

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.11 + phase: 0
(265 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
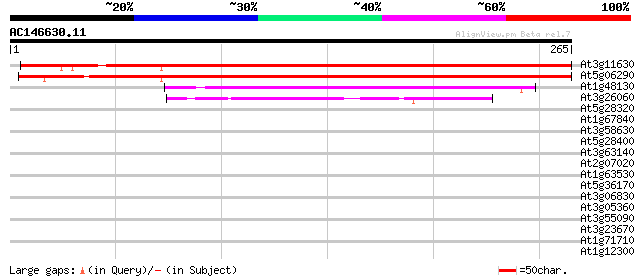
Score E
Sequences producing significant alignments: (bits) Value
At3g11630 putative 2-cys peroxiredoxin 400 e-112
At5g06290 2-cys peroxiredoxin-like protein 396 e-111
At1g48130 1-Cys peroxiredoxin (AtPER1) 94 1e-19
At3g26060 putative peroxiredoxin 74 6e-14
At5g28320 putative protein 32 0.27
At1g67840 unknown protein 32 0.27
At3g58630 unknown protein 30 1.0
At5g28400 putative protein 30 1.3
At3g63140 mRNA binding protein precursor - like 30 1.7
At2g07020 putative protein kinase 30 1.7
At1g63530 unknown protein 29 2.3
At5g36170 translation releasing factor RF-2 29 3.0
At3g06830 pectin methylesterase like protein 28 3.9
At3g05360 putative disease resistance protein 28 3.9
At3g55090 ABC transporter - like protein 28 5.1
At3g23670 hypothetical protein 27 8.7
At1g71710 RIBOSOMAL PROTEIN, putative (At1g71710) 27 8.7
At1g12300 hypothetical protein 27 8.7
>At3g11630 putative 2-cys peroxiredoxin
Length = 266
Score = 400 bits (1027), Expect = e-112
Identities = 210/265 (79%), Positives = 231/265 (86%), Gaps = 8/265 (3%)
Query: 6 APSASLLSSNPNILFSPK--LSSPP--HLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRR 61
A S +L+SS + +F K LSSP L +LS P+AS SL R+ + ++SRR
Sbjct: 5 ASSTTLISSPSSRVFPAKSSLSSPSVSFLRTLSSPSASASL---RSGFARRSSLSSTSRR 61
Query: 62 SFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT 120
SF V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLS+YIGKKYVILFFYPLDFTFVCPT
Sbjct: 62 SFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSDYIGKKYVILFFYPLDFTFVCPT 121
Query: 121 EITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISK 180
EITAFSDRH+EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPL+SDVTKSISK
Sbjct: 122 EITAFSDRHSEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLISDVTKSISK 181
Query: 181 SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC 240
S+GVLI DQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQY+QENPDEVC
Sbjct: 182 SFGVLIHDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETMRTLQALQYIQENPDEVC 241
Query: 241 PAGWKPGEKSMKPDPKLSKEYFSAV 265
PAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 242 PAGWKPGEKSMKPDPKLSKEYFSAI 266
>At5g06290 2-cys peroxiredoxin-like protein
Length = 273
Score = 396 bits (1018), Expect = e-111
Identities = 209/268 (77%), Positives = 230/268 (84%), Gaps = 9/268 (3%)
Query: 5 SAPSASLLSSN------PNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTS 58
S+ S +LLSS+ + L SP +S P + S S +S+SL +SL ++
Sbjct: 8 SSSSTTLLSSSRVLLPSKSSLLSPTVSFPRIIPSSSA--SSSSLCSGFSSLGSLTTNRSA 65
Query: 59 SRRSFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 117
SRR+F V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV
Sbjct: 66 SRRNFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 125
