

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.9 + phase: 0
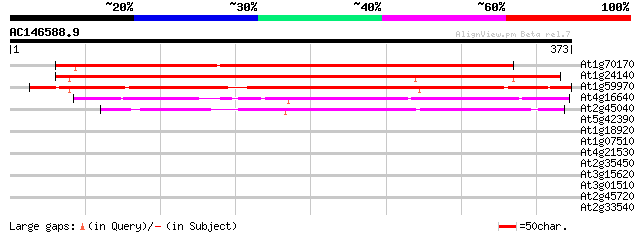
(373 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g70170 predicted GPI-anchored protein 387 e-108
At1g24140 metalloproteinase like protein, predicted GPI-anchor 377 e-105
At1g59970 predicted GPI-anchored protein 350 5e-97
At4g16640 proteinase-like protein, predicted GPI-anchored protei... 267 8e-72
At2g45040 metalloproteinase like protein ,predicted GPI-anchored... 253 2e-67
At5g42390 pitrilysin 31 1.3
At1g18920 hypothetical protein 30 1.6
At1g07510 unknown protein 30 1.6
At4g21530 hypothetical protein 30 2.8
At2g35450 unknown protein 30 2.8
At3g15620 6-4 photolyase (UVR3) 29 3.6
At3g01510 unknown protein 28 6.2
At2g45720 unknown protein 28 6.2
At2g33540 CTD phosphatase-like 3 (CPL3) 28 6.2
>At1g70170 predicted GPI-anchored protein
Length = 378
Score = 387 bits (993), Expect = e-108
Identities = 186/315 (59%), Positives = 235/315 (74%), Gaps = 11/315 (3%)
Query: 31 SWIPPGTLPVGP---------WDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSN 81
+W P + V P WDAF NFTGC G+N +GL +K YFQ FGYIP + N
Sbjct: 20 AWFFPNSTAVPPSLRNTTRVFWDAFSNFTGCHHGQNVDGLYRIKKYFQRFGYIPETFSGN 79
Query: 82 FSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTE 141
F+DDFDD L+ A++ YQ NFNLNVTGELD +T++ +++PRCG D++NGT+ M+ G+ +
Sbjct: 80 FTDDFDDILKAAVELYQTNFNLNVTGELDALTIQHIVIPRCGNPDVVNGTSLMHGGR-RK 138
Query: 142 TTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFATAFARWSE 201
T + S+ H V +T+FPG+PRWP +++LTYAF P N LTE VKSVF+ AF RWS+
Sbjct: 139 TFEVNFSRTHLHAVKRYTLFPGEPRWPRNRRDLTYAFDPKNPLTEEVKSVFSRAFGRWSD 198
Query: 202 VTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVV 261
VT L FT + +S +DI IGF+ GDHGDGEPFDG LGTLAHAFSP +G+FHLDA E+WVV
Sbjct: 199 VTALNFTLSESFSTSDITIGFYTGDHGDGEPFDGVLGTLAHAFSPPSGKFHLDADENWVV 258
Query: 262 SGDV-SKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRG 320
SGD+ S S+ AVDLESVAVHEIGHLLGLGHSS EE+IM+PTI++ +KV L +DDV G
Sbjct: 259 SGDLDSFLSVTAAVDLESVAVHEIGHLLGLGHSSVEESIMYPTITTGKRKVDLTNDDVEG 318
Query: 321 IQYLYGTNPSFNGST 335
IQYLYG NP+FNG+T
Sbjct: 319 IQYLYGANPNFNGTT 333
>At1g24140 metalloproteinase like protein, predicted GPI-anchor
Length = 384
Score = 377 bits (969), Expect = e-105
Identities = 191/344 (55%), Positives = 242/344 (69%), Gaps = 8/344 (2%)
Query: 31 SWIPPGTL---PVGPWDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKS-NFSDDF 86
S IPP L PW++F NFTGC G+ Y+GL LK YFQHFGYI + S NF+DDF
Sbjct: 28 SAIPPQLLRNATGNPWNSFLNFTGCHAGKKYDGLYMLKQYFQHFGYITETNLSGNFTDDF 87
Query: 87 DDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNS 146
DD L+ A++ YQ+NF LNVTG LD++TL+ V++PRCG D++NGT+TM++G+ T S +
Sbjct: 88 DDILKNAVEMYQRNFQLNVTGVLDELTLKHVVIPRCGNPDVVNGTSTMHSGRKTFEVSFA 147
Query: 147 DSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFATAFARWSEVTTLK 206
RFH V H++ FPG+PRWP +++LTYAF P N LTE VKSVF+ AF RW EVT L
Sbjct: 148 GRGQRFHAVKHYSFFPGEPRWPRNRRDLTYAFDPRNALTEEVKSVFSRAFTRWEEVTPLT 207
