

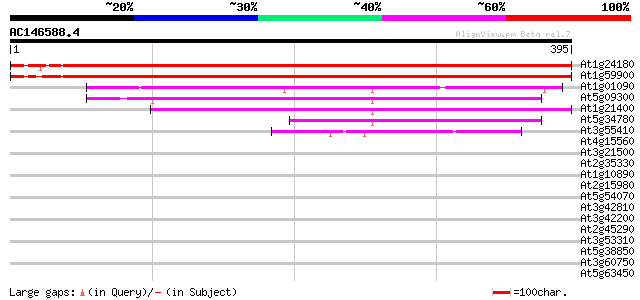
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.4 - phase: 0
(395 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g24180 pyruvate dehydrogenase E1 alpha subunit 627 e-180
At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative 624 e-179
At1g01090 pyruvate dehydrogenase E1 alpha subunit 237 1e-62
At5g09300 branched-chain alpha keto-acid dehydrogenase E1 alpha ... 149 3e-36
At1g21400 branched-chain alpha keto-acid dehydrogenase E1 alpha ... 145 3e-35
At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein 96 3e-20
At3g55410 2-oxoglutarate dehydrogenase, E1 subunit - like protein 44 1e-04
At4g15560 1-D-deoxyxylulose 5-phosphate synthase, putative 41 0.001
At3g21500 1-D-deoxyxylulose 5-phosphate synthase, putative 37 0.014
At2g35330 unknown protein 34 0.12
At1g10890 unknown protein 32 0.47
At2g15980 F7H1.1 32 0.61
At5g54070 putative protein 32 0.79
At3g42810 putative protein 31 1.0
At3g42200 putative protein 31 1.0
At2g45290 putative transketolase precursor 31 1.4
At3g53310 unknown protein 30 1.8
At5g38850 disease resistance protein - like 30 2.3
At3g60750 transketolase - like protein 30 2.3
At5g63450 cytochrome P450-like protein 30 3.0
>At1g24180 pyruvate dehydrogenase E1 alpha subunit
Length = 393
Score = 627 bits (1617), Expect = e-180
Identities = 309/397 (77%), Positives = 348/397 (86%), Gaps = 6/397 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYF--LSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTV 58
MALSR SS +T LKP L S+IR R +S+ S+ +T+ET++PFTSH CE PS +V
Sbjct: 1 MALSRLSSRS--NTFLKPAITALPSSIR-RHVSTDSSP-ITIETAVPFTSHLCESPSRSV 56
Query: 59 QTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCV 118
+T++ E+++FF DM MRRMEIAADSLYKAKLIRGFCHLYDGQEA+AVGMEAAI +KD +
Sbjct: 57 ETSSEEILAFFRDMARMRRMEIAADSLYKAKLIRGFCHLYDGQEALAVGMEAAITKKDAI 116
Query: 119 ITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIP 178
IT+YRDHCTF+ RGG LV+ FSELMGRK GCS GKGGSMHFY+K+ FYGGHGIVGAQIP
Sbjct: 117 ITSYRDHCTFIGRGGKLVDAFSELMGRKTGCSHGKGGSMHFYKKDASFYGGHGIVGAQIP 176
Query: 179 LGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTA 238
LG GLAF QKYNKD VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGTA
Sbjct: 177 LGCGLAFAQKYNKDEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTA 236
Query: 239 EWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHS 298
WRSAKSPAY+KRGDY PGLKVDGMD LAVKQACKFAKEHALKNGP+ILEMDTYRYHGHS
Sbjct: 237 TWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGPIILEMDTYRYHGHS 296
Query: 299 MSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKA 358
MSDPGSTYRTRDEISGVRQ RDPIERVRKL+L HDI+TEKELKD+EKE RK+VD+A+A+A
Sbjct: 297 MSDPGSTYRTRDEISGVRQVRDPIERVRKLLLTHDIATEKELKDMEKEIRKEVDDAVAQA 356