Query: 118 CPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKS 177
CPTEITAFSDR+ EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSD+TKS
Sbjct: 126 CPTEITAFSDRYEEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDITKS 185
Query: 178 ISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPD 237
ISKS+GVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQYVQENPD
Sbjct: 186 ISKSFGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETMRTLQALQYVQENPD 245
Query: 238 EVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
EVCPAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 246 EVCPAGWKPGEKSMKPDPKLSKEYFSAI 273
>At1g48130 1-Cys peroxiredoxin (AtPER1)
Length = 216
Score = 93.6 bits (231), Expect = 1e-19
Identities = 53/176 (30%), Positives = 88/176 (49%), Gaps = 5/176 (2%)
Query: 74 VGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFA 133
+G+ P+ E E D K KL +Y + +LF +P DFT VC TE+ A + EF
Sbjct: 6 LGDTVPNLEVETTHD----KFKLHDYFANSWTVLFSHPGDFTPVCTTELGAMAKYAHEFD 61
Query: 134 ELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQGIAL 193
+ ++LG+S D V SH W++ +NYP+++D K I ++ P +
Sbjct: 62 KRGVKLLGLSCDDVQSHKDWIKDIEAFNHGSKVNYPIIADPNKEIIPQLNMIDPIENGPS 121
Query: 194 RGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC-PAGWKPGE 248
R L I+ + I+ S + GR++DE R L +L ++ +++ P WKP +
Sbjct: 122 RALHIVGPDSKIKLSFLYPSTTGRNMDEVLRALDSLLMASKHNNKIATPVNWKPDQ 177
>At3g26060 putative peroxiredoxin
Length = 216
Score = 74.3 bits (181), Expect = 6e-14
Identities = 53/157 (33%), Positives = 78/157 (48%), Gaps = 16/157 (10%)
Query: 75 GNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFAE 134
G AAPDF + DQ V L +Y GK V+L+FYP D T C + AF D + +F +
Sbjct: 72 GQAAPDFTLK---DQNGKPVSLKKYKGKP-VVLYFYPADETPGCTKQACAFRDSYEKFKK 127
Query: 135 LNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQ---GI 191
E++G+S D SH A+ + L Y L+SD + K +GV P +
Sbjct: 128 AGAEVIGISGDDSASHKAFASKYK-------LPYTLLSDEGNKVRKDWGV--PGDLFGAL 178
Query: 192 ALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQA 228
R +++DK GV+Q N + +DET + L+A
Sbjct: 179 PGRQTYVLDKNGVVQLIYNNQFQPEKHIDETLKFLKA 215
>At5g28320 putative protein
Length = 967
Score = 32.3 bits (72), Expect = 0.27
Identities = 31/125 (24%), Positives = 50/125 (39%), Gaps = 12/125 (9%)
Query: 129 HAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGV--LI 186
H+E + E LG S DS + L+ +G + D +P K V L+
Sbjct: 372 HSEGDDDKDERLGTSTDSENTELSAFAVPMLNGAMVDSGFPNHEMAASDKKKVSNVVPLV 431
Query: 187 PDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTL----------QALQYVQENP 236
P G+ D+ ++++ST + RSV E K L Q L + ++
Sbjct: 432 PTDGVIQASDVTKDQLSMMKNSTGRKSRVIRSVKEAKEFLSRRSGEKELTQELSQMAQDS 491
Query: 237 DEVCP 241
DE+ P
Sbjct: 492 DEIFP 496
>At1g67840 unknown protein
Length = 611
Score = 32.3 bits (72), Expect = 0.27
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query: 1 MACCSAPSASLLSSNPNILFSPKLSSP-PHLSSLSIPNASNS 41
