

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.13 + phase: 0
(314 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
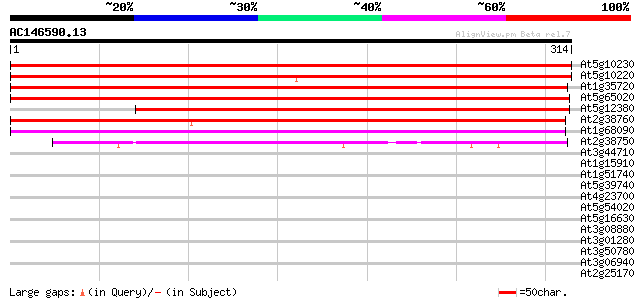
Score E
Sequences producing significant alignments: (bits) Value
At5g10230 annexin -like protein 297 4e-81
At5g10220 annexin -like protein 297 6e-81
At1g35720 Ca2+-dependent membrane-binding protein annexin 296 1e-80
At5g65020 annexin 291 4e-79
At5g12380 annexin - like protein 254 4e-68
At2g38760 putative annexin 234 6e-62
At1g68090 calcium-binding protein annexin 5 (ANN5) 194 5e-50
At2g38750 putative annexin 157 9e-39
At3g44710 unknown protein 29 2.9
At1g15910 29 2.9
At1g51740 unknown protein 29 3.8
At5g39740 ribosomal protein L5 - like 28 4.9
At4g23700 putative Na+/H+-exchanging protein 28 4.9
At5g54020 CHP-rich zinc finger protein-like 28 6.5
At5g16630 unknown protein 28 6.5
At3g08880 hypothetical protein 28 6.5
At3g01280 putative porin 28 6.5
At3g50780 unknown protein (At3g50780) 28 8.4
At3g06940 putative mudrA protein 28 8.4
At2g25170 putative chromodomain-helicase-DNA-binding protein 28 8.4
>At5g10230 annexin -like protein
Length = 316
Score = 297 bits (761), Expect = 4e-81
Identities = 159/315 (50%), Positives = 213/315 (67%), Gaps = 1/315 (0%)
Query: 1 MATLIAPMNHS-PKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P P+EDA+ L+KA KGWGT+E II+I+ RNA QR IR Y Y +D
Sbjct: 1 MASLKVPATVPLPEEDAEQLYKAFKGWGTNERMIISILAHRNATQRSFIRAVYAANYNKD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELSG+FE+A+ W +PA+R A LA + K K+ V+VEIA EL
Sbjct: 61 LLKELDRELSGDFERAVMLWTFEPAERDAYLAKESTKMFTKNNWVLVEIACTRSALELFN 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
+ AY RYK SLEEDVA HTSG R+LLV LVS+FRYDG E+N LA+ EA ILHE +K
Sbjct: 121 AKQAYQARYKTSLEEDVAYHTSGDIRKLLVPLVSTFRYDGDEVNMTLARSEAKILHEKIK 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
K +++IRIL TRSK Q+ AT N Y+++ G S+SK L ++ +++++ + I+C+
Sbjct: 181 EKAYADDDLIRILTTRSKAQISATLNHYKNNFGTSMSKYLKEDSENEYIQLLKAVIKCLT 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+KY+EKVLR A+ ++GTDE GLTRVV TRAE D++ IKE Y +RNSV L+ +AK+
Sbjct: 241 YPEKYFEKVLRQAINKLGTDEWGLTRVVTTRAEFDMERIKEEYIRRNSVPLDRAIAKDTH 300
Query: 300 GDYKKFLLTLLGKGH 314
GDY+ LL LLG H
Sbjct: 301 GDYEDILLALLGHDH 315
>At5g10220 annexin -like protein
Length = 318
Score = 297 bits (760), Expect = 6e-81
Identities = 163/317 (51%), Positives = 211/317 (66%), Gaps = 3/317 (0%)
Query: 1 MATLIAPMNHS-PKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P N P+ED++ L KA KGWGT+E II+I+ RNA QR IR Y Y +D
Sbjct: 1 MASLKIPANIPLPEEDSEQLHKAFKGWGTNEGMIISILAHRNATQRSFIRAVYAANYNKD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELSG+FE+ + W LDP +R A LAN + K K+ V+VEIA E
Sbjct: 61 LLKELDGELSGDFERVVMLWTLDPTERDAYLANESTKLFTKNIWVLVEIACTRPSLEFFK 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDG--VEINPILAKHEADILHEA 177
