

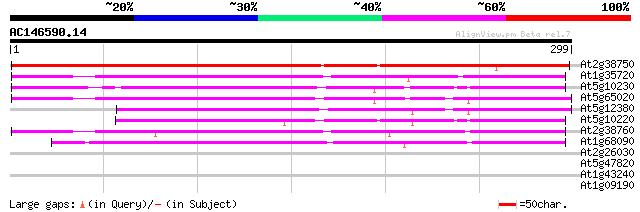
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.14 - phase: 1 /pseudo
(299 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g38750 putative annexin 337 6e-93
At1g35720 Ca2+-dependent membrane-binding protein annexin 177 5e-45
At5g10230 annexin -like protein 167 6e-42
At5g65020 annexin 161 5e-40
At5g12380 annexin - like protein 160 6e-40
At5g10220 annexin -like protein 157 9e-39
At2g38760 putative annexin 156 1e-38
At1g68090 calcium-binding protein annexin 5 (ANN5) 117 8e-27
At2g26030 unknown protein 30 1.2
At5g47820 kinesin-like protein 28 4.6
At1g43240 hypothetical protein 28 4.6
At1g09190 hypothetical protein 28 7.8
>At2g38750 putative annexin
Length = 319
Score = 337 bits (863), Expect = 6e-93
Identities = 173/299 (57%), Positives = 222/299 (73%), Gaps = 4/299 (1%)
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAV 61
GVDE +LI+ LGK R+ +RK + FF+ED ER F++ +DH VR LK EF RF AV
Sbjct: 21 GVDENALISTLGKSQKEHRKLFRKASKSFFVEDEERAFEKCHDHFVRHLKLEFSRFNTAV 80
Query: 62 VLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDV 121
V+W+MHPWERDARL K+ALKKG +Y +++E++CTRS+E+LLGARKAYHSLFD S+EED+
Sbjct: 81 VMWAMHPWERDARLVKKALKKGEEAYNLIVEVSCTRSAEDLLGARKAYHSLFDQSMEEDI 140
Query: 122 ASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRIL 181
ASH+HG RKLLV LVSAYRYEG KVKDD+AKS+AK L+ A+ ++ + VE DEV+RIL
Sbjct: 141 ASHVHGPQRKLLVGLVSAYRYEGNKVKDDSAKSDAKILAEAVASS-GEEAVEKDEVVRIL 199
Query: 182 ATRSKLXLQAVYTAL*RDFWEES*RGIINK*TLFKETVQCLCTPQVYFSKVLDAALKNDV 241
TRSKL LQ +Y + G ++K +L E + CL P +YFSK+LDA+L D
Sbjct: 200 TTRSKLHLQHLYKHF-NEIKGSDLLGGVSKSSLLNEALICLLKPALYFSKILDASLNKDA 258
Query: 242 NKNIKKSLTRVIVTRAD--IDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTLIAR 298
+K KK LTRV VTRAD +M EIK EYNNLYG +L Q+I+E KGNY+DFLLTL+++
Sbjct: 259 DKTTKKWLTRVFVTRADHSDEMNEIKEEYNNLYGETLAQRIQEKIKGNYRDFLLTLLSK 317
>At1g35720 Ca2+-dependent membrane-binding protein annexin
Length = 317
Score = 177 bits (450), Expect = 5e-45
Identities = 111/302 (36%), Positives = 170/302 (55%), Gaps = 24/302 (7%)
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVR-FKNA 60
G +E +I++L +R+ R+ + ED ++ L E F+ A
Sbjct: 28 GTNEDLIISILAHRSAEQRKVIRQAYHETYGED-----------LLKTLDKELSNDFERA 76
Query: 61 VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED 120
++LW++ P ERDA LA EA K+ + S VL+E+ACTR+S +LL AR+AYH+ + S+EED
Sbjct: 77 ILLWTLEPGERDALLANEATKRWTSSNQVLMEVACTRTSTQLLHARQAYHARYKKSLEED 136