M + S +LLSSNPN+ FS + +P P SL + NAS+S
Sbjct: 1 MLLSAIASQTLLSSNPNLHFSNSIPNPRPSNPSLKLLNASSS 42
>At3g58630 unknown protein
Length = 321
Score = 30.4 bits (67), Expect = 1.0
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 7/73 (9%)
Query: 9 ASLLSSNPNILFSPK--LSSPPHLSSLSIPNASN-----SLPKLRTSLPLSLNRFTSSRR 61
A + SNP SP S+ L S P +SN ++P R SLP+S+N + R
Sbjct: 103 ARVSESNPGAYISPWPFFSALDDLLRESFPTSSNPDSTDNIPHQRLSLPMSINPVPVAPR 162
Query: 62 SFVVRASSELPLV 74
S + R + P +
Sbjct: 163 SAIPRRPATSPAI 175
>At5g28400 putative protein
Length = 996
Score = 30.0 bits (66), Expect = 1.3
Identities = 29/116 (25%), Positives = 46/116 (39%), Gaps = 12/116 (10%)
Query: 138 EILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGV--LIPDQGIALRG 195
E LG S DS + L+ +G + D +P K V L+P G+
Sbjct: 392 ERLGTSTDSENTELSAFAVPMLNGAMVDSGFPNHEMAASDKKKVSNVVPLVPTDGVIQAS 451
Query: 196 LFIIDKEGVIQHSTINNLGIGRSVDETKRTL----------QALQYVQENPDEVCP 241
D+ ++++ST + RSV E K L Q L + ++ DE+ P
Sbjct: 452 DVTKDQLSMMKNSTGRKSRVIRSVKEAKEFLSRRSGEKELTQELSQMAQDSDEIFP 507
>At3g63140 mRNA binding protein precursor - like
Length = 406
Score = 29.6 bits (65), Expect = 1.7
Identities = 23/71 (32%), Positives = 35/71 (48%), Gaps = 15/71 (21%)
Query: 14 SNPNILFSPKLSSP-------PHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRS---- 62
S+ ++ FS K +SP P L S+P++S+S L +S S + T S R+
Sbjct: 5 SSSSLFFSSKTTSPISNLLIPPSLHRFSLPSSSSSFSSLSSSSSSSSSLLTFSLRTSRRL 64
Query: 63 ----FVVRASS 69
F V+ASS
Sbjct: 65 SPQKFTVKASS 75
>At2g07020 putative protein kinase
Length = 620
Score = 29.6 bits (65), Expect = 1.7
Identities = 41/170 (24%), Positives = 72/170 (42%), Gaps = 20/170 (11%)
Query: 34 SIPNASNSLPKLRTSLP-LSLNRFTSSRRSFVVRASSELPLVGNAAPDFEAEAVFDQEFI 92
S+ +A++S P L T P L ++ +F RA L V + Q+ I
Sbjct: 162 SVRSATSSPPPLCTIRPQLPARSSNANNNNFSPRAQRRLQSVQSI-----------QDEI 210
Query: 93 KVKLSEYIGKKYVILFFYPL----DFTFVCPTEITA--FSDRHAEFAELNTEILGVSVDS 146
++K S Y+ K+Y + L D +FV + F D + +A ++ S+D
Sbjct: 211 EIK-SPYLRKEYDQGTYQALVSDSDLSFVSSDRPSMDWFEDNRSNYATSSSSSEKQSIDL 269
Query: 147 VFSHLAWVQTDRKSGGLGDLN-YPLVSDVTKSISKSYGVLIPDQGIALRG 195
S+ A+ + ++SG L L+ Y + T S S+ + G +G
Sbjct: 270 CSSYSAFSTSSQESGRLSSLSMYSQIEQGTTKFSDSHKIGEGSYGTVYKG 319
>At1g63530 unknown protein
Length = 499
Score = 29.3 bits (64), Expect = 2.3
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 5/63 (7%)
Query: 6 APSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVV 65
A AS + S PN +FSP S +S S+P+ RT+ +L FT F
Sbjct: 313 AAHASPIGSKPNAVFSPSTVSTSLFASPSVPDHHQ-----RTNFSTTLGNFTGPAFGFFP 367
Query: 66 RAS 68
A+
Sbjct: 368 AAA 370
>At5g36170 translation releasing factor RF-2
Length = 456
Score = 28.9 bits (63), Expect = 3.0
Identities = 18/32 (56%), Positives = 22/32 (68%), Gaps = 2/32 (6%)
Query: 41 SLPKLRTSLPLSLNRFTSSRR--SFVVRASSE 70
+LP LR SLPLSL F+SS R S +V A+ E
Sbjct: 31 NLPLLRLSLPLSLPNFSSSSRFNSPIVFAAQE 62
>At3g06830 pectin methylesterase like protein
Length = 568