+ AYH RYK SLEEDVA HTSG R+LLV LVS+FRYDG E+N LA+ EA LH+
Sbjct: 121 TKQAYHVRYKTSLEEDVAYHTSGNIRKLLVPLVSTFRYDGNADEVNVKLARSEAKTLHKK 180
Query: 178 VKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRC 237
+ K E++IRIL TRSK Q+ AT N ++D G SI+K L +++DD+++ + AI+C
Sbjct: 181 ITEKAYTDEDLIRILTTRSKAQINATLNHFKDKFGSSINKFLKEDSNDDYVQLLKTAIKC 240
Query: 238 IDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKE 297
+ +KY+EKVLR A+ R+GTDE LTRVV TRAE DL+ IKE Y +RNSV L+ +A +
Sbjct: 241 LTYPEKYFEKVLRRAINRMGTDEWALTRVVTTRAEVDLERIKEEYLRRNSVPLDRAIAND 300
Query: 298 ISGDYKKFLLTLLGKGH 314
SGDYK LL LLG H
Sbjct: 301 TSGDYKDMLLALLGHDH 317
>At1g35720 Ca2+-dependent membrane-binding protein annexin
Length = 317
Score = 296 bits (757), Expect = 1e-80
Identities = 159/314 (50%), Positives = 211/314 (66%), Gaps = 2/314 (0%)
Query: 1 MATL-IAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL ++ +P +DA+ L A +GWGT+E II+I+ R+A QR+ IRQAY + Y ED
Sbjct: 1 MATLKVSDSVPAPSDDAEQLRTAFEGWGTNEDLIISILAHRSAEQRKVIRQAYHETYGED 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELS +FE+A+ W L+P +R A+LAN A K V++E+A +LL
Sbjct: 61 LLKTLDKELSNDFERAILLWTLEPGERDALLANEATKRWTSSNQVLMEVACTRTSTQLLH 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
R AYH RYK SLEEDVA HT+G R+LLV LV+S+RY+G E+N LAK EA ++HE +K
Sbjct: 121 ARQAYHARYKKSLEEDVAHHTTGDFRKLLVSLVTSYRYEGDEVNMTLAKQEAKLVHEKIK 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDD-FLKAVHVAIRCI 238
+K N E+VIRIL TRSK Q+ ATFNRY+DDHG I K L DD FL + I+C+
Sbjct: 181 DKHYNDEDVIRILSTRSKAQINATFNRYQDDHGEEILKSLEEGDDDDKFLALLRSTIQCL 240
Query: 239 DDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEI 298
+ Y+ VLR A+ + GTDE LTR+V TRAE DLK I E Y +RNS+ LE + K+
Sbjct: 241 TRPELYFVDVLRSAINKTGTDEGALTRIVTTRAEIDLKVIGEEYQRRNSIPLEKAITKDT 300
Query: 299 SGDYKKFLLTLLGK 312
GDY+K L+ LLG+
Sbjct: 301 RGDYEKMLVALLGE 314
>At5g65020 annexin
Length = 317
Score = 291 bits (744), Expect = 4e-79
Identities = 156/315 (49%), Positives = 212/315 (66%), Gaps = 2/315 (0%)
Query: 1 MATLIAPMNHS-PKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P N P++DA+ L KA GWGT+E II+I+ RNA QR IR Y Y ED
Sbjct: 1 MASLKVPSNVPLPEDDAEQLHKAFSGWGTNEKLIISILAHRNAAQRSLIRSVYAATYNED 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELS +FE+A+ W LDP +R A LA + K K+ V+VEIA EL+
Sbjct: 61 LLKALDKELSSDFERAVMLWTLDPPERDAYLAKESTKMFTKNNWVLVEIACTRPALELIK 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
V+ AY RYK S+EEDVA HTSG R+LL+ LVS+FRY+G ++N +LA+ EA ILHE V
Sbjct: 121 VKQAYQARYKKSIEEDVAQHTSGDLRKLLLPLVSTFRYEGDDVNMMLARSEAKILHEKVS 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASD-DFLKAVHVAIRCI 238
K + ++ IRIL TRSK QL AT N Y +++G +I+K L E+ D D++K + I C+
Sbjct: 181 EKSYSDDDFIRILTTRSKAQLGATLNHYNNEYGNAINKNLKEESDDNDYMKLLRAVITCL 240
Query: 239 DDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEI 298