Query: 207 FTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVS 266
FT +S +DI IGF++G+HGDGEPFDG + TLAHAFSP G FHLD E+W+VSG+
Sbjct: 208 FTRVERFSTSDISIGFYSGEHGDGEPFDGPMRTLAHAFSPPTGHFHLDGEENWIVSGEGG 267
Query: 267 KS--SLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYL 324
S+ AVDLESVAVHEIGHLLGLGHSS E +IM+PTI + +KV L DDV G+QYL
Sbjct: 268 DGFISVSEAVDLESVAVHEIGHLLGLGHSSVEGSIMYPTIRTGRRKVDLTTDDVEGVQYL 327
Query: 325 YGTNPSFNG--STVISSPERNIGNGGCSLTSLWSPWRLFSLLTF 366
YG NP+FNG S S+ +R+ G+ G S S L +LL +
Sbjct: 328 YGANPNFNGSRSPPPSTQQRDTGDSGAPGRSDGSRSVLTNLLQY 371
>At1g59970 predicted GPI-anchored protein
Length = 360
Score = 350 bits (899), Expect = 5e-97
Identities = 192/366 (52%), Positives = 234/366 (63%), Gaps = 24/366 (6%)
Query: 14 FSLTLLIVSARLFPDVPSWIPPGTL---PVGPWDAFRNFTGCRQGENYNGLSNLKNYFQH 70
F T+ +SA+ + +V S IPP W+ F GC GEN NGLS LK YF+
Sbjct: 11 FFFTVNPISAKFYTNVSS-IPPLQFLNATQNAWETFSKLAGCHIGENINGLSKLKQYFRR 69
Query: 71 FGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIING 130
FGYI + N +DDFDD LQ AI TYQKNFNL VTG+LD TLRQ++ PRCG D+I+G
Sbjct: 70 FGYITTT--GNCTDDFDDVLQSAINTYQKNFNLKVTGKLDSSTLRQIVKPRCGNPDLIDG 127
Query: 131 TTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKS 190
+ MN GK TT ++ FPG+PRWP+ K++LTYAF P N LT+ VK
Sbjct: 128 VSEMNGGKILRTTEK------------YSFFPGKPRWPKRKRDLTYAFAPQNNLTDEVKR 175
Query: 191 VFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGR 250
VF+ AF RW+EVT L FT + ADI IGFF+G+HGDGEPFDG++GTLAHA SP G
Sbjct: 176 VFSRAFTRWAEVTPLNFTRSESILRADIVIGFFSGEHGDGEPFDGAMGTLAHASSPPTGM 235
Query: 251 FHLDAAEDWVVS-GDVSKSSLP--TAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSR 307
HLD EDW++S G++S+ LP T VDLESVAVHEIGHLLGLGHSS E+AIMFP IS
Sbjct: 236 LHLDGDEDWLISNGEISRRILPVTTVVDLESVAVHEIGHLLGLGHSSVEDAIMFPAISGG 295
Query: 308 MKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWRLFSLLTFV 367
+KV LA DD+ GIQ+LYG NP NG S P R + G W W + SL +
Sbjct: 296 DRKVELAKDDIEGIQHLYGGNP--NGDGGGSKPSRESQSTGGDSVRRWRGW-MISLSSIA 352
Query: 368 LSHFLL 373
FL+
Sbjct: 353 TCIFLI 358
>At4g16640 proteinase-like protein, predicted GPI-anchored protein
(by homology)
Length = 364
Score = 267 bits (682), Expect = 8e-72
Identities = 154/334 (46%), Positives = 196/334 (58%), Gaps = 25/334 (7%)
Query: 43 WDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFN 102
W F + G + +G+S LK Y FGY+ FSD FD L+ AI YQ+N
Sbjct: 52 WHDFSRLVDVQIGSHVSGVSELKRYLHRFGYV-NDGSEIFSDVFDGPLESAISLYQENLG 110
Query: 103 LNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFP 162
L +TG LD T+ + LPRCGV+D T T N+D HT +H+T F
Sbjct: 111 LPITGRLDTSTVTLMSLPRCGVSD-------------THMTINNDF---LHTTAHYTYFN 154
Query: 163 GQPRWPEGKQELTYAFFPGNEL----TETVKSVFATAFARWSEVTTLKFTETTLYSGADI 218
G+P+W + LTYA ++L +E VK+VF AF++WS V + F E ++ AD+
Sbjct: 155 GKPKW--NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWSSVIPVSFEEVDDFTTADL 212
Query: 219 KIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLES 278
KIGF+ GDHGDG PFDG LGTLAHAF+P NGR HLDAAE W+V D+ K S AVDLES