Query: 121 VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI 180
VA H G+ RKLLV+LV++YRYEG +V AK EAK + IK+ D++VIRI
Sbjct: 137 VAHHTTGDFRKLLVSLVTSYRYEGDEVNMTLAKQEAKLVHEKIKDKH----YNDEDVIRI 192
Query: 181 LATRSKLXLQAVYTAL*RDFWEES*RGIINK------*TLFKETVQCLCTPQVYFSKVLD 234
L+TRSK + A + D EE + + L + T+QCL P++YF VL
Sbjct: 193 LSTRSKAQINATFNRYQDDHGEEILKSLEEGDDDDKFLALLRSTIQCLTRPELYFVDVLR 252
Query: 235 AALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLT 294
+A+ + +LTR++ TRA+ID+K I EY + L + I + +G+Y+ L+
Sbjct: 253 SAINK--TGTDEGALTRIVTTRAEIDLKVIGEEYQRRNSIPLEKAITKDTRGDYEKMLVA 310
Query: 295 LI 296
L+
Sbjct: 311 LL 312
>At5g10230 annexin -like protein
Length = 316
Score = 167 bits (423), Expect = 6e-42
Identities = 116/303 (38%), Positives = 169/303 (55%), Gaps = 27/303 (8%)
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVRFKNAV 61
G +E+ +I++L + +R R + + +D ++ R L +F R AV
Sbjct: 28 GTNERMIISILAHRNATQRSFIRAVYAANYNKDLLKELDRE-------LSGDFER---AV 77
Query: 62 VLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDV 121
+LW+ P ERDA LAKE+ K + + VL+EIACTRS+ EL A++AY + + S+EEDV
Sbjct: 78 MLWTFEPAERDAYLAKESTKMFTKNNWVLVEIACTRSALELFNAKQAYQARYKTSLEEDV 137
Query: 122 ASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRIL 181
A H G+ RKLLV LVS +RY+G +V A+SEAK L IK + DD++IRIL
Sbjct: 138 AYHTSGDIRKLLVPLVSTFRYDGDEVNMTLARSEAKILHEKIK----EKAYADDDLIRIL 193
Query: 182 ATRSKLXLQAVY--------TAL*RDFWEES*RGIINK*TLFKETVQCLCTPQVYFSKVL 233
TRSK + A T++ + E+S I L K ++CL P+ YF KVL
Sbjct: 194 TTRSKAQISATLNHYKNNFGTSMSKYLKEDSENEYIQ---LLKAVIKCLTYPEKYFEKVL 250
Query: 234 DAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLL 293
A+ N + + + LTRV+ TRA+ DM+ IK EY V L + I + G+Y+D LL
Sbjct: 251 RQAI-NKLGTD-EWGLTRVVTTRAEFDMERIKEEYIRRNSVPLDRAIAKDTHGDYEDILL 308
Query: 294 TLI 296
L+
Sbjct: 309 ALL 311
>At5g65020 annexin
Length = 317
Score = 161 bits (407), Expect = 5e-40
Identities = 112/309 (36%), Positives = 165/309 (53%), Gaps = 32/309 (10%)
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVR-FKNA 60
G +EK +I++L + +R R + + ED ++ L E F+ A
Sbjct: 28 GTNEKLIISILAHRNAAQRSLIRSVYAATYNED-----------LLKALDKELSSDFERA 76
Query: 61 VVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEED 120
V+LW++ P ERDA LAKE+ K + + VL+EIACTR + EL+ ++AY + + SIEED
Sbjct: 77 VMLWTLDPPERDAYLAKESTKMFTKNNWVLVEIACTRPALELIKVKQAYQARYKKSIEED 136
Query: 121 VASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRI 180