+K++EKVLR ++ ++GTDE GLTRVV TR E D++ IKE Y +RNS+ L+ +AK+
Sbjct: 241 TYPEKHFEKVLRLSINKMGTDEWGLTRVVTTRTEVDMERIKEEYQRRNSIPLDRAIAKDT 300
Query: 299 SGDYKKFLLTLLGKG 313
SGDY+ L+ LLG G
Sbjct: 301 SGDYEDMLVALLGHG 315
>At5g12380 annexin - like protein
Length = 257
Score = 254 bits (649), Expect = 4e-68
Identities = 127/243 (52%), Positives = 177/243 (72%)
Query: 71 NFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKN 130
+ ++A+ W+LDP +R A+LAN+A++ DY V+VEIA + P+++LA R AY YK+
Sbjct: 12 SLQRAICLWVLDPPERDALLANLALQKPIPDYKVLVEIACMRSPEDMLAARRAYRCLYKH 71
Query: 131 SLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIR 190
SLEED+A+ T G R+LLV +VS+++YDG EI+ +LA+ EA ILH+ + K + EE IR
Sbjct: 72 SLEEDLASRTIGDIRRLLVAMVSAYKYDGEEIDEMLAQSEAAILHDEILGKAVDHEETIR 131
Query: 191 ILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDDHKKYYEKVLR 250
+L TRS QL A FNRY+D +G SI+K LLN ++++L A+ AIRCI + +YY KVLR
Sbjct: 132 VLSTRSSMQLSAIFNRYKDIYGTSITKDLLNHPTNEYLSALRAAIRCIKNPTRYYAKVLR 191
Query: 251 GALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISGDYKKFLLTLL 310
++ +GTDED L RV++TRAEKDL +I LY+KRN+V L+ +AKE SGDYK FLL LL
Sbjct: 192 NSINTVGTDEDALNRVIVTRAEKDLTNITGLYFKRNNVSLDQAIAKETSGDYKAFLLALL 251
Query: 311 GKG 313
G G
Sbjct: 252 GHG 254
>At2g38760 putative annexin
Length = 321
Score = 234 bits (596), Expect = 6e-62
Identities = 133/318 (41%), Positives = 201/318 (62%), Gaps = 7/318 (2%)
Query: 1 MATLIAPMN-HSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MAT+ P SP +D++ L +A++GWGTDE AII ++GQR+ QR++IR+++++IY +D
Sbjct: 1 MATIRVPNEVPSPAQDSETLKQAIRGWGTDEKAIIRVLGQRDQSQRRKIRESFREIYGKD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINK-----DYHVIVEIASVLQP 114
LI L SELSG+F KA+ W DPA+R A L N + K + VIVEI+ P
Sbjct: 61 LIDVLSSELSGDFMKAVVSWTYDPAERDARLVNKILNKEKKKKSLENLKVIVEISCTTSP 120
Query: 115 QELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADIL 174
L+AVR AY + + +SLEE +A+ +LLV L S+FRYD + +A EA +L
Sbjct: 121 NHLIAVRKAYCSLFDSSLEEHIASSLPFPLAKLLVTLASTFRYDKDRTDAEVATIEAAML 180
Query: 175 HEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASD-DFLKAVHV 233
EA++ K+ + + V+ IL TRS QL+ TF Y+ ++G +I K + D D + V
Sbjct: 181 REAIEKKQLDHDHVLYILGTRSIYQLRETFVAYKKNYGVTIDKDVDGCPGDADLRSLLKV 240
Query: 234 AIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDT 293
AI CID +K++ KV+R +++ GTDED LTR ++TRAE DL ++ Y+ + +++
Sbjct: 241 AIFCIDTPEKHFAKVVRDSIEGFGTDEDSLTRAIVTRAEIDLMKVRGEYFNMYNTSMDNA 300
Query: 294 VAKEISGDYKKFLLTLLG 311
+ +ISGDYK F++TLLG
Sbjct: 301 ITGDISGDYKDFIITLLG 318
>At1g68090 calcium-binding protein annexin 5 (ANN5)
Length = 316
Score = 194 bits (493), Expect = 5e-50
Identities = 112/313 (35%), Positives = 178/313 (56%), Gaps = 2/313 (0%)
Query: 1 MATLIAPMN-HSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MAT+ PM SP+ DAD L+KA KG G D S II I+ RNA QR I Q Y+ + +D