Query: 359 KESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
KES +PD S+LFTN+YVK GVE+ GADRKE+K TLP
Sbjct: 357 KESPIPDASELFTNMYVKDCGVESFGADRKELKVTLP 393
>At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative
Length = 389
Score = 624 bits (1610), Expect = e-179
Identities = 302/395 (76%), Positives = 347/395 (87%), Gaps = 6/395 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 60
MALSR SS + + +P+ SA R IS+ +T +T+ETS+PFT+H C+PPS +V++
Sbjct: 1 MALSRLSSRS--NIITRPF---SAAFSRLISTDTTP-ITIETSLPFTAHLCDPPSRSVES 54
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
++ EL+ FF M LMRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEAAI +KD +IT
Sbjct: 55 SSQELLDFFRTMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKDAIIT 114
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
AYRDHC FL RGG+L EVFSELMGR+ GCSKGKGGSMHFY+KE FYGGHGIVGAQ+PLG
Sbjct: 115 AYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQVPLG 174
Query: 181 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 240
G+AF QKYNK+ VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGTAEW
Sbjct: 175 CGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTAEW 234
Query: 241 RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS 300
R+AKSP+YYKRGDY PGLKVDGMD AVKQACKFAK+HAL+ GP+ILEMDTYRYHGHSMS
Sbjct: 235 RAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGPIILEMDTYRYHGHSMS 294
Query: 301 DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE 360
DPGSTYRTRDEISGVRQERDPIER++KLVL+HD++TEKELKD+EKE RK+VD+AIAKAK+
Sbjct: 295 DPGSTYRTRDEISGVRQERDPIERIKKLVLSHDLATEKELKDMEKEIRKEVDDAIAKAKD 354
Query: 361 SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
MP+PS+LFTNVYVKG G E+ G DRKEVKA+LP
Sbjct: 355 CPMPEPSELFTNVYVKGFGTESFGPDRKEVKASLP 389
>At1g01090 pyruvate dehydrogenase E1 alpha subunit
Length = 428
Score = 237 bits (604), Expect = 1e-62
Identities = 136/348 (39%), Positives = 195/348 (55%), Gaps = 17/348 (4%)
Query: 55 STTVQTTASELMSFFNDMVLMRRME-IAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAIN 113
+T++ T E + + DM+L R E + A Y+ K+ GF HLY+GQEAV+ G +
Sbjct: 74 NTSLLITKEEGLELYEDMILGRSFEDMCAQMYYRGKMF-GFVHLYNGQEAVSTGFIKLLT 132
Query: 114 RKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIV 173
+ D V++ YRDH L +G + V SEL G+ GC +G+GGSMH + KE GG +
Sbjct: 133 KSDSVVSTYRDHVHALSKGVSARAVMSELFGKVTGCCRGQGGSMHMFSKEHNMLGGFAFI 192
Query: 174 GAQIPLGVGLAFGQKYNKD------PNVTFTLYGDGAANQGQLFEALNIAALWDLPAILV 227
G IP+ G AF KY ++ +VT +GDG N GQ FE LN+AAL+ LP I V
Sbjct: 193 GEGIPVATGAAFSSKYRREVLKQDCDDVTVAFFGDGTCNNGQFFECLNMAALYKLPIIFV 252
Query: 228 CENNHYGMGTAEWRSAKSPAYYKRGDY--APGLKVDGMDVLAVKQACKFAKEHALK-NGP 284
ENN + +G + R+ P +K+G PG+ VDGMDVL V++ K A A + GP