Sbjct: 213 KIGFYAGDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWIVDDDL-KGSSEVAVDLES 271
Query: 279 VAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVIS 338
VA HEIGHLLGLGHSS+E A+M+P++ R KKV L DDV G+ LYG NP ++
Sbjct: 272 VATHEIGHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAGVLKLYGPNPKLRLDSLTQ 331
Query: 339 SPERNIGNGGCSLTSLWSPWRLFSLLTFVLSHFL 372
S E +I NG S L + + LL L FL
Sbjct: 332 S-EDSIKNGTVSHRFLSGNFIGYVLLVVGLILFL 364
>At2g45040 metalloproteinase like protein ,predicted GPI-anchored
protein (by homology)
Length = 342
Score = 253 bits (645), Expect = 2e-67
Identities = 145/315 (46%), Positives = 188/315 (59%), Gaps = 34/315 (10%)
Query: 61 LSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLP 120
+ +K + Q +GY+P++ +S+ D ++A+ YQKN L +TG+ D TL Q++LP
Sbjct: 50 IPEIKRHLQQYGYLPQNKESD-----DVSFEQALVRYQKNLGLPITGKPDSDTLSQILLP 104
Query: 121 RCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQ-ELTYAFF 179
RCG D + T FHT + FPG+PRW +LTYAF
Sbjct: 105 RCGFPDDVEPKTAP-----------------FHTGKKYVYFPGRPRWTRDVPLKLTYAFS 147
Query: 180 PGN---ELTET-VKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDG 235
N L T ++ VF AF +W+ V + F ET Y ADIKIGFFNGDHGDGEPFDG
Sbjct: 148 QENLTPYLAPTDIRRVFRRAFGKWASVIPVSFIETEDYVIADIKIGFFNGDHGDGEPFDG 207
Query: 236 SLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSE 295
LG LAH FSP NGR HLD AE W V D KSS+ AVDLESVAVHEIGH+LGLGHSS
Sbjct: 208 VLGVLAHTFSPENGRLHLDKAETWAVDFDEEKSSV--AVDLESVAVHEIGHVLGLGHSSV 265
Query: 296 EEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFN-GSTVISSPERNIGNGGCSLTSL 354
++A M+PT+ R KKV L DDV G+Q LYGTNP+F S + S N+ +G + +
Sbjct: 266 KDAAMYPTLKPRSKKVNLNMDDVVGVQSLYGTNPNFTLNSLLASETSTNLADG----SRI 321
Query: 355 WSPWRLFSLLTFVLS 369
S ++S L+ V++
Sbjct: 322 RSQGMIYSTLSTVIA 336
>At5g42390 pitrilysin
Length = 1265
Score = 30.8 bits (68), Expect = 1.3
Identities = 33/131 (25%), Positives = 51/131 (38%), Gaps = 24/131 (18%)
Query: 218 IKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVS------------GDV 265
+ I NGD EP SL +L +L++ +D V+S GD
Sbjct: 907 LMIAMLNGDERFVEPTPKSLQSL-----------NLESVKDAVMSHFVGDNMEVSIVGDF 955
Query: 266 SKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLY 325
S+ + + L+ + + H SE PT + ++V L D D R Y+
Sbjct: 956 SEEEIERCI-LDYLGTVKASHDSAKPPGSEPILFRQPTAGLQFQQVFLKDTDERACAYIA 1014
Query: 326 GTNPSFNGSTV 336
G P+ G TV
Sbjct: 1015 GPAPNRWGFTV 1025
>At1g18920 hypothetical protein
Length = 980
Score = 30.4 bits (67), Expect = 1.6
Identities = 19/62 (30%), Positives = 32/62 (50%), Gaps = 2/62 (3%)
Query: 251 FHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKK 310
F AE ++GD +S AV+L S E L+ HS+ E+ ++F + +R+K
Sbjct: 55 FRAQLAELQFLAGDTVRSGSDLAVELRSK--FEFLKLVYKYHSAAEDEVIFSALDTRVKN 112
Query: 311 VV 312
+V
Sbjct: 113 IV 114
>At1g07510 unknown protein
Length = 813
Score = 30.4 bits (67), Expect = 1.6
Identities = 17/56 (30%), Positives = 28/56 (49%)
Query: 252 HLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSR 307
H D+A D V+ G K+ + + ++ +VA HE GH + E ++ TI R