Sbjct: 1 MATMKIPMTVPSPRVDADQLFKAFKGRGCDTSVIINILAHRNATQRALIEQEYETKFSDD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L KRL SEL G+ +KA+ W+ + +R A + +++ D+ I EI +L
Sbjct: 61 LRKRLHSELHGHLKKAVLLWMPEAVERDASILKRSLRGAVTDHKAIAEIICTRSGSQLRQ 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
++ Y N + LEED+ + SG H+++L+ +++ RY+G EI+ +++A L AV
Sbjct: 121 IKQVYSNTFGVKLEEDIESEASGNHKRVLLAYLNTTRYEGPEIDNASVENDARTLKSAVA 180
Query: 180 NK-KGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCI 238
K K + + +I+I RS+T L A + YR +G + K + +E +F + ++C
Sbjct: 181 RKHKSDDQTLIQIFTDRSRTHLVAVRSTYRSMYGKELGKAIRDETRGNFEHVLLTILQCA 240
Query: 239 DDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEI 298
++ Y+ K LR ++K +GTD+ L R+V+TRAE D++ I Y KR L + V +
Sbjct: 241 ENSCFYFAKALRKSMKGLGTDDTALIRIVVTRAEVDMQFIITEYRKRYKKTLYNAVHSDT 300
Query: 299 SGDYKKFLLTLLG 311
+ Y+ FLL+LLG
Sbjct: 301 TSHYRTFLLSLLG 313
>At2g38750 putative annexin
Length = 319
Score = 157 bits (396), Expect = 9e-39
Identities = 104/306 (33%), Positives = 165/306 (52%), Gaps = 25/306 (8%)
Query: 25 GWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED-----------LIKRLESELSGNFE 73
G G DE+A+I+ +G+ R+ R+A + + ED ++ L+ E S F
Sbjct: 19 GMGVDENALISTLGKSQKEHRKLFRKASKSFFVEDEERAFEKCHDHFVRHLKLEFS-RFN 77
Query: 74 KAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLE 133
A+ W + P +R A L A+K + Y++IVE++ ++LL R AYH+ + S+E
Sbjct: 78 TAVVMWAMHPWERDARLVKKALKKGEEAYNLIVEVSCTRSAEDLLGARKAYHSLFDQSME 137
Query: 134 EDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNI---EEVIR 190
ED+A+H G R+LLVGLVS++RY+G ++ AK +A IL EAV + +EV+R
Sbjct: 138 EDIASHVHGPQRKLLVGLVSAYRYEGNKVKDDSAKSDAKILAEAVASSGEEAVEKDEVVR 197
Query: 191 ILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDDHKKYYEKVLR 250
IL TRSK L+ + + + G LL S L ++ A+ C+ Y+ K+L
Sbjct: 198 ILTTRSKLHLQHLYKHFNEIKG----SDLLGGVSKSSL--LNEALICLLKPALYFSKILD 251
Query: 251 GALKRIG--TDEDGLTRVVITRAE--KDLKDIKELYYKRNSVHLEDTVAKEISGDYKKFL 306
+L + T + LTRV +TRA+ ++ +IKE Y L + ++I G+Y+ FL
Sbjct: 252 ASLNKDADKTTKKWLTRVFVTRADHSDEMNEIKEEYNNLYGETLAQRIQEKIKGNYRDFL 311
Query: 307 LTLLGK 312
LTLL K
Sbjct: 312 LTLLSK 317
>At3g44710 unknown protein
Length = 504
Score = 29.3 bits (64), Expect = 2.9
Identities = 14/43 (32%), Positives = 26/43 (59%), Gaps = 1/43 (2%)
Query: 235 IRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKD 277
++ I++HK + K+ KR G D +GL + V + E+D++D
Sbjct: 82 LQMIEEHKLRFLKIFMDEAKRKGVDTNGLIKAV-SVLEEDIRD 123
>At1g15910
Length = 634
Score = 29.3 bits (64), Expect = 2.9
Identities = 51/238 (21%), Positives = 99/238 (41%), Gaps = 37/238 (15%)
Query: 62 KRLESELSGNFEKAMYRWI---------------LDPADRYAVLANVAIKSINKDYHVIV 106
K+ +E SG+ E Y W L + +++++ K++ V+
Sbjct: 198 KKEWTERSGDSESKAYGWCARADDFESQGPIGEYLSKEGQLRTVSDISQKNVQDRNTVLE 257
Query: 107 EIASVLQP--QELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINP 164