Sbjct: 253 VENNLWAIGMSHLRATSDPEIWKKGPAFGMPGVHVDGMDVLKVREVAKEAVTRARRGEGP 312
Query: 285 LILEMDTYRYHGHSMSDPGSTYRTRDEISGVR-QERDPIERVRKLVLAHDISTEKELKDI 343
++E +TYR+ GHS++DP RD + RDPI ++K ++ + ++ E ELK I
Sbjct: 313 TLVECETYRFRGHSLADPD---ELRDAAEKAKYAARDPIAALKKYLIENKLAKEAELKSI 369
Query: 344 EKEARKQVDEAIAKAKESQMPDPSDLFTNVYV--KGLGVEACGADRKE 389
EK+ + V+EA+ A S P S L NV+ KG G+ G R E
Sbjct: 370 EKKIDELVEEAVEFADASPQPGRSQLLENVFADPKGFGIGPDGRYRCE 417
>At5g09300 branched-chain alpha keto-acid dehydrogenase E1 alpha
subunit-like protein
Length = 472
Score = 149 bits (375), Expect = 3e-36
Identities = 91/326 (27%), Positives = 164/326 (49%), Gaps = 10/326 (3%)
Query: 55 STTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYD---GQEAVAVGMEAA 111
S VQ + + ++DMV ++ M D+++ +G Y G+EA+ + AA
Sbjct: 120 SQFVQVSEEVAVKIYSDMVTLQIM----DNIFYEAQRQGRLSFYATAIGEEAINIASAAA 175
Query: 112 INRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHG 171
+ +D + YR+ L RG TL E ++ G K KG+ +H+ + ++
Sbjct: 176 LTPQDVIFPQYREPGVLLWRGFTLQEFANQCFGNKSDYGKGRQMPVHYGSNKLNYFTVSA 235
Query: 172 IVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENN 231
+ Q+P VG A+ K +K T +GDG ++G ALNIAA+ + P + +C NN
Sbjct: 236 TIATQLPNAVGAAYSLKMDKKDACAVTYFGDGGTSEGDFHAALNIAAVMEAPVLFICRNN 295
Query: 232 HYGMGTAEWRSAKSPAYYKRGDY--APGLKVDGMDVLAVKQACKFAKEHALK-NGPLILE 288
+ + T +S +G ++VDG D LA+ A A+E A++ P+++E
Sbjct: 296 GWAISTPTSDQFRSDGVVVKGRAYGIRSIRVDGNDALAMYSAVHTAREMAIREQRPILIE 355
Query: 289 MDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEAR 348
TYR HS SD + YR+ EI + R+P+ R R + ++ ++K D+ +
Sbjct: 356 ALTYRVGHHSTSDDSTRYRSAGEIEWWNKARNPLSRFRTWIESNGWWSDKTESDLRSRIK 415
Query: 349 KQVDEAIAKAKESQMPDPSDLFTNVY 374
K++ EA+ A++++ P+ ++F++VY
Sbjct: 416 KEMLEALRVAEKTEKPNLQNMFSDVY 441
>At1g21400 branched-chain alpha keto-acid dehydrogenase E1 alpha
subunit
Length = 472
Score = 145 bits (367), Expect = 3e-35
Identities = 92/300 (30%), Positives = 149/300 (49%), Gaps = 4/300 (1%)
Query: 100 GQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHF 159
G+EA+ + AA++ D V+ YR+ L RG TL E ++ G K KG+ +H+
Sbjct: 164 GEEAINIASAAALSPDDVVLPQYREPGVLLWRGFTLEEFANQCFGNKADYGKGRQMPIHY 223
Query: 160 YRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAAL 219
++ + Q+P G+ + K +K T T GDG ++G LN AA+
Sbjct: 224 GSNRLNYFTISSPIATQLPQAAGVGYSLKMDKKNACTVTFIGDGGTSEGDFHAGLNFAAV 283
Query: 220 WDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDY--APGLKVDGMDVLAVKQACKFAKE 277
+ P + +C NN + + T +S +G ++VDG D LAV A + A+E
Sbjct: 284 MEAPVVFICRNNGWAISTHISEQFRSDGIVVKGQAYGIRSIRVDGNDALAVYSAVRSARE 343
Query: 278 HAL-KNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDIST 336
A+ + P+++EM TYR HS SD + YR DEI + R+P+ R RK V + +
Sbjct: 344 MAVTEQRPVLIEMMTYRVGHHSTSDDSTKYRAADEIQYWKMSRNPVNRFRKWVEDNGWWS 403
