

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.6 - phase: 0 /pseudo
(670 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
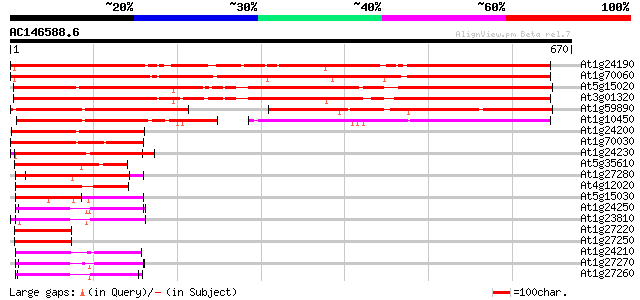
Score E
Sequences producing significant alignments: (bits) Value
At1g24190 hypothetical protein 677 0.0
At1g70060 hypothetical protein 667 0.0
At5g15020 transcriptional regulatory - like protein 614 e-176
At3g01320 unknown protein 598 e-171
At1g59890 hypothetical protein 291 1e-78
At1g10450 unknown protein 282 4e-76
At1g24200 hypothetical protein 209 5e-54
At1g70030 unknown protein 168 8e-42
At1g24230 hypothetical protein 126 4e-29
At5g35610 putative protein 107 2e-23
At1g27280 hypothetical protein 105 8e-23
At4g12020 putative disease resistance protein 104 2e-22
At5g15030 putative protein 100 4e-21
At1g24250 hypothetical protein 98 2e-20
At1g23810 hypothetical protein 95 1e-19
At1g27220 putative protein 89 1e-17
At1g27250 hypothetical protein 88 2e-17
At1g24210 unknown protein 88 2e-17
At1g27270 hypothetical protein 86 5e-17
At1g27260 hypothetical protein 86 9e-17
>At1g24190 hypothetical protein
Length = 1263
Score = 677 bits (1748), Expect = 0.0
Identities = 364/662 (54%), Positives = 471/662 (70%), Gaps = 51/662 (7%)
Query: 1 MTGG---QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKEL 57
M GG QKLTTNDAL+YLKAV++ FQ+ + KYDEFLEVMK+FK+QR+DTAGVI RVKEL
Sbjct: 1 MVGGGSAQKLTTNDALAYLKAVKDKFQDQRGKYDEFLEVMKNFKSQRVDTAGVITRVKEL 60
Query: 58 FKGHKDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYK 117
FKGH++LILGFNTFLPKG+ ITL +D QP KK VEFEEAI+FV KIK RFQG+DRVYK
Sbjct: 61 FKGHQELILGFNTFLPKGFEITLQPEDGQPPLKKRVEFEEAISFVNKIKTRFQGDDRVYK 120
Query: 118 TFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLL 177
+FLDILNMYR++ K IT VYQEV+ LF+DH DLL EFTHFLPDTS A A S + +
Sbjct: 121 SFLDILNMYRRDSKSITEVYQEVAILFRDHSDLLVEFTHFLPDTS--ATASIPSVKTSV- 177
Query: 178 RDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRR 237
R+R ++ DK+++ T H D D +H D + +R + + +KE R +++
Sbjct: 178 RERGVSLA-------DKKDRIITPHPDHDYGTEHIDQDRERPIKKENKEHMRGTNKENEH 230
Query: 238 EERDRRERERDDRDYDNDGNLERLPHKKKSVH-RATDPGTAEPLHDADEKLDLLPNSSTC 296
RD RD++ E+ +KK+ +H R DP + + + +P+SST
Sbjct: 231 ---------RDARDFEPHSKKEQFLNKKQKLHIRGDDPAE---ISNQSKLSGAVPSSSTY 278
Query: 297 EDKSSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGK 356
++K ++KS S LA +++VKEKL N +YQEFL+CL+++S+EII+R EL +LVG+L+G
Sbjct: 279 DEKGAMKSY-SQDLAIVDRVKEKL-NASEYQEFLRCLNLFSKEIISRPELQSLVGNLIGV 336
Query: 357 YADIMEGFDDFVTQCEKNE-------------GFLAGVMNKKSLWNEGHGQKPLKVEEKD 403
Y D+M+ F +F+ QCEKNE G L+G++ KKSLW+EG +P ++D
Sbjct: 337 YPDLMDSFIEFLVQCEKNEKRQICNLLNLLAEGLLSGILTKKSLWSEGKYPQPSLDNDRD 396
Query: 404 RDRGRDDGVKERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNV 463
++ RDDG+++RD E +K+ I ++ LS E ++ L N
Sbjct: 397 QEHKRDDGLRDRDHEKERLEKAAANLKWAKPISELDLSNC-----EQCTPSYRLLPKN-- 449
Query: 464 LPATVSYPKIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDM 523
P +++ QKTE+G VLNDHWVSVTSGSEDYSF HMRKNQYEESLF+CEDDRFELDM
Sbjct: 450 YPISIAS---QKTEIGKLVLNDHWVSVTSGSEDYSFSHMRKNQYEESLFKCEDDRFELDM 506
Query: 524 LLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLK 583
LLES+N+T+K VEE++ K+N + + + PIR+E+HL+ALNLRCIERLYGDHGLDVM+VLK
Sbjct: 507 LLESVNSTTKHVEELLTKINSNELKTNSPIRVEDHLTALNLRCIERLYGDHGLDVMDVLK 566
Query: 584 KNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLS 643
KN SLALPVILTRLKQKQEEWARCR DF+KVWAEIYAKNY+KSLDHRSFYFKQQD+K LS
Sbjct: 567 KNVSLALPVILTRLKQKQEEWARCRSDFDKVWAEIYAKNYYKSLDHRSFYFKQQDSKKLS 626
Query: 644 TK 645
K
Sbjct: 627 MK 628
>At1g70060 hypothetical protein
Length = 1386
Score = 667 bits (1720), Expect = 0.0
Identities = 363/680 (53%), Positives = 483/680 (70%), Gaps = 47/680 (6%)
Query: 1 MTGG---QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKEL 57
M GG QKLTTNDAL+YLKAV++ FQ+ ++KYDEFLEVMKDFKAQR+DT GVI RVKEL
Sbjct: 1 MVGGSSAQKLTTNDALAYLKAVKDKFQDKRDKYDEFLEVMKDFKAQRVDTTGVILRVKEL 60
Query: 58 FKGHKDLILGFNTFLPKGYAITLPSDDEQPLQ-KKPVEFEEAINFVGKIKNRFQGNDRVY 116
FKG+++LILGFNTFLPKG+ ITL +D+QP KKPVEFEEAI+FV KIK RFQG+DRVY
Sbjct: 61 FKGNRELILGFNTFLPKGFEITLRPEDDQPAAPKKPVEFEEAISFVNKIKTRFQGDDRVY 120
Query: 117 KTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPL 176
K+FLDILNMYRKE K IT VY EV+ LF+DH DLL EFTHFLPDTS A+ + S + P+
Sbjct: 121 KSFLDILNMYRKENKSITEVYHEVAILFRDHHDLLGEFTHFLPDTSATASTN-DSVKVPV 179
