THHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGA 818
H+ GLFLSQ+KYA +++ A M+ C +TP+ +P+ + N + +PS++RSLAG
Sbjct: 1089 QFHSQGLFLSQQKYAEDLLATAAMSNCSPVATPLPLQPERTPNQTELFDNPSYFRSLAGK 1148
Query: 819 L 819
L
Sbjct: 1149 L 1149
>At4g27200 putative protein
Length = 819
Score = 501 bits (1290), Expect = e-142
Identities = 293/739 (39%), Positives = 402/739 (53%), Gaps = 58/739 (7%)
Query: 334 ILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKI 393
+L +K S S R+ G C+P NK S CVFLGY ++ Y+C + ++
Sbjct: 1 MLVRKRHSTSSWRI-GSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRL 59
Query: 394 IISRHVIFDETQFPFAKLH---NPQPYT-----YGFMDDGPSPYVIHHLTSQ-------- 437
ISRHVIFDE+ +PF+ + +PQP T + D P+P +S+
Sbjct: 60 YISRHVIFDESVYPFSHTYKHLHPQPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFTSAD 119
Query: 438 -PSLGQPAQHDLPNTQP----------TTQPTTPEEQH-------------AHSPPSSSP 473
P L Q LP P TTQ + + +HS + S
Sbjct: 120 FPPLPQRKTPLLPTLVPISSVSHASNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSD 179
Query: 474 NPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRD 533
+ A+ + P +V VTR++ GI KP + + P P+ +AL+
Sbjct: 180 SEETIQQASVNVHQTPASTNVHPMVTRAKVGISKPNPRYVFLSHKVSYPEPKTVTAALKH 239
Query: 534 PNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGK 593
P W AM +E + +TW LVP D++V+ S W+F K +DG + KAR+V G
Sbjct: 240 PGWTGAMTEEIGNCSETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGF 299
Query: 594 TQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMG 653
Q+ G+D ET+SPVV+ T+R VL LA + W I Q+DVKNAFLHG+LKETVYM QP
Sbjct: 300 LQEEGIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAA 359
Query: 654 FRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAY 713
RD HVCLL KS+YGLKQ+PRAW+ +F+ ++ GF SD SLFIY +
Sbjct: 360 NRD-----HVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCRKSDPSLFIYAHNNNLIL 414
Query: 714 ILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHY-FLGIAVTHHTGGLFLSQRKY 772
+LL S L ++++ L+ EF M D+G FLGI V GLF+SQ+KY
Sbjct: 415 LLL----------SQTL-TSLLAALNKEFRMTDMGQHSLTFLGIQVQRQQNGLFMSQQKY 463
Query: 773 AAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYA 832
A +++ A M C TP+ + + +DP+++RS+AG LQYLT TRPDI +A
Sbjct: 464 AEDLLIAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFA 523
Query: 833 VQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRR 892
V VC MH P H LKRI+RYI+GT+ G+ S + L +Y+D+DWG C TRR
Sbjct: 524 VNFVCQKMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYSDSDWGNCKQTRR 583
Query: 893 STSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKA 952
S G C F+G NL+SWS+K+ T+SRSS EAEY+ +++ SE WL LL EL P+
Sbjct: 584 SVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPDT 643
Query: 953 SLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKG 1012
++CDN+SA+YL+ NP H RTKH ++D H VRE+VA + V H+P QIADIFTK
Sbjct: 644 PELFCDNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTKS 703
Query: 1013 LPLVLFEDFRNNLSVRQPP 1031
LP F R L V PP
Sbjct: 704 LPYEAFIHLRGKLGVTLPP 722
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,991,207
Number of Sequences: 26719
Number of extensions: 1178725
Number of successful extensions: 9961
Number of sequences better than 10.0: 383
Number of HSP's better than 10.0 without gapping: 224
Number of HSP's successfully gapped in prelim test: 169
Number of HSP's that attempted gapping in prelim test: 4571
Number of HSP's gapped (non-prelim): 1903
length of query: 1038
length of database: 11,318,596
effective HSP length: 109
effective length of query: 929
effective length of database: 8,406,225
effective search space: 7809383025
effective search space used: 7809383025
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146632.4