Query: 337 EKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVY-VKGLGVEACGADRKEVKATLP 395
E++ + ARKQ+ +AI A++ + ++LF +VY VK +E KE+ P
Sbjct: 404 EEDESKLRSNARKQLLQAIQAAEKWEKQPLTELFNDVYDVKPKNLEEQELGLKELVKKQP 463
>At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein
Length = 365
Score = 95.9 bits (237), Expect = 3e-20
Identities = 58/180 (32%), Positives = 93/180 (51%), Gaps = 3/180 (1%)
Query: 198 TLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDY--A 255
T GDG ++G LN AA+ + P + +C NN + + T +S +G
Sbjct: 32 TFIGDGGTSEGDFHAGLNFAAVMEAPVVFICRNNGWAISTHISEQFRSDGIVVKGQAYGI 91
Query: 256 PGLKVDGMDVLAVKQACKFAKEHAL-KNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISG 314
++VDG D LAV A A+E A+ + P+++EM YR HS SD + YR DEI
Sbjct: 92 RSIRVDGNDALAVYSAVCSAREMAVTEQRPVLIEMMIYRVGHHSTSDDSTKYRAADEIQY 151
Query: 315 VRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVY 374
+ R+ + R RK V + +E++ + ARKQ+ +AI A++ + ++LF +VY
Sbjct: 152 WKMSRNSVNRFRKSVEDNGWWSEEDESKLRSNARKQLLQAIQAAEKWEKQPLTELFNDVY 211
>At3g55410 2-oxoglutarate dehydrogenase, E1 subunit - like protein
Length = 1017
Score = 44.3 bits (103), Expect = 1e-04
Identities = 41/185 (22%), Positives = 89/185 (47%), Gaps = 12/185 (6%)
Query: 185 FGQKYNKDPNVTFTLYGDGA-ANQGQLFEALNIAALWDLPA---ILVCENNHYGMGTAEW 240
+ ++ N+ ++GDG+ A QG ++E L+++AL + I + NN T +
Sbjct: 382 YSNDLDRTKNLGILIHGDGSFAGQGVVYETLHLSALPNYTTGGTIHIVVNNQVAF-TTDP 440
Query: 241 RSAKSPAY---YKRGDYAPGLKVDGMDVLAVKQACKFAKE-HALKNGPLILEMDTYRYHG 296
R+ +S Y + AP V+G DV AV AC+ A E + +++++ YR G
Sbjct: 441 RAGRSSQYCTDVAKALSAPIFHVNGDDVEAVVHACELAAEWRQTFHSDVVVDLVCYRRFG 500
Query: 297 HS-MSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAI 355
H+ + +P T ++ ++ ++ K +L +++++ I+++ ++E
Sbjct: 501 HNEIDEPSFTQPKMYKV--IKNHPSTLQIYHKKLLECGEVSQQDIDRIQEKVNTILNEEF 558
Query: 356 AKAKE 360
+K+
Sbjct: 559 VASKD 563
>At4g15560 1-D-deoxyxylulose 5-phosphate synthase, putative
Length = 717
Score = 40.8 bits (94), Expect = 0.001
Identities = 27/92 (29%), Positives = 38/92 (40%), Gaps = 12/92 (13%)
Query: 152 GKGGSMHFYRKEGGFYG------------GHGIVGAQIPLGVGLAFGQKYNKDPNVTFTL 199
G+ G M R+ G G G G I G+G+A G+ N +
Sbjct: 156 GRRGKMPTMRQTNGLSGFTKRGESEHDCFGTGHSSTTISAGLGMAVGRDLKGKNNNVVAV 215
Query: 200 YGDGAANQGQLFEALNIAALWDLPAILVCENN 231
GDGA GQ +EA+N A D I++ +N
Sbjct: 216 IGDGAMTAGQAYEAMNNAGYLDSDMIVILNDN 247
>At3g21500 1-D-deoxyxylulose 5-phosphate synthase, putative
Length = 628
Score = 37.4 bits (85), Expect = 0.014
Identities = 25/92 (27%), Positives = 38/92 (41%), Gaps = 12/92 (13%)
Query: 152 GKGGSMHFYRKEGGFYG------------GHGIVGAQIPLGVGLAFGQKYNKDPNVTFTL 199
G+ G M R+ G G G G + G+G+A G+ N ++
Sbjct: 134 GRRGKMKTIRQTNGLSGYTKRRESEHDSFGTGHSSTTLSAGLGMAVGRDLKGMNNSVVSV 193