Query: 177 LRDRS-SAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRR-EREK 234
RDR ++ T RQ+ +DK+++ T H +R L ++ D + +R +++ KE+ RR +++
Sbjct: 180 -RDRGIKSLPTMRQIDLDKKDRIITSHPNRALKTENMDVDHERSLLKDSKEEVRRIDKKN 238
Query: 235 DRREERDRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGT--AEPLHDADEKLDLLPN 292
D ++RDR+ D R D+D + E + KK + R D ++ + D+ +P+
Sbjct: 239 DFMDDRDRK----DYRGLDHDSHKEHFFNSKKKLIRKDDDSAEMSDQAREGDKFSGAIPS 294
Query: 293 SSTCEDKSSLKSLC---SPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLAL 349
SST ++K + S LAF+++VK KL D+ QEFL+CL++YS+EII++ EL +L
Sbjct: 295 SSTYDEKGFIIDFLESHSQELAFVDRVKAKLDTADN-QEFLRCLNLYSKEIISQPELQSL 353
Query: 350 VGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVMNK----------------KSLWNEGHG 393
V DL+G Y D+M+ F F+ QC+KN+G L+G+++K +SLW+EG
Sbjct: 354 VSDLIGVYPDLMDAFKVFLAQCDKNDGLLSGIVSKSKSSTFYNILLTYLFGQSLWSEGKC 413
Query: 394 QKPLKVEEKDRDRGRD--DGVKERDRELRERDKSTGISNKDVSIPKVSLSKDKTS----- 446
+P K +KD DR R+ + +ERDRE +K I ++ LS +
Sbjct: 414 PQPTKSLDKDTDREREKIERYRERDREKERLEKVAASQKWAKPISELDLSNCEQCTPSYR 473
Query: 447 -M*ESL*MNWTFLTVNNVLPATVSYPKIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKN 505
+ ++L ++ F+ + +P QK E+G++VLNDHWVSVTSGSEDYSFKHMRKN
Sbjct: 474 RLPKNLNVHTYFVLLQYPIPIAS-----QKMEIGSQVLNDHWVSVTSGSEDYSFKHMRKN 528
Query: 506 QYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLR 565
QYEESLF+CEDDRFELDMLLES+ + + +VEE++ K+N + + D PI IE+HL+ALNLR
Sbjct: 529 QYEESLFKCEDDRFELDMLLESVISATNRVEELLAKINSNELKTDTPICIEDHLTALNLR 588
Query: 566 CIERLYGDHGLDVMEVLKKNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHK 625
CIERLY DHGLDV+++LKKNA LALPVILTRLKQKQEEWARCR +FNKVWA+IY KNYH+
Sbjct: 589 CIERLYSDHGLDVLDLLKKNAYLALPVILTRLKQKQEEWARCRTEFNKVWADIYTKNYHR 648
Query: 626 SLDHRSFYFKQQDTKSLSTK 645
SLDHRSFYFKQQD+K+LSTK
Sbjct: 649 SLDHRSFYFKQQDSKNLSTK 668
>At5g15020 transcriptional regulatory - like protein
Length = 1343
Score = 614 bits (1584), Expect = e-176
Identities = 338/652 (51%), Positives = 441/652 (66%), Gaps = 47/652 (7%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
QKLTT+DAL+YLK V+EMFQ+ ++KYD FLEVMKDFKAQ+ DT+GVI RVKELFKGH +L
Sbjct: 46 QKLTTDDALTYLKEVKEMFQDQRDKYDMFLEVMKDFKAQKTDTSGVISRVKELFKGHNNL 105
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILN 124
I GFNTFLPKG+ ITL DD + KK VEFEEAI+FV KIK RFQ N+ VYK+FL+ILN
Sbjct: 106 IFGFNTFLPKGFEITL--DDVEAPSKKTVEFEEAISFVNKIKTRFQHNELVYKSFLEILN 163
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGA-AAAHFASARNPLLRDRSSA 183
MYRK+ K IT VY EVS LF+DH DLLEEFT FLPD+ A ++ DR S
Sbjct: 164 MYRKDNKDITEVYNEVSTLFEDHSDLLEEFTRFLPDSLAPHTEAQLLRSQAQRYDDRGSG 223
Query: 184 MTTGRQMHVDK---REKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREER 240
R+M ++K RE+T DRD SVD D D+ +++ ++QR+R +KD RE
Sbjct: 224 PPLVRRMFMEKDRRRERTVASRGDRDHSVDRSDLNDDKSMVKMHRDQRKRV-DKDNRE-- 280
Query: 241 DRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKS 300
RR R+ +D + + D NL+ K+KS R E + ++ +K+
Sbjct: 281 -RRSRDLEDGEAEQD-NLQHFSEKRKSSRRM-------------EGFEAYSGPASHSEKN 325
Query: 301 SLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADI 360
+LKS+ + F EKVKE+L + DDYQ FLKCL+++S II R++L LV D+LGK+ D+
Sbjct: 326 NLKSMYNQAFLFCEKVKERLCSQDDYQAFLKCLNMFSNGIIQRKDLQNLVSDVLGKFPDL 385
Query: 361 MEGFDDFVTQCEKNEGF--LAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRD-DGVKERDR 417
M+ F+ F +CE +GF LAGVM+KKSL +E + + +K EEKDR+ RD + KE++R
Sbjct: 386 MDEFNQFFERCESIDGFQHLAGVMSKKSLGSEENLSRSVKGEEKDREHKRDVEAAKEKER 445
Query: 418 ELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPKIQ-KT 476
+DK G S +++ + + LP P ++ +
Sbjct: 446 S---KDKYMGKSIQELDLSDCERCTPSYRL----------------LPPDYPIPSVRHRQ 486
Query: 477 ELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKKVE 536
+ GA VLNDHWVSVTSGSEDYSFKHMR+NQYEESLFRCEDDRFELDMLLES+ + +K E
Sbjct: 487 KSGAAVLNDHWVSVTSGSEDYSFKHMRRNQYEESLFRCEDDRFELDMLLESVGSAAKSAE 546
Query: 537 EIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVILTR 596
E++ + D I + RIE+H +ALNLRCIERLYGDHGLDV ++++KN + ALPVILTR
Sbjct: 547 ELLNIIIDKKISFEGSFRIEDHFTALNLRCIERLYGDHGLDVTDLIRKNPAAALPVILTR 606
Query: 597 LKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTKGKI 648
LKQKQ+EW +CRE FN VWA++YAKN++KSLDHRSFYFKQQD+K+LS K +
Sbjct: 607 LKQKQDEWTKCREGFNVVWADVYAKNHYKSLDHRSFYFKQQDSKNLSAKALV 658