E++ ++ ++L V+++Y NR SL+ V H+ +F + ++
Sbjct: 258 ELSDMIAMTNEDLNKVQYSY-NRTAMSLQR-VLDEKKNLHQ--------AFADETKKMQQ 307
Query: 165 ILAKHEADILHEAVKNKKGN-IEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEA 223
+ +H IL++ K K N ++ +R L +R+K K D K+
Sbjct: 308 MSLRHIQKILYD--KEKLSNELDRKMRDLESRAKQLEKHEALTELDRQKLDEDKR----K 361
Query: 224 SDDFLKAVHVAIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKEL 281
SD K++ +A R + KK E VLR + ED L ++++ + D K E+
Sbjct: 362 SDAMNKSLQLASR---EQKKADESVLRLVEEHQRQKEDALNKILLLEKQLDTKQTLEM 416
>At1g51740 unknown protein
Length = 310
Score = 28.9 bits (63), Expect = 3.8
Identities = 19/69 (27%), Positives = 32/69 (45%), Gaps = 4/69 (5%)
Query: 161 EINPILAKHEADI--LHEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKK 218
E+ + KH D LH + +K +IE+ + I K Q+ N R++ + SK
Sbjct: 60 ELELFMLKHRKDYVDLHRTTEQEKDSIEQEVAAFIKACKEQIDILINSIRNEE--ANSKG 117
Query: 219 LLNEASDDF 227
L +D+F
Sbjct: 118 WLGLPADNF 126
>At5g39740 ribosomal protein L5 - like
Length = 301
Score = 28.5 bits (62), Expect = 4.9
Identities = 17/61 (27%), Positives = 30/61 (48%), Gaps = 7/61 (11%)
Query: 183 GNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDDHK 242
G++ +++L +L+ F+ Y I K + E+ ++ K VH AIR +HK
Sbjct: 201 GHVSNYMKLLGEDEPEKLQTHFSAY-------IKKGVEAESIEEMYKKVHAAIRAEPNHK 253
Query: 243 K 243
K
Sbjct: 254 K 254
>At4g23700 putative Na+/H+-exchanging protein
Length = 820
Score = 28.5 bits (62), Expect = 4.9
Identities = 27/133 (20%), Positives = 57/133 (42%), Gaps = 4/133 (3%)
Query: 94 AIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSL---EEDVAAHTSGYHRQLLVG 150
A + IN+ ++ V +++ E + H +N L +D + + S ++V
Sbjct: 474 ASRGINRKENLSVYAMHLMELSERSSAILMAHKVRRNGLPFWNKDKSENNSSSSDMVVVA 533
Query: 151 LVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDD 210
+ R V + P+ A +HE + + + IL +L T+ R+D
Sbjct: 534 FEAFRRLSRVSVRPMTAISPMATIHEDICQSAERKKTAMVILPFHKHVRLDRTWETTRND 593
Query: 211 HGFSISKKLLNEA 223
+ + I+KK++ E+
Sbjct: 594 YRW-INKKVMEES 605
>At5g54020 CHP-rich zinc finger protein-like
Length = 556
Score = 28.1 bits (61), Expect = 6.5
Identities = 18/60 (30%), Positives = 27/60 (45%), Gaps = 4/60 (6%)
Query: 173 ILHEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVH 232
ILHE K N+ +R L+ K L+ +F R+R F + +D F+ A H
Sbjct: 313 ILHE----KCANLPRKMRHLLHNHKLSLEVSFTRFRRSDNFFYTCIACQTHTDGFIYACH 368
>At5g16630 unknown protein
Length = 865
Score = 28.1 bits (61), Expect = 6.5
Identities = 25/99 (25%), Positives = 43/99 (43%), Gaps = 10/99 (10%)
Query: 47 QIRQAYQDIYQEDLIKRLESELSGNFEKAMYRW------ILDPA---DRYAVLAN-VAIK 96
+ + + Y E+ K+ E E N +A RW IL +RYA +N V K
Sbjct: 731 EFKDTILEAYAEEQEKKEEEERRRNEAQAASRWYQLLSSILTRERLKNRYANNSNDVEAK 790
Query: 97 SINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLEED 135
S+ + +V+ +V P++ + +R + S ED