Query: 200 YGDGAANQGQLFEALNIAALWDLPAILVCENN 231
GDGA GQ +EA+N A I++ +N
Sbjct: 194 IGDGAMTAGQAYEAMNNAGYLHSNMIVILNDN 225
>At2g35330 unknown protein
Length = 738
Score = 34.3 bits (77), Expect = 0.12
Identities = 21/57 (36%), Positives = 35/57 (60%), Gaps = 5/57 (8%)
Query: 311 EISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARK---QVDE--AIAKAKESQ 362
E+ +R ER+ I+RV+K + ST K+L ++E RK QVD+ A+ +A E++
Sbjct: 436 ELKSLRSEREEIQRVKKGKQTREDSTLKKLSEMENALRKASGQVDKANAVVRALENE 492
>At1g10890 unknown protein
Length = 592
Score = 32.3 bits (72), Expect = 0.47
Identities = 21/62 (33%), Positives = 32/62 (50%), Gaps = 7/62 (11%)
Query: 302 PGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKES 361
P +T + +G + +R+ ER R+ E ELK IE+E K+V+EAI K E
Sbjct: 48 PAATLESAKNRNGEKLKREEEERKRR-------QREAELKLIEEETVKRVEEAIRKKVEE 100
Query: 362 QM 363
+
Sbjct: 101 SL 102
>At2g15980 F7H1.1
Length = 498
Score = 32.0 bits (71), Expect = 0.61
Identities = 36/111 (32%), Positives = 45/111 (40%), Gaps = 24/111 (21%)
Query: 75 MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITA--------YRDHC 126
M+R AD L L+ G C DGQ V EAA KD V A Y
Sbjct: 377 MKRKGFEADGLTIEALVEGLCDDRDGQRVV----EAADIVKDAVREAMFYPSRNCYELLV 432
Query: 127 TFLCRGGTL---VEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVG 174
LC G + + + +E++G KG S YR F G+GIVG
Sbjct: 433 KRLCEDGKMDRALNIQAEMVG------KGFKPSQETYR---AFIDGYGIVG 474
>At5g54070 putative protein
Length = 331
Score = 31.6 bits (70), Expect = 0.79
Identities = 19/58 (32%), Positives = 29/58 (49%), Gaps = 7/58 (12%)
Query: 315 VRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTN 372
+++E + E V+KL L D T+K L D+E+ E +A A P+P L N
Sbjct: 251 IQRELEAAEFVKKLKLLQDQETQKNLLDVER-------EFMAMAATEHNPEPDILVNN 301
>At3g42810 putative protein
Length = 154
Score = 31.2 bits (69), Expect = 1.0
Identities = 24/90 (26%), Positives = 47/90 (51%), Gaps = 9/90 (10%)
Query: 272 CKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLA 331
C F +H L+N + T+++ ++ + T D+ + QE + R ++V
Sbjct: 51 CGFTAKHRLRN-----DEHTFKWVDEALLNEIETLI--DKTREIEQELKEL-RTERMVFE 102
Query: 332 HDISTEKELKDIEKEARKQVDEAIAKAKES 361
+S + E+K IE E ++V+EA+++AK S
Sbjct: 103 RMVSEKLEIK-IENEVLEKVEEALSEAKAS 131
>At3g42200 putative protein
Length = 134
Score = 31.2 bits (69), Expect = 1.0
Identities = 25/92 (27%), Positives = 47/92 (50%), Gaps = 9/92 (9%)
Query: 272 CKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLA 331
C FA +H L+N + T+++ ++ + T D+ QE R ++V
Sbjct: 51 CGFAAKHRLRN-----DEHTFKWVDKALLNEIETLT--DKTREFEQELKE-HRTERMVFE 102
Query: 332 HDISTEKELKDIEKEARKQVDEAIAKAKESQM 363
+S + E+K IE E ++V+EA+++AK S +
Sbjct: 103 RMVSEKLEIK-IENEVLEKVEEALSEAKASSI 133
>At2g45290 putative transketolase precursor