>At3g01320 unknown protein
Length = 1324
Score = 598 bits (1542), Expect = e-171
Identities = 344/655 (52%), Positives = 433/655 (65%), Gaps = 55/655 (8%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
QKLTTNDALSYL+ V+EMFQ+ +EKYD FLEVMKDFKAQR DT GVI RVKELFKGH +L
Sbjct: 51 QKLTTNDALSYLREVKEMFQDQREKYDRFLEVMKDFKAQRTDTGGVIARVKELFKGHNNL 110
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILN 124
I GFNTFLPKGY ITL +D+ L KK VEFE+AINFV KIK RF+ ++ VYK+FL+ILN
Sbjct: 111 IYGFNTFLPKGYEITLIEEDDA-LPKKTVEFEQAINFVNKIKMRFKHDEHVYKSFLEILN 169
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT--SGAAAAHFASARNPLLRDRSS 182
MYRKE K I VY EVS LFQ H DLLE+FT FLP + S +AA H S DR S
Sbjct: 170 MYRKENKEIKEVYNEVSILFQGHLDLLEQFTRFLPASLPSHSAAQHSRSQAQQY-SDRGS 228
Query: 183 AMTTGRQMHVDK---REKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREE 239
QM V+K RE+ L D SV+ D D+ +++ +EQR+R +E
Sbjct: 229 DPPLLHQMQVEKERRRERAVALRGD--YSVERYDLNDDKTMVKIQREQRKRLD----KEN 282
Query: 240 RDRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDK 299
R RR R+ DDR+ D NL P K+KS RA E L+ S++ +K
Sbjct: 283 RARRGRDLDDREAGQD-NLHHFPEKRKSSRRA-------------EALEAYSGSASHSEK 328
Query: 300 SSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYAD 359
+LKS+ F EKVK++L + DDYQ FLKCL+I+S II R++L LV DLLGK+ D
Sbjct: 329 DNLKSMYKQAFVFCEKVKDRLCSQDDYQTFLKCLNIFSNGIIQRKDLQNLVSDLLGKFPD 388
Query: 360 IMEGFDDFVTQCEKNEGF-------LAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRD-DG 411
+M+ F+ F +CE G LAGVM+KK +E +P+KVEEK+ + + +
Sbjct: 389 LMDEFNQFFERCESITGTEIHGFQRLAGVMSKKLFSSEEQLSRPMKVEEKESEHKPELEA 448
Query: 412 VKERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYP 471
VKE ++ +E SI ++ LS + +LPA P
Sbjct: 449 VKETEQCKKEY--------MGKSIQELDLSDCECCT-----------PSYRLLPADYPIP 489
Query: 472 -KIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINA 530
Q++ELGA+VLNDHWVSVTSGSEDYSFKHMR+NQYEESLFRCEDDRFELDMLLES+++
Sbjct: 490 IASQRSELGAEVLNDHWVSVTSGSEDYSFKHMRRNQYEESLFRCEDDRFELDMLLESVSS 549
Query: 531 TSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLAL 590
++ E ++ + + I RIE+H +ALNLRCIERLYGDHGLDV+++L KN + AL
Sbjct: 550 AARSAESLLNIITEKKISFSGSFRIEDHFTALNLRCIERLYGDHGLDVIDILNKNPATAL 609
Query: 591 PVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
PVILTRLKQKQ EW +CR+DF+KVWA +YAKN++KSLDHRSFYFKQQD+K+LS K
Sbjct: 610 PVILTRLKQKQGEWKKCRDDFDKVWANVYAKNHYKSLDHRSFYFKQQDSKNLSAK 664
>At1g59890 hypothetical protein
Length = 1125
Score = 291 bits (744), Expect = 1e-78
Identities = 165/354 (46%), Positives = 222/354 (62%), Gaps = 25/354 (7%)
Query: 310 LAFLEKVKEKLKNPDD-YQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
+ F+ ++K + D Y++FL L++Y +E + E+ V L + D++ F F+
Sbjct: 131 IEFVNRIKARFGGDDRAYKKFLDILNMYRKETKSINEVYQEVTLLFQDHEDLLGEFVHFL 190
Query: 369 TQCEKNEGFLAGVMNKKSLWNEGH----GQKPLKVEEKDRDRGRDDGVKERDRELRERDK 424
+ + + ++ + + G P E+K + R R D E + + D+
Sbjct: 191 PDFRGSVSVNDPLFQRNTIPRDRNSTFPGMHPKHFEKKIK-RSRHDEYTELSDQREDGDE 249
Query: 425 STGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPKIQ---------- 474
+ + D+ K LS + + L + + T SY ++
Sbjct: 250 NLVAYSADIGSQKNLLSTNHMAK------AINELDLTDCAQCTPSYRRLPDDYPIQIPSY 303
Query: 475 KTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKK 534
+ LG KVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLES++A K+
Sbjct: 304 RNSLGEKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVSAAIKR 363
Query: 535 VEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVIL 594
VE ++EK+N++ I + PI I EHLS +CIERLYGD+GLDVM+ LKKN+ +ALPVIL
Sbjct: 364 VESLLEKINNNTISIETPICIREHLSG---KCIERLYGDYGLDVMDFLKKNSHIALPVIL 420
Query: 595 TRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTKGKI 648
TRLKQKQEEWARCR DF KVWAE+YAKN+HKSLDHRSFYFKQQD+K+LSTKG +
Sbjct: 421 TRLKQKQEEWARCRADFRKVWAEVYAKNHHKSLDHRSFYFKQQDSKNLSTKGLV 474
Score = 235 bits (599), Expect = 7e-62
Identities = 127/213 (59%), Positives = 153/213 (71%), Gaps = 5/213 (2%)
Query: 2 TGGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGH 61
TGG LTT DAL+YLKAV++MFQ++KEKY+ FL VMKDFKAQR+DT GVI RVK+LFKG+
Sbjct: 38 TGG--LTTVDALTYLKAVKDMFQDNKEKYETFLGVMKDFKAQRVDTNGVIARVKDLFKGY 95
Query: 62 KDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLD 121
DL+LGFNTFLPKGY ITL +DE+P KKPV+F+ AI FV +IK RF G+DR YK FLD
Sbjct: 96 DDLLLGFNTFLPKGYKITLQPEDEKP--KKPVDFQVAIEFVNRIKARFGGDDRAYKKFLD 153