VA H G+ RKLL+ LVS +RYEG V A+SEAK L + ++ DD+ IRI
Sbjct: 137 VAQHTSGDLRKLLLPLVSTFRYEGDDVNMMLARSEAKILHEKV----SEKSYSDDDFIRI 192
Query: 181 LATRSKLXLQAVY--------TAL*RDFWEES*RGIINK*TLFKETVQCLCTPQVYFSKV 232
L TRSK L A A+ ++ EES K L + + CL P+ +F KV
Sbjct: 193 LTTRSKAQLGATLNHYNNEYGNAINKNLKEESDDNDYMK--LLRAVITCLTYPEKHFEKV 250
Query: 233 LDAALKNDVNK--NIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKD 290
L+ +NK + LTRV+ TR ++DM+ IK EY + L + I + G+Y+D
Sbjct: 251 ----LRLSINKMGTDEWGLTRVVTTRTEVDMERIKEEYQRRNSIPLDRAIAKDTSGDYED 306
Query: 291 FLLTLIARG 299
L+ L+ G
Sbjct: 307 MLVALLGHG 315
>At5g12380 annexin - like protein
Length = 257
Score = 160 bits (406), Expect = 6e-40
Identities = 98/249 (39%), Positives = 145/249 (57%), Gaps = 15/249 (6%)
Query: 58 KNAVVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSI 117
+ A+ LW + P ERDA LA AL+K Y VL+EIAC RS E++L AR+AY L+ HS+
Sbjct: 14 QRAICLWVLDPPERDALLANLALQKPIPDYKVLVEIACMRSPEDMLAARRAYRCLYKHSL 73
Query: 118 EEDVASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEV 177
EED+AS G+ R+LLVA+VSAY+Y+G ++ + A+SEA L + I V+ +E
Sbjct: 74 EEDLASRTIGDIRRLLVAMVSAYKYDGEEIDEMLAQSEAAILHDEILG----KAVDHEET 129
Query: 178 IRILATRSKLXLQAVYTAL*RDFWEES*RGIINK*T-----LFKETVQCLCTPQVYFSKV 232
IR+L+TRS + L A++ + + ++N T + ++C+ P Y++KV
Sbjct: 130 IRVLSTRSSMQLSAIFNRYKDIYGTSITKDLLNHPTNEYLSALRAAIRCIKNPTRYYAKV 189
Query: 233 LDAALKNDVNK--NIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKD 290
L+N +N + +L RVIVTRA+ D+ I Y VSL Q I + G+YK
Sbjct: 190 ----LRNSINTVGTDEDALNRVIVTRAEKDLTNITGLYFKRNNVSLDQAIAKETSGDYKA 245
Query: 291 FLLTLIARG 299
FLL L+ G
Sbjct: 246 FLLALLGHG 254
>At5g10220 annexin -like protein
Length = 318
Score = 157 bits (396), Expect = 9e-39
Identities = 101/248 (40%), Positives = 143/248 (56%), Gaps = 15/248 (6%)
Query: 57 FKNAVVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHS 116
F+ V+LW++ P ERDA LA E+ K + + VL+EIACTR S E ++AYH + S
Sbjct: 73 FERVVMLWTLDPTERDAYLANESTKLFTKNIWVLVEIACTRPSLEFFKTKQAYHVRYKTS 132
Query: 117 IEEDVASHIHGNDRKLLVALVSAYRYEGT--KVKDDTAKSEAKTLSNAIKNAQNKPIVED 174
+EEDVA H GN RKLLV LVS +RY+G +V A+SEAKTL I + D
Sbjct: 133 LEEDVAYHTSGNIRKLLVPLVSTFRYDGNADEVNVKLARSEAKTLHKKI----TEKAYTD 188
Query: 175 DEVIRILATRSKLXLQAVYTAL*RDFWEES*RGIINK*T------LFKETVQCLCTPQVY 228
+++IRIL TRSK + A +D + S + + + L K ++CL P+ Y
Sbjct: 189 EDLIRILTTRSKAQINATLNHF-KDKFGSSINKFLKEDSNDDYVQLLKTAIKCLTYPEKY 247