Length = 741
Score = 30.8 bits (68), Expect = 1.4
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 11/73 (15%)
Query: 171 GIVGAQIPLGVGLAFGQK-----YNKDPN-----VTFTLYGDGAANQGQLFEALNIAALW 220
G +G I VGLA +K +NK N T+++ GDG +G E ++A W
Sbjct: 192 GPLGQGIANAVGLALAEKHLAARFNKPDNEIVDHYTYSILGDGCQMEGISNEVCSLAGHW 251
Query: 221 DLPAILV-CENNH 232
L ++ ++NH
Sbjct: 252 GLGKLIAFYDDNH 264
>At3g53310 unknown protein
Length = 286
Score = 30.4 bits (67), Expect = 1.8
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 19/122 (15%)
Query: 273 KFAKEHALKNGPLILEMDTYRYHGH-----SMSDPGSTYRTRDEISGV---RQERDPIER 324
KFA+EH LKNG E T+ Y GH S+ D + R EI + + D +
Sbjct: 71 KFAEEHELKNG----EFMTFVYDGHRTFEVSVFDRWGSKEVRAEIQAIPLSDSDSDSVVE 126
Query: 325 VRK--LVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTN--VYVKGLGV 380
K + D E E +D + + DE I+ +S P + T+ V+ L V
Sbjct: 127 DEKDSTDVVEDDDDEDEDEDEDDDGSFDEDEEIS---QSLYPIDEETATDAAVFEGNLDV 183
Query: 381 EA 382
EA
Sbjct: 184 EA 185
>At5g38850 disease resistance protein - like
Length = 986
Score = 30.0 bits (66), Expect = 2.3
Identities = 14/44 (31%), Positives = 28/44 (62%)
Query: 73 VLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKD 116
+L+RR+EI+ D +A+L G+ L++ ++A+ + + N KD
Sbjct: 394 ILIRRLEISLDRDNEAQLRVGYDSLHENEQALFLSIAVFFNYKD 437
>At3g60750 transketolase - like protein
Length = 741
Score = 30.0 bits (66), Expect = 2.3
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 11/73 (15%)
Query: 171 GIVGAQIPLGVGLAFGQK-----YNKDP-----NVTFTLYGDGAANQGQLFEALNIAALW 220
G +G I VGLA +K +NK + T+ + GDG +G EA ++A W
Sbjct: 192 GPLGQGIANAVGLALAEKHLAARFNKPDAEVVDHYTYAILGDGCQMEGISNEACSLAGHW 251
Query: 221 DLPAILV-CENNH 232
L ++ ++NH
Sbjct: 252 GLGKLIAFYDDNH 264
>At5g63450 cytochrome P450-like protein
Length = 510
Score = 29.6 bits (65), Expect = 3.0
Identities = 23/77 (29%), Positives = 35/77 (44%), Gaps = 6/77 (7%)
Query: 311 EISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKES-----QMPD 365
EIS R+ +P+ V K+ ++ +EK L++ K V E I K+S + D
Sbjct: 219 EISA-RRATEPVYAVWKVKRFLNVGSEKRLREAIKTVHLSVSEIIRAKKKSLDIGGDVSD 277
Query: 366 PSDLFTNVYVKGLGVEA 382
DL + G G EA
Sbjct: 278 KQDLLSRFLAAGHGEEA 294
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,962,990
Number of Sequences: 26719
Number of extensions: 397465
Number of successful extensions: 1305
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1278
Number of HSP's gapped (non-prelim): 31
length of query: 395
length of database: 11,318,596
effective HSP length: 101
effective length of query: 294
effective length of database: 8,619,977
effective search space: 2534273238
effective search space used: 2534273238
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146588.4