Query: 122 ILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHF-ASARNPLLRDR 180
ILNMYRKE K I VYQEV+ LFQDH DLL EF HFLPD G+ + + RN + RDR
Sbjct: 154 ILNMYRKETKSINEVYQEVTLLFQDHEDLLGEFVHFLPDFRGSVSVNDPLFQRNTIPRDR 213
Query: 181 SSAMTTGRQMHVDKREKTTTLHADRDLSVDHPD 213
+S H +K+ K + +LS D
Sbjct: 214 NSTFPGMHPKHFEKKIKRSRHDEYTELSDQRED 246
Score = 32.7 bits (73), Expect = 0.67
Identities = 17/60 (28%), Positives = 33/60 (54%), Gaps = 1/60 (1%)
Query: 310 LAFLEKVKEKLK-NPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
L +L+ VK+ + N + Y+ FL + + + + ++A V DL Y D++ GF+ F+
Sbjct: 47 LTYLKAVKDMFQDNKEKYETFLGVMKDFKAQRVDTNGVIARVKDLFKGYDDLLLGFNTFL 106
>At1g10450 unknown protein
Length = 1173
Score = 282 bits (722), Expect = 4e-76
Identities = 177/398 (44%), Positives = 234/398 (58%), Gaps = 43/398 (10%)
Query: 286 KLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKNPDD-YQEFLKCLHIYSREIITRQ 344
K+ LLP E+K ++ + F+ K+K + + + Y+ FL L++Y +E +
Sbjct: 150 KITLLPE----EEKPKIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKEKKSIS 205
Query: 345 ELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVM--NKKSLWNEGHGQKPLKVEEK 402
E+ V L + D++ F +F+ C ++ + +K + H K K K
Sbjct: 206 EVYEEVTMLFKGHEDLLMEFVNFLPNCPESAPSTKNAVPRHKGTATTAMHSDKKRKQRCK 265
Query: 403 DRDRG-----RDDGVKE------------------RDRELRE---------RDKSTGISN 430
D R+DG + RD E RE +KS +
Sbjct: 266 LEDYSGHSDQREDGDENLVTCSADSPVGEGQPGYFRDYENREDTETDTADRTEKSAASGS 325
Query: 431 KDVSIPKVSLSKDKTSM*ESL*MNWTFLTVN-NVLPA--TVSYPKIQKTELGAKVLNDHW 487
+D+ K + T + E T T + +LP V P + T LG K LNDH
Sbjct: 326 QDIGNHKSTTKYVGTPINELDLSECTQCTPSYRLLPKDYAVEIPSYRNT-LGKKTLNDHL 384
Query: 488 VSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKVNDDII 547
VSVTSGSEDYSF HMRKNQYEESLFRCEDDR+E+DMLL S+++ K+VE ++EK+N++ I
Sbjct: 385 VSVTSGSEDYSFSHMRKNQYEESLFRCEDDRYEMDMLLGSVSSAIKQVEILLEKMNNNTI 444
Query: 548 PGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVILTRLKQKQEEWARC 607
D I IE+HLSA+NLRCIERLYGD+GLDVM++LKKN ALPVILTRLKQKQEEWARC
Sbjct: 445 SVDSTICIEKHLSAMNLRCIERLYGDNGLDVMDLLKKNMHSALPVILTRLKQKQEEWARC 504
Query: 608 REDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
DF KVWAE+YAKN+HKSLDHRSFYFKQQD+K+LSTK
Sbjct: 505 HSDFQKVWAEVYAKNHHKSLDHRSFYFKQQDSKNLSTK 542
Score = 201 bits (512), Expect = 8e-52
Identities = 118/255 (46%), Positives = 160/255 (62%), Gaps = 24/255 (9%)
Query: 9 TNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGF 68
T DAL+YLKAV+++F ++KEKY+ FLE+MK+FKAQ IDT GVIER+K LFKG++DL+LGF
Sbjct: 82 TIDALTYLKAVKDIFHDNKEKYESFLELMKEFKAQTIDTNGVIERIKVLFKGYRDLLLGF 141
Query: 69 NTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRK 128
NTFLPKGY ITL ++E+P K V+F++AI FV KIK RF ++ YK FLDILN+YRK
Sbjct: 142 NTFLPKGYKITLLPEEEKP--KIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRK 199
Query: 129 ELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGR 188
E K I+ VY+EV+ LF+ H DLL EF +FLP+ +A S +N + R + +A T
Sbjct: 200 EKKSISEVYEEVTMLFKGHEDLLMEFVNFLPNCPESA----PSTKNAVPRHKGTATTA-- 253
Query: 189 QMHVDKREKTT------TLHADR---------DLSVDHPDPELDRGVMRTDKEQRRRERE 233
MH DK+ K + H+D+ S D P E G R + + E +
Sbjct: 254 -MHSDKKRKQRCKLEDYSGHSDQREDGDENLVTCSADSPVGEGQPGYFRDYENREDTETD 312
Query: 234 KDRREERDRRERERD 248
R E+ +D
Sbjct: 313 TADRTEKSAASGSQD 327
>At1g24200 hypothetical protein
Length = 196
Score = 209 bits (531), Expect = 5e-54
Identities = 105/159 (66%), Positives = 123/159 (77%), Gaps = 3/159 (1%)
Query: 3 GGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHK 62
G K TTNDAL YL+AV+ FQ +EKYDEFL++M D+K QRID +GVI R+KEL K +
Sbjct: 6 GAHKPTTNDALKYLRAVKAKFQGQREKYDEFLQIMIDYKTQRIDISGVIIRMKELLKEQQ 65
Query: 63 DLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDI 122
L+LGFN FLP GY IT EQP QKKPVE EAI+F+ KIK RFQG+DRVYK+ LDI
Sbjct: 66 GLLLGFNAFLPNGYMIT---HHEQPSQKKPVELGEAISFINKIKTRFQGDDRVYKSVLDI 122
Query: 123 LNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT 161
LNMYRK+ KPITAVY+EV+ LF DH +LL EFTHFLP T
Sbjct: 123 LNMYRKDRKPITAVYREVAILFLDHNNLLVEFTHFLPAT 161
>At1g70030 unknown protein
Length = 160
Score = 168 bits (426), Expect = 8e-42
Identities = 86/159 (54%), Positives = 110/159 (69%), Gaps = 2/159 (1%)
Query: 1 MTGGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKG 60
M +KLTT DAL +L V+ +Q+++E YD FL +MKDF+ QR T VI +VKELFKG