Query: 229 FSKVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNY 288
F KVL A+ N + + + +LTRV+ TRA++D++ IK EY V L + I G+Y
Sbjct: 248 FEKVLRRAI-NRMGTD-EWALTRVVTTRAEVDLERIKEEYLRRNSVPLDRAIANDTSGDY 305
Query: 289 KDFLLTLI 296
KD LL L+
Sbjct: 306 KDMLLALL 313
>At2g38760 putative annexin
Length = 321
Score = 156 bits (394), Expect = 1e-38
Identities = 107/307 (34%), Positives = 162/307 (51%), Gaps = 29/307 (9%)
Query: 2 GVDEKSLIAVLGKWDPLERETYRKKTSHFFIEDHERQFQRWNDHCVRLLKHEFVR-FKNA 60
G DEK++I VLG+ D +R R+ + +D + +L E F A
Sbjct: 28 GTDEKAIIRVLGQRDQSQRRKIRESFREIYGKD-----------LIDVLSSELSGDFMKA 76
Query: 61 VVLWSMHPWERDARLA-----KEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDH 115
VV W+ P ERDARL KE KK + V++EI+CT S L+ RKAY SLFD
Sbjct: 77 VVSWTYDPAERDARLVNKILNKEKKKKSLENLKVIVEISCTTSPNHLIAVRKAYCSLFDS 136
Query: 116 SIEEDVASHIHGNDRKLLVALVSAYRYEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDD 175
S+EE +AS + KLLV L S +RY+ + + A EA L AI+ Q ++ D
Sbjct: 137 SLEEHIASSLPFPLAKLLVTLASTFRYDKDRTDAEVATIEAAMLREAIEKKQ----LDHD 192
Query: 176 EVIRILATRSKLXLQAVYTAL*RDFW------EES*RGIINK*TLFKETVQCLCTPQVYF 229
V+ IL TRS L+ + A +++ + G + +L K + C+ TP+ +F
Sbjct: 193 HVLYILGTRSIYQLRETFVAYKKNYGVTIDKDVDGCPGDADLRSLLKVAIFCIDTPEKHF 252
Query: 230 SKVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYK 289
+KV+ +++ + SLTR IVTRA+ID+ +++ EY N+Y S+ I G+YK
Sbjct: 253 AKVVRDSIEGFGTD--EDSLTRAIVTRAEIDLMKVRGEYFNMYNTSMDNAITGDISGDYK 310
Query: 290 DFLLTLI 296
DF++TL+
Sbjct: 311 DFIITLL 317
>At1g68090 calcium-binding protein annexin 5 (ANN5)
Length = 316
Score = 117 bits (293), Expect = 8e-27
Identities = 81/280 (28%), Positives = 140/280 (49%), Gaps = 13/280 (4%)
Query: 23 YRKKTSHFFIE-DHERQFQRWNDHCVRLLKHEFVRFKNAVVLWSMHPWERDARLAKEALK 81
+R T IE ++E +F +D RL K AV+LW ERDA + K +L+
Sbjct: 40 HRNATQRALIEQEYETKFS--DDLRKRLHSELHGHLKKAVLLWMPEAVERDASILKRSLR 97
Query: 82 KGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVASHIHGNDRKLLVALVSAYR 141
+ + EI CTRS +L ++ Y + F +EED+ S GN +++L+A ++ R
Sbjct: 98 GAVTDHKAIAEIICTRSGSQLRQIKQVYSNTFGVKLEEDIESEASGNHKRVLLAYLNTTR 157
Query: 142 YEGTKVKDDTAKSEAKTLSNAIKNAQNKPIVEDDEVIRILATRSKLXLQAVYTAL*RDFW 201
YEG ++ + + +++A+TL +A+ +D +I+I RS+ L AV + +
Sbjct: 158 YEGPEIDNASVENDARTLKSAVARKHKS---DDQTLIQIFTDRSRTHLVAVRSTYRSMYG 214
Query: 202 EES*RGII-----NK*TLFKETVQCLCTPQVYFSKVLDAALKNDVNKNIKKSLTRVIVTR 256
+E + I N + +QC YF+K L ++K + +L R++VTR