Sbjct: 791 SLEVNSETVVKAKNVKAPEKQRVAKRGEKSRVRKSRNED 829
>At3g08880 hypothetical protein
Length = 107
Score = 28.1 bits (61), Expect = 6.5
Identities = 12/30 (40%), Positives = 19/30 (63%)
Query: 45 RQQIRQAYQDIYQEDLIKRLESELSGNFEK 74
+++ +AY DI ED I+RL+ EL E+
Sbjct: 69 KKKTEEAYSDIAAEDEIERLQKELDEEMER 98
>At3g01280 putative porin
Length = 276
Score = 28.1 bits (61), Expect = 6.5
Identities = 20/58 (34%), Positives = 27/58 (46%)
Query: 57 QEDLIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQP 114
+EDLI L G+ A Y I++P AV A V+ K +KD + V L P
Sbjct: 166 KEDLIASLTVNDKGDLLNASYYHIVNPLFNTAVGAEVSHKLSSKDSTITVGTQHSLDP 223
>At3g50780 unknown protein (At3g50780)
Length = 520
Score = 27.7 bits (60), Expect = 8.4
Identities = 28/137 (20%), Positives = 60/137 (43%), Gaps = 23/137 (16%)
Query: 148 LVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIRILITRSKTQLKATFNRY 207
++ + + +GV + P+L + ++ + + IE V+R +S+ ++K
Sbjct: 241 VISSILRLKTEGVGVTPVLKRVASNAVDPPKETLSRIIELVLRSKEEKSRREMK------ 294
Query: 208 RDDHGFSISKKLLNE-----ASDDFLKAVHVAIR-CIDDHKKYYEKVLRGA-----LKRI 256
SI KLL E +D+F ++ + + C+D +++ G K+I
Sbjct: 295 ------SIVLKLLREQNGANVADNFNDTIYSSCQTCLDSVLSLFKQASEGEKPETDTKQI 348
Query: 257 GTDEDGLTRVVITRAEK 273
+ D LT ++ AE+
Sbjct: 349 AVEADNLTWLLDVLAER 365
>At3g06940 putative mudrA protein
Length = 749
Score = 27.7 bits (60), Expect = 8.4
Identities = 33/143 (23%), Positives = 59/143 (41%), Gaps = 20/143 (13%)
Query: 30 ESAIIAIM-----GQRNAVQRQQIRQAYQDIYQEDLIKRLESELSGNFEKAMYRWILD-- 82
ES II + G +NA+ + + A+ L ++L +L G F R++L+
Sbjct: 430 ESRIITFVADFQNGLKNAIAQVFEKDAHHAYCLGQLAEKLNVDLKGQFSHEARRYMLNDF 489
Query: 83 --------PADRYAVLANVAIKSINKD-YHVIVEIASVLQPQELLAVRHAYHNRYKNSLE 133
P Y L N IKSI+ D Y+ ++E L + +N+ ++
Sbjct: 490 YSVAYATTPVGYYLALEN--IKSISPDAYNWVIESEPHHWANALF--QGERYNKMNSNFG 545
Query: 134 EDVAAHTSGYHRQLLVGLVSSFR 156
+D + S H + ++ FR
Sbjct: 546 KDFYSWVSEAHEFPITQMIDEFR 568
>At2g25170 putative chromodomain-helicase-DNA-binding protein
Length = 1359
Score = 27.7 bits (60), Expect = 8.4
Identities = 13/59 (22%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query: 46 QQIRQAYQDIYQEDLIKRLESELSGNFEKAMYRWIL-DPADRYAVLANVAIKSINKDYH 103
++ ++ ++DI QE+ I RL L+ + + + + ++ D + ++ V + S+ K+Y+
Sbjct: 477 EEFQEEFKDINQEEQISRLHKMLAPHLLRRVKKDVMKDMPPKKELILRVDLSSLQKEYY 535
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,726,523
Number of Sequences: 26719
Number of extensions: 273731
Number of successful extensions: 804
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 753
Number of HSP's gapped (non-prelim): 20
length of query: 314
length of database: 11,318,596
effective HSP length: 99
effective length of query: 215
effective length of database: 8,673,415
effective search space: 1864784225
effective search space used: 1864784225
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146590.13