Sbjct: 1 MVANKKLTTTDALDFLHLVKTKYQDNREIYDSFLTIMKDFRGQRAKTCDVISKVKELFKG 60
Query: 61 HKDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFL 120
+L+LGFNTFLP G+ ITL SDDE K F+EA FV K+K RFQ ND V+ +FL
Sbjct: 61 QPELLLGFNTFLPTGFEITL-SDDELTSNSKFAHFDEAYEFVNKVKTRFQNND-VFNSFL 118
Query: 121 DILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLP 159
++L ++KE K + +YQEV+ LFQ H DLLEEF FLP
Sbjct: 119 EVLKTHKKENKSVAELYQEVAILFQGHRDLLEEFHLFLP 157
Score = 32.3 bits (72), Expect = 0.88
Identities = 18/57 (31%), Positives = 30/57 (52%)
Query: 312 FLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
F+ KVK + +N D + FL+ L + +E + EL V L + D++E F F+
Sbjct: 100 FVNKVKTRFQNNDVFNSFLEVLKTHKKENKSVAELYQEVAILFQGHRDLLEEFHLFL 156
>At1g24230 hypothetical protein
Length = 245
Score = 126 bits (316), Expect = 4e-29
Identities = 66/169 (39%), Positives = 103/169 (60%), Gaps = 5/169 (2%)
Query: 6 KLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLI 65
K T +D SY+ +++E F+++ KY +FLE++ D+ A+R+D + R+ EL K H++L+
Sbjct: 81 KETFHDVRSYIYSLKESFRDEPAKYAQFLEILNDYSARRVDAPSAVARMTELMKDHRNLV 140
Query: 66 LGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGND-RVYKTFLDILN 124
LGF+ L G T P + E K+ NF+ K+K RFQGND VY++FL+IL
Sbjct: 141 LGFSVLLSTGDTKTTPLEAEPDNNKR----IRVANFISKLKARFQGNDGHVYESFLEILT 196
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASAR 173
MY++ K + +YQEV AL Q H DL+ EF++ T+G + + A R
Sbjct: 197 MYQQGNKSVNDLYQEVVALLQGHEDLVMEFSNVFKRTTGPSGSKSARDR 245
Score = 83.6 bits (205), Expect = 3e-16
Identities = 53/162 (32%), Positives = 81/162 (49%), Gaps = 19/162 (11%)
Query: 1 MTGGQ---KLTTNDALSYLKAVREMFQNDKE-KYDEFLEVMKDFKAQRIDTAGVIERVKE 56
M GG T ++A SY+ AV+E F D+ KY EFL++M D +A R+D A V+ R++E
Sbjct: 1 MVGGSLSPAFTIDEATSYINAVKEAFGADQPAKYREFLDIMLDLRANRVDLATVVPRMRE 60
Query: 57 LFKGHKDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVY 116
L K H +L+L FN FLP T F + +++ +K F+ Y
Sbjct: 61 LLKDHVNLLLRFNAFLPAEAKET---------------FHDVRSYIYSLKESFRDEPAKY 105
Query: 117 KTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFL 158
FL+ILN Y + ++ L +DH +L+ F+ L
Sbjct: 106 AQFLEILNDYSARRVDAPSAVARMTELMKDHRNLVLGFSVLL 147
Score = 36.2 bits (82), Expect = 0.061
Identities = 20/65 (30%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Query: 96 EEAINFVGKIKNRFQGNDRV-YKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEF 154
+EA +++ +K F + Y+ FLDI+ R + V + L +DH +LL F
Sbjct: 13 DEATSYINAVKEAFGADQPAKYREFLDIMLDLRANRVDLATVVPRMRELLKDHVNLLLRF 72
Query: 155 THFLP 159
FLP
Sbjct: 73 NAFLP 77
>At5g35610 putative protein
Length = 155
Score = 107 bits (267), Expect = 2e-23
Identities = 63/146 (43%), Positives = 90/146 (61%), Gaps = 15/146 (10%)
Query: 6 KLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLI 65
K+T ++ YLK V+ QN +E Y FL+VM + AQRID +GV VKELFK ++ I
Sbjct: 5 KITKSNPRKYLKIVKNKLQNKREIYVRFLQVMTAYSAQRIDPSGVKSVVKELFKEDQEPI 64
Query: 66 LGFNTFLPKGYAITLPSDD---------EQPLQKK--PVEFEEAINFVGKIKNRFQGNDR 114
GFNTFLPKG+ I D EQ KK +E+ EA++FV K+K+ +DR
Sbjct: 65 SGFNTFLPKGFEIKPECDQNGFKIKLECEQTPPKKYVDIEYSEALDFVRKVKD----DDR 120
Query: 115 VYKTFLDILNMYRKELKPITAVYQEV 140
+YK+F+ I++MY+K+ K + V +EV
Sbjct: 121 IYKSFVTIMDMYKKKNKSLDEVCREV 146
Score = 32.3 bits (72), Expect = 0.88
Identities = 15/55 (27%), Positives = 26/55 (47%)
Query: 105 IKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLP 159
+KN+ Q +Y FL ++ Y + + V V LF++ + + F FLP
Sbjct: 18 VKNKLQNKREIYVRFLQVMTAYSAQRIDPSGVKSVVKELFKEDQEPISGFNTFLP 72
Score = 28.9 bits (63), Expect = 9.7
Identities = 19/71 (26%), Positives = 38/71 (52%), Gaps = 8/71 (11%)
Query: 312 FLEKVKEKLKNPDD-YQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV-- 368
+L+ VK KL+N + Y FL+ + YS + I + ++V +L + + + GF+ F+
Sbjct: 14 YLKIVKNKLQNKREIYVRFLQVMTAYSAQRIDPSGVKSVVKELFKEDQEPISGFNTFLPK 73
Query: 369 -----TQCEKN 374
+C++N
Sbjct: 74 GFEIKPECDQN 84
>At1g27280 hypothetical protein
Length = 225
Score = 105 bits (262), Expect = 8e-23
Identities = 57/137 (41%), Positives = 84/137 (60%), Gaps = 1/137 (0%)
Query: 8 TTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILG 67
T +DA+SY+ V+E F ++ KY EF ++ D + + ID AG I RV+EL K HK+L++
Sbjct: 78 TIDDAVSYINTVKEAFHDEPAKYYEFFQLFYDIRYRLIDVAGGITRVEELLKAHKNLLVR 137
Query: 68 FNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDR-VYKTFLDILNMY 126
N FLP L EQ + + +F+GK+K RFQG+DR VY++FL+IL MY