Sbjct: 215 KELGKAIRDETRGNFEHVLLTILQCAENSCFYFAKALRKSMKGLGTDD--TALIRIVVTR 272
Query: 257 ADIDMKEIKAEYNNLYGVSLPQKIEETAKGNYKDFLLTLI 296
A++DM+ I EY Y +L + +Y+ FLL+L+
Sbjct: 273 AEVDMQFIITEYRKRYKKTLYNAVHSDTTSHYRTFLLSLL 312
>At2g26030 unknown protein
Length = 442
Score = 30.4 bits (67), Expect = 1.2
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 5/81 (6%)
Query: 215 FKETVQCLCTPQVYFSKVLDAALKNDVNKNIKKSLTRVIVTRADIDMKEIKAEYNNLYGV 274
F+ +V C YFS +DA +N N +S T V+ + M +I +E+N +G
Sbjct: 214 FEYSVSC-----TYFSVEIDAPRLEYLNFNDDQSDTIVVKNMTSLSMIDIDSEFNVKFGG 268
Query: 275 SLPQKIEETAKGNYKDFLLTL 295
S + + + +DFL +
Sbjct: 269 SRLEPGDLRKRDIIRDFLTAI 289
>At5g47820 kinesin-like protein
Length = 1035
Score = 28.5 bits (62), Expect = 4.6
Identities = 26/100 (26%), Positives = 43/100 (43%), Gaps = 14/100 (14%)
Query: 80 LKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVASHIHGNDRKLLVALVSA 139
+ KG ++ G +I A + GA Y D + + + GN R +++A +S
Sbjct: 294 INKGLLALGNVIS-ALGDEKKRKDGAHVPYR---DSKLTRLLQDSLGGNSRTVMIACISP 349
Query: 140 YRYEGTKVKDDTAKSEAKTL--SNAIKNAQNKPIVEDDEV 177
D A+ TL +N +N +NKP+V D V
Sbjct: 350 --------ADINAEETLNTLKYANRARNIRNKPVVNRDPV 381
>At1g43240 hypothetical protein
Length = 698
Score = 28.5 bits (62), Expect = 4.6
Identities = 13/47 (27%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query: 51 KHEFVRFKNAVVLWSMHPWERDARLAKEALKKGSISYGVLIEIACTR 97
KH F++F N H W + L+ L+ ++Y +L+E C +
Sbjct: 4 KHTFIQFDNGYYS-EEHEWISNNSLSGVLLRTSKVTYSMLVEKICKK 49
>At1g09190 hypothetical protein
Length = 999
Score = 27.7 bits (60), Expect = 7.8
Identities = 18/74 (24%), Positives = 32/74 (42%), Gaps = 5/74 (6%)
Query: 73 ARLAKEALKKGSISYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVASHIHGNDRKL 132
A++AK L + Y +E AC + ++ +K YH H I+ H+H +
Sbjct: 361 AKIAKVILDDKDLEY---LEAACFLHNIGIITGKKGYHKQSYHIIKN--GDHLHSYTAEE 415
Query: 133 LVALVSAYRYEGTK 146
+ + RY+ K
Sbjct: 416 IELIAMLVRYQRKK 429
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,183,721
Number of Sequences: 26719
Number of extensions: 239932
Number of successful extensions: 866
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 817
Number of HSP's gapped (non-prelim): 14
length of query: 299
length of database: 11,318,596
effective HSP length: 99
effective length of query: 200
effective length of database: 8,673,415
effective search space: 1734683000
effective search space used: 1734683000
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146590.14