Score = 28.5 bits (62), Expect = 3.9
Identities = 17/57 (29%), Positives = 24/57 (41%), Gaps = 3/57 (5%)
Query: 190 GIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWKP 246
G+ L G I I ++N +GR E RT+ + D + PAGW P
Sbjct: 448 GLVLHGCHITGDPAYIPMKSVNKAYLGRPWKEFSRTIIMKTTID---DVIDPAGWLP 501
>At3g05360 putative disease resistance protein
Length = 786
Score = 28.5 bits (62), Expect = 3.9
Identities = 22/84 (26%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Query: 34 SIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSELPLVGNAAPDFEAEAVFDQ---- 89
+IP + +L KL +SL +S N+FT SF++ + L + A+ F++ D
Sbjct: 174 NIPTSFTNLTKL-SSLDISSNQFTLENFSFILPNLTSLSSLNVASNHFKSTLPSDMSGLH 232
Query: 90 --EFIKVKLSEYIGKKYVILFFYP 111
++ V+ + ++G LF P
Sbjct: 233 NLKYFDVRENSFVGTFPTSLFTIP 256
>At3g55090 ABC transporter - like protein
Length = 720
Score = 28.1 bits (61), Expect = 5.1
Identities = 16/51 (31%), Positives = 26/51 (50%)
Query: 163 LGDLNYPLVSDVTKSISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNL 213
LG+L Y + + S+S+S G+ I G ++ ++GV Q S N L
Sbjct: 643 LGELTYGMKLRLLDSVSRSIGMRISSSTCLTTGADVLKQQGVTQLSKWNCL 693
>At3g23670 hypothetical protein
Length = 1263
Score = 27.3 bits (59), Expect = 8.7
Identities = 19/64 (29%), Positives = 32/64 (49%), Gaps = 1/64 (1%)
Query: 7 PSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVR 66
PS + S I S + + PP L+SL IP+ +S KL++ LP +R +
Sbjct: 23 PSLTKSKSQRKIKSSKENAPPPDLNSL-IPDHRSSPAKLKSPLPPRPPSSNPLKRKLIAE 81
Query: 67 ASSE 70
A+++
Sbjct: 82 ATAD 85
>At1g71710 RIBOSOMAL PROTEIN, putative (At1g71710)
Length = 664
Score = 27.3 bits (59), Expect = 8.7
Identities = 24/94 (25%), Positives = 42/94 (44%), Gaps = 9/94 (9%)
Query: 92 IKVKLSEYIGKKYVILFFYPLDFTFVC--PTEITAFSDRHAEFAELNTEILGVSVDSVFS 149
+ V + YIG K + ++ TF C T +TA +R + + N ++ + +VF
Sbjct: 441 VGVGVMGYIGNKGAVSVSMSINQTFFCFINTHLTA-GEREVDQIKRNADVHEIHKRTVFH 499
Query: 150 HLA------WVQTDRKSGGLGDLNYPLVSDVTKS 177
++ + + LGDLNY L S K+
Sbjct: 500 SVSALGLPKLIYDHERIIWLGDLNYRLSSSYEKT 533
>At1g12300 hypothetical protein
Length = 637
Score = 27.3 bits (59), Expect = 8.7
Identities = 16/57 (28%), Positives = 30/57 (52%), Gaps = 2/57 (3%)
Query: 90 EFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFAE-LNTEILGVSVD 145
+F++ +L E G + L P + +F C +AFSDR+ + E L + ++ + D
Sbjct: 15 KFVQPRLLE-TGTLRIALINCPNELSFCCERGFSAFSDRNLSYRERLRSGLVDIKAD 70
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,885,569
Number of Sequences: 26719
Number of extensions: 242846
Number of successful extensions: 830
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 821
Number of HSP's gapped (non-prelim): 18
length of query: 265
length of database: 11,318,596
effective HSP length: 97
effective length of query: 168
effective length of database: 8,726,853
effective search space: 1466111304
effective search space used: 1466111304
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146630.11