Sbjct: 138 LNAFLPPEAQRILHLKIEQRAASDINKRKRVASFIGKLKERFQGDDRHVYESFLEILTMY 197
Query: 127 RKELKPITAVYQEVSAL 143
++ K + +YQEV L
Sbjct: 198 QEGNKSVNDLYQEVGFL 214
Score = 50.1 bits (118), Expect = 4e-06
Identities = 34/147 (23%), Positives = 65/147 (44%), Gaps = 10/147 (6%)
Query: 19 VREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGFNTFLP----K 74
+ E++ ++ EK++ L +++D+ R D A + + EL + H +L++ F F P +
Sbjct: 1 MEEVYHDEPEKFNHILHLIRDYFNHRDDRARITACMGELMRDHLNLLVRFFDFFPAEASE 60
Query: 75 GYA--ITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKP 132
G A T+ + P ++A++++ +K F Y F + R L
Sbjct: 61 GLAQLQTIAATSVSP----DSTIDDAVSYINTVKEAFHDEPAKYYEFFQLFYDIRYRLID 116
Query: 133 ITAVYQEVSALFQDHGDLLEEFTHFLP 159
+ V L + H +LL FLP
Sbjct: 117 VAGGITRVEELLKAHKNLLVRLNAFLP 143
>At4g12020 putative disease resistance protein
Length = 1895
Score = 104 bits (259), Expect = 2e-22
Identities = 58/136 (42%), Positives = 85/136 (61%), Gaps = 12/136 (8%)
Query: 7 LTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLIL 66
+TT DAL+YLKAV++ F+ D EKYD FLEV+ D K Q +DT+GVI R+K+LFKGH DL+L
Sbjct: 304 VTTGDALNYLKAVKDKFE-DSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLL 362
Query: 67 GFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMY 126
GFNT+L K Y IT+ +D+ P I+F+ K++ ++ + +T NM
Sbjct: 363 GFNTYLSKEYQITILPEDDFP-----------IDFLDKVEGPYEMTYQQAQTVQANANMQ 411
Query: 127 RKELKPITAVYQEVSA 142
+ P ++ Q S+
Sbjct: 412 PQTEYPSSSAVQSFSS 427
Score = 39.7 bits (91), Expect = 0.006
Identities = 19/62 (30%), Positives = 38/62 (60%), Gaps = 1/62 (1%)
Query: 97 EAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTH 156
+A+N++ +K++F+ +++ Y TFL++LN + + + V + LF+ H DLL F
Sbjct: 308 DALNYLKAVKDKFEDSEK-YDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 157 FL 158
+L
Sbjct: 367 YL 368
Score = 33.5 bits (75), Expect = 0.39
Identities = 15/61 (24%), Positives = 36/61 (58%)
Query: 310 LAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFVT 369
L +L+ VK+K ++ + Y FL+ L+ + + ++A + DL + D++ GF+ +++
Sbjct: 310 LNYLKAVKDKFEDSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNTYLS 369
Query: 370 Q 370
+
Sbjct: 370 K 370
>At5g15030 putative protein
Length = 271
Score = 99.8 bits (247), Expect = 4e-21
Identities = 48/79 (60%), Positives = 60/79 (75%)
Query: 7 LTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLIL 66
LTT+DAL+YLK ++++F + K KY FLE+M DFKAQR DT+ VI RVK+L KGH LIL
Sbjct: 192 LTTDDALAYLKEIKDVFHDQKYKYHLFLEIMSDFKAQRTDTSVVIARVKDLLKGHNHLIL 251
Query: 67 GFNTFLPKGYAITLPSDDE 85
FN FLP G+ ITL +DE
Sbjct: 252 VFNKFLPHGFEITLDDEDE 270
Score = 55.1 bits (131), Expect = 1e-07
Identities = 46/181 (25%), Positives = 84/181 (45%), Gaps = 32/181 (17%)
Query: 7 LTTNDALSYLKAVREMF--QNDKEKYDEFLEVMKDFKAQRI-----DTAGVIERVKELFK 59
+T +DA +YL+ V+ F ++++KY F +V+ DFKAQRI D+A +K+L
Sbjct: 82 MTISDARAYLQQVKNTFIDHDERDKYAMFRKVLFDFKAQRIVADRSDSARFCSHLKDL-- 139
Query: 60 GHKDLILGFNTFLPK-------------GYAITLPSDDEQPLQKKP--------VEFEEA 98
DL F+ + + + + P ++ Q KP + ++A
Sbjct: 140 --SDLFQIFSERMKRIGDKEIDGSGSQNQHPVGSPRNELQGQSPKPGNGNTKDALTTDDA 197
Query: 99 INFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFL 158
+ ++ +IK+ F Y FL+I++ ++ + + V V L + H L+ F FL
Sbjct: 198 LAYLKEIKDVFHDQKYKYHLFLEIMSDFKAQRTDTSVVIARVKDLLKGHNHLILVFNKFL 257
Query: 159 P 159
P
Sbjct: 258 P 258
>At1g24250 hypothetical protein
Length = 252
Score = 97.8 bits (242), Expect = 2e-20
Identities = 61/154 (39%), Positives = 82/154 (52%), Gaps = 24/154 (15%)
Query: 8 TTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILG 67
T +DA SYL AV+E F ++ KY E L+++KDFKA+R+D A VI RV+EL K H +L+ G
Sbjct: 117 TIDDATSYLIAVKEAFHDEPAKYGEMLKLLKDFKARRVDAACVIARVEELMKDHLNLLFG 176
Query: 68 FNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGN-DRVYKTFLDILNMY 126
F FL +F K+K RFQG+ +V + L I+ MY
Sbjct: 177 FCVFL-----------------------SATTSFTTKLKARFQGDGSQVVDSVLQIMRMY 213
Query: 127 RKELKPITAVYQEVSALFQDHGDLLEEFTHFLPD 160
+ K YQEV AL Q H DL+ E + L D
Sbjct: 214 GEGNKSKHDAYQEVVALVQGHDDLVMELSQILTD 247
Score = 57.4 bits (137), Expect = 3e-08
Identities = 43/172 (25%), Positives = 70/172 (40%), Gaps = 20/172 (11%)
Query: 11 DALSYLKAVREMFQN-DKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGFN 69
D +Y+ AV Q + ++ F+ + + F A RI R+++L K H L LG N
Sbjct: 14 DVKAYVNAVEVALQEMEPARFGMFVRLFRGFTAPRIGMPTFSARMQDLLKDHPSLCLGLN 73