Sbjct: 562 HFDSAIDRVIGGLEKKNRVISKLERRTVAYHESGHAVAGWFLEHAEPLLKVTIVPR 617
>At4g21530 hypothetical protein
Length = 493
Score = 29.6 bits (65), Expect = 2.8
Identities = 17/44 (38%), Positives = 22/44 (49%), Gaps = 8/44 (18%)
Query: 75 PRSPK--------SNFSDDFDDDLQEAIKTYQKNFNLNVTGELD 110
PR PK S+F DD +D L E T + FN+ TG+ D
Sbjct: 126 PRPPKMPGLVAGDSSFMDDGEDSLAELSNTSFRKFNILCTGDRD 169
>At2g35450 unknown protein
Length = 129
Score = 29.6 bits (65), Expect = 2.8
Identities = 18/52 (34%), Positives = 25/52 (47%), Gaps = 10/52 (19%)
Query: 136 AGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTET 187
AG D+ETTS++ + H H W ++ TY +FPG E T T
Sbjct: 49 AGSDSETTSSTSRVIDSHL--HI--------WASPQEAETYPYFPGQEPTLT 90
>At3g15620 6-4 photolyase (UVR3)
Length = 556
Score = 29.3 bits (64), Expect = 3.6
Identities = 25/101 (24%), Positives = 41/101 (39%), Gaps = 11/101 (10%)
Query: 213 YSGADIKIGFFNGDHGDG--EPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGD------ 264
Y D+K+ + G P +L AH G+ L V+G+
Sbjct: 136 YQALDVKVKDYASSTGVEVFSPVSHTLFNPAHIIEKNGGKPPLSYQSFLKVAGEPSCAKS 195
Query: 265 ---VSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFP 302
+S SSLP D+ ++ + E+ L LG+ +E+A P
Sbjct: 196 ELVMSYSSLPPIGDIGNLGISEVPSLEELGYKDDEQADWTP 236
>At3g01510 unknown protein
Length = 716
Score = 28.5 bits (62), Expect = 6.2
Identities = 28/109 (25%), Positives = 38/109 (34%), Gaps = 16/109 (14%)
Query: 179 FPGNELTETVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLG 238
FPGN L ++S A ++ SE +T L F H DG S
Sbjct: 98 FPGNCLWSPLRS---PAASKHSESSTSTPKRCRLKK-------FLTRSHSDGGVLPSSAS 147
Query: 239 TLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHL 287
+ F R H D GD SK S PT +++G +
Sbjct: 148 KRCYTFKDLKRRSHSDG------GGDTSKRSSPTVKVKNKTVSYKVGEV 190
>At2g45720 unknown protein
Length = 553
Score = 28.5 bits (62), Expect = 6.2
Identities = 30/97 (30%), Positives = 47/97 (47%), Gaps = 11/97 (11%)
Query: 261 VSGDVSKSSLPTAVDLESVAVHEIGHLLGLGH-SSEEEAI--MFPTISSRMKKVVLA--- 314
V G+V+K + DLE+ +V E+ L +GH S+ +A+ + + K V+ A
Sbjct: 131 VLGEVTKPLSSSTQDLETFSVRELLARLQIGHLESKRKALEQLVEVMKEDEKAVITALGR 190
Query: 315 DDDVRGIQYLYGTNPSF--NGSTVISSPERNIGNGGC 349
+ +Q L T+PS N TVI S +GGC
Sbjct: 191 TNVASLVQLLTATSPSVRENAVTVICSLAE---SGGC 224
>At2g33540 CTD phosphatase-like 3 (CPL3)
Length = 1241
Score = 28.5 bits (62), Expect = 6.2
Identities = 10/27 (37%), Positives = 18/27 (66%)
Query: 86 FDDDLQEAIKTYQKNFNLNVTGELDDM 112
++ D ++A+ TYQ+ F LN + DD+
Sbjct: 408 YESDARKAVSTYQQKFGLNSVFKTDDL 434
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,022,225
Number of Sequences: 26719
Number of extensions: 410563
Number of successful extensions: 986
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 960
Number of HSP's gapped (non-prelim): 16
length of query: 373
length of database: 11,318,596
effective HSP length: 101
effective length of query: 272
effective length of database: 8,619,977
effective search space: 2344633744
effective search space used: 2344633744
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146588.9