Query: 70 TFLPKGYAITLPSDDEQPLQK-----KPV--------------EFEEAINFVGKIKNRFQ 110
LP Y +T+P + + K PV ++A +++ +K F
Sbjct: 74 VLLPPEYQLTIPPEASEEFHKVVGRSVPVPPKVVGRSLPRPEPTIDDATSYLIAVKEAFH 133
Query: 111 GNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTS 162
Y L +L ++ V V L +DH +LL F FL T+
Sbjct: 134 DEPAKYGEMLKLLKDFKARRVDAACVIARVEELMKDHLNLLFGFCVFLSATT 185
>At1g23810 hypothetical protein
Length = 241
Score = 95.1 bits (235), Expect = 1e-19
Identities = 57/156 (36%), Positives = 82/156 (52%), Gaps = 24/156 (15%)
Query: 8 TTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILG 67
T +DA SYL AV+E F ++ KY+E L+++ DFKA+R++ A VI RV+EL K H +L+ G
Sbjct: 106 TIDDATSYLIAVKEAFHDEPAKYEEMLKLLNDFKARRVNAASVIARVEELMKDHSNLLFG 165
Query: 68 FNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGN-DRVYKTFLDILNMY 126
F FL +F K+K +FQG+ +V + L I+ MY
Sbjct: 166 FCVFL-----------------------SATTSFTTKLKAKFQGDGSQVVDSVLQIMRMY 202
Query: 127 RKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTS 162
+ K YQE+ AL Q H DL+ E + D S
Sbjct: 203 GEGNKSKHDAYQEIVALVQGHDDLVMELSQIFTDPS 238
Score = 65.9 bits (159), Expect = 7e-11
Identities = 46/174 (26%), Positives = 76/174 (43%), Gaps = 12/174 (6%)
Query: 1 MTGGQKLTTN---DALSYLKAVREMFQN-DKEKYDEFLEVMKDFKAQRIDTAGVIERVKE 56
M GG K + D +Y+ AV+ + + KY EFL + + A+R+ A R+++
Sbjct: 1 MAGGSKSPASSLEDGKAYVNAVKVALEEAEPAKYQEFLRLFHEVIARRMGMATFSARMQD 60
Query: 57 LFKGHKDLILGFNTFLPKGYAITLPSDDEQPLQK--------KPVEFEEAINFVGKIKNR 108
L K H L LG N L Y +P + + K ++A +++ +K
Sbjct: 61 LLKDHPSLCLGLNVMLAPEYQRAIPPEASEEFHKVVGRSVPRPEPTIDDATSYLIAVKEA 120
Query: 109 FQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTS 162
F Y+ L +LN ++ +V V L +DH +LL F FL T+
Sbjct: 121 FHDEPAKYEEMLKLLNDFKARRVNAASVIARVEELMKDHSNLLFGFCVFLSATT 174
>At1g27220 putative protein
Length = 90
Score = 88.6 bits (218), Expect = 1e-17
Identities = 39/69 (56%), Positives = 52/69 (74%)
Query: 6 KLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLI 65
K T +DA +Y++ V+ F ND +KYD+F+ +MK+FKA++ID IE VKEL KGH+DLI
Sbjct: 9 KATVDDAYAYIRTVKSTFHNDPDKYDDFMAIMKNFKARKIDRNTCIEEVKELLKGHRDLI 68
Query: 66 LGFNTFLPK 74
GFN FLPK
Sbjct: 69 SGFNAFLPK 77
Score = 34.7 bits (78), Expect = 0.18
Identities = 17/70 (24%), Positives = 31/70 (44%)
Query: 90 KKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGD 149
K ++A ++ +K+ F + Y F+ I+ ++ +EV L + H D
Sbjct: 7 KPKATVDDAYAYIRTVKSTFHNDPDKYDDFMAIMKNFKARKIDRNTCIEEVKELLKGHRD 66
Query: 150 LLEEFTHFLP 159
L+ F FLP
Sbjct: 67 LISGFNAFLP 76
>At1g27250 hypothetical protein
Length = 137
Score = 87.8 bits (216), Expect = 2e-17
Identities = 40/69 (57%), Positives = 53/69 (75%)
Query: 6 KLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLI 65
+ T +DA +YL+AVR F ND +KYD+F+ VM +FKA+RID G I+ V++L KGH+DLI
Sbjct: 16 RATKDDAYAYLRAVRAKFHNDSKKYDDFVAVMTNFKARRIDRDGCIKEVEQLLKGHRDLI 75
Query: 66 LGFNTFLPK 74
GFN FLPK
Sbjct: 76 SGFNAFLPK 84
Score = 32.7 bits (73), Expect = 0.67
Identities = 14/64 (21%), Positives = 31/64 (47%)
Query: 96 EEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFT 155
++A ++ ++ +F + + Y F+ ++ ++ +EV L + H DL+ F
Sbjct: 20 DDAYAYLRAVRAKFHNDSKKYDDFVAVMTNFKARRIDRDGCIKEVEQLLKGHRDLISGFN 79
Query: 156 HFLP 159
FLP
Sbjct: 80 AFLP 83
>At1g24210 unknown protein
Length = 155
Score = 87.8 bits (216), Expect = 2e-17
Identities = 53/152 (34%), Positives = 80/152 (51%), Gaps = 22/152 (14%)
Query: 7 LTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLIL 66
LT +DA SY+ AV+E F ++ KY EF++++ R+D VI RV+EL K H+DL+L
Sbjct: 10 LTKDDAHSYIIAVKETFHDEPTKYQEFIKLLNGVCDHRVDKYSVIARVEELMKDHQDLLL 69
Query: 67 GFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGND-RVYKTFLDILNM 125
GF+ FLP PV E+ F+ K+K RFQ D V ++ M
Sbjct: 70 GFSVFLP------------------PVSVED---FINKLKTRFQSLDTHVVGAIRGLMKM 108
Query: 126 YRKELKPITAVYQEVSALFQDHGDLLEEFTHF 157
+++ + V +EV + H DL+E+F F
Sbjct: 109 FKEGKMSVKEVQEEVIDVLFYHEDLIEDFLRF 140
Score = 35.0 bits (79), Expect = 0.14
Identities = 20/67 (29%), Positives = 32/67 (46%)
Query: 96 EEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFT 155
++A +++ +K F Y+ F+ +LN +V V L +DH DLL F+
Sbjct: 13 DDAHSYIIAVKETFHDEPTKYQEFIKLLNGVCDHRVDKYSVIARVEELMKDHQDLLLGFS 72
Query: 156 HFLPDTS 162
FLP S
Sbjct: 73 VFLPPVS 79
Score = 31.6 bits (70), Expect = 1.5
Identities = 17/59 (28%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Query: 311 AFLEKVKEKLKN-PDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
+++ VKE + P YQEF+K L+ + + ++A V +L+ + D++ GF F+
Sbjct: 17 SYIIAVKETFHDEPTKYQEFIKLLNGVCDHRVDKYSVIARVEELMKDHQDLLLGFSVFL 75
>At1g27270 hypothetical protein
Length = 241
Score = 86.3 bits (212), Expect = 5e-17
Identities = 57/154 (37%), Positives = 80/154 (51%), Gaps = 24/154 (15%)
Query: 8 TTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILG 67
T +DA SYL AV+E F ++ KY E +++ D KA+RI+ A VI R++EL K H +L+LG
Sbjct: 110 TMDDATSYLNAVKEAFHDEPAKYMEITKLLTDLKARRINAASVIARMEELLKDHLNLLLG 169
Query: 68 FNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGN-DRVYKTFLDILNMY 126
F FL P ++ F+ K+K RF G+ +V + L IL M+
Sbjct: 170 FCVFL-------------SPTRR----------FITKLKARFLGDGSQVVDSVLQILRMH 206
Query: 127 RKELKPITAVYQEVSALFQDHGDLLEEFTHFLPD 160
+ K QEV AL Q H DLL E + D
Sbjct: 207 SEGNKSKDEASQEVRALIQGHEDLLMELSEIFSD 240
Score = 52.0 bits (123), Expect = 1e-06
Identities = 38/164 (23%), Positives = 65/164 (39%), Gaps = 13/164 (7%)
Query: 11 DALSYLKAVREMFQN-DKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGFN 69
D ++Y AV+ Q+ + EKY EF+ + D+ A R + ++EL + H +L G N
Sbjct: 14 DGMAYFDAVKVALQDTEPEKYQEFVRIFIDYTANRFGFETLSASLQELLRDHANLCHGSN 73
Query: 70 TFLPKGYAITLPSDDEQPLQKKPV------------EFEEAINFVGKIKNRFQGNDRVYK 117
LP +P + V ++A +++ +K F Y
Sbjct: 74 VLLPFRLQTAIPPEASAEFHINKVVGRSVPPAVAVPTMDDATSYLNAVKEAFHDEPAKYM 133
Query: 118 TFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT 161
+L + +V + L +DH +LL F FL T
Sbjct: 134 EITKLLTDLKARRINAASVIARMEELLKDHLNLLLGFCVFLSPT 177
>At1g27260 hypothetical protein
Length = 222
Score = 85.5 bits (210), Expect = 9e-17
Identities = 51/147 (34%), Positives = 76/147 (51%), Gaps = 24/147 (16%)
Query: 8 TTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILG 67
T DA SYL +V+ F ++ KY+E L+++ D +A+R+D A I V+EL K H+ L+ G
Sbjct: 90 TIEDATSYLNSVKRAFHDEPAKYEELLKLLNDIEARRVDAASFIASVEELMKDHQTLLNG 149
Query: 68 FNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGN-DRVYKTFLDILNMY 126
F+ FL + F+ K+K +FQG+ V + L IL MY
Sbjct: 150 FSVFL-----------------------SAEMKFIRKLKAKFQGDGSHVADSVLQILRMY 186
Query: 127 RKELKPITAVYQEVSALFQDHGDLLEE 153
+ K + +QEV L QDH DL+ E
Sbjct: 187 SEGNKSKSEAFQEVVPLVQDHEDLVME 213
Score = 83.6 bits (205), Expect = 3e-16
Identities = 49/153 (32%), Positives = 77/153 (50%), Gaps = 4/153 (2%)
Query: 10 NDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGFN 69
N A +Y+ AVR+ F+++ KY +FL +++D +A+RID A + EL K H DL+LGFN
Sbjct: 2 NKAFAYIIAVRDRFRDEPAKYRQFLSLLRDRRARRIDKATFFVGLVELIKDHLDLLLGFN 61
Query: 70 TFLPKGYAITLPSDDEQPLQKKPV----EFEEAINFVGKIKNRFQGNDRVYKTFLDILNM 125
LP + I + Q + + V E+A +++ +K F Y+ L +LN
Sbjct: 62 ALLPARFQIPITPAGFQNVVGRSVPPETTIEDATSYLNSVKRAFHDEPAKYEELLKLLND 121
Query: 126 YRKELKPITAVYQEVSALFQDHGDLLEEFTHFL 158
+ V L +DH LL F+ FL
Sbjct: 122 IEARRVDAASFIASVEELMKDHQTLLNGFSVFL 154
Score = 30.0 bits (66), Expect = 4.4
Identities = 17/63 (26%), Positives = 29/63 (45%)
Query: 97 EAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTH 156
+A ++ +++RF+ Y+ FL +L R + + L +DH DLL F
Sbjct: 3 KAFAYIIAVRDRFRDEPAKYRQFLSLLRDRRARRIDKATFFVGLVELIKDHLDLLLGFNA 62
Query: 157 FLP 159
LP
Sbjct: 63 LLP 65
Score = 28.9 bits (63), Expect = 9.7
Identities = 20/81 (24%), Positives = 36/81 (43%), Gaps = 11/81 (13%)
Query: 290 LPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKN-PDDYQEFLKCLHIYSREIITRQELLA 348
+P +T ED +S +L VK + P Y+E LK L+ + +A
Sbjct: 85 VPPETTIEDATS----------YLNSVKRAFHDEPAKYEELLKLLNDIEARRVDAASFIA 134
Query: 349 LVGDLLGKYADIMEGFDDFVT 369
V +L+ + ++ GF F++
Sbjct: 135 SVEELMKDHQTLLNGFSVFLS 155
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,640,023
Number of Sequences: 26719
Number of extensions: 733603
Number of successful extensions: 5286
Number of sequences better than 10.0: 198
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 101
Number of HSP's that attempted gapping in prelim test: 3485
Number of HSP's gapped (non-prelim): 827
length of query: 670
length of database: 11,318,596
effective HSP length: 106
effective length of query: 564
effective length of database: 8,486,382
effective search space: 4786319448
effective search space used: 4786319448
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146588.6


