

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146587.10 + phase: 0 /pseudo
(1157 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
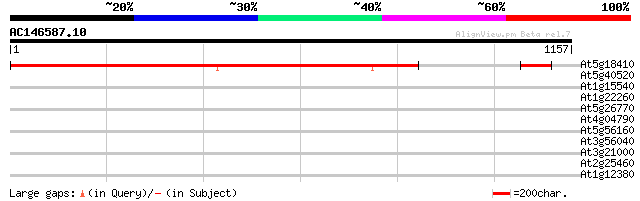
Score E
Sequences producing significant alignments: (bits) Value
At5g18410 putative protein 1422 0.0
At5g40520 putative protein 34 0.55
At1g15540 hypothetical protein 32 1.6
At1g22260 hypothetical protein 32 2.1
At5g26770 unknown protein 32 2.7
At4g04790 hypothetical protein 31 4.7
At5g56160 putative protein 30 8.0
At3g56040 unknown protein 30 8.0
At3g21000 unknown protein 30 8.0
At2g25460 SYNC1 protein 30 8.0
At1g12380 hypothetical protein 30 8.0
>At5g18410 putative protein
Length = 1235
Score = 1422 bits (3681), Expect = 0.0
Identities = 713/854 (83%), Positives = 774/854 (90%), Gaps = 13/854 (1%)
Query: 1 MAVPVEEAIAALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKA 60
MAVPVEEAIAALSTFSLEDEQPEVQGP V VS ER AT+SPIEY DVAAYRLSLSEDTKA
Sbjct: 1 MAVPVEEAIAALSTFSLEDEQPEVQGPAVMVSAERAATDSPIEYSDVAAYRLSLSEDTKA 60
Query: 61 LNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSRLREI 120
LNQL++L QEGKEMAS+LYTYRSCVKALPQLP+SMK SQADLYLETYQVLDLEMSRLREI
Sbjct: 61 LNQLNTLIQEGKEMASILYTYRSCVKALPQLPESMKHSQADLYLETYQVLDLEMSRLREI 120
Query: 121 QRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASIPNDF 180
QRWQ+SAS+KLA DMQRFSRPERRINGPT++HLWSMLKL DVLVQLDHLKNAKASIPNDF
Sbjct: 121 QRWQSSASAKLAADMQRFSRPERRINGPTVTHLWSMLKLLDVLVQLDHLKNAKASIPNDF 180
Query: 181 SWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLLHIFS 240
SWYKRTFTQVS QWQDTD+MREELDDLQIFLSTRWAILLNLHVEMFRVNKYP + I
Sbjct: 181 SWYKRTFTQVSAQWQDTDTMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPFFQTFITI 240
Query: 241 LSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESLYKRVKI 300
+ VEDILQVLIVF VESLELDFALLFPER+ILLRVLPVLVVL T SEKD+E+LYKRVK+
Sbjct: 241 VWLVEDILQVLIVFIVESLELDFALLFPERYILLRVLPVLVVLATPSEKDTEALYKRVKL 300
Query: 301 NRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPPLNHIGA 360
NRLINIFKN+ VIPAFPDLHLSPAAI+KELS YF KFSSQTRLLTL APHELPPLNHIGA
Sbjct: 301 NRLINIFKNDPVIPAFPDLHLSPAAILKELSVYFQKFSSQTRLLTLPAPHELPPLNHIGA 360
Query: 361 VRAEHDDFTIRFASAMNQSTDGSDVDWSKEVKGNMYDMIVEGFQLLSRWTARIWEQCAWK 420
+RAEHDDFTIRFAS+MNQS DG+ +W +EVKGNMYDM+VEGFQLLSRWTARIWEQCAWK
Sbjct: 361 LRAEHDDFTIRFASSMNQSNDGAYTEWCREVKGNMYDMVVEGFQLLSRWTARIWEQCAWK 420
Query: 421 FSRPCKD------ASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQRCDTLVAD 474
FSRPC+D AS S+SDYEKVVR+NY+AEERKALVELV IKSVGSM+QRCDTLVAD
Sbjct: 421 FSRPCRDAGETPEASGSYSDYEKVVRFNYTAEERKALVELVGYIKSVGSMLQRCDTLVAD 480
Query: 475 ALWETIHAEVQDFVQNTLASMLRTTFRKKKDLSRILSDMRTLSADWMANTNKSESELQSS 534
ALWETIHAEVQDFVQNTLA+MLRTTFRKKKDLSRILSDMRTLSADWMANT + E E+ SS
Sbjct: 481 ALWETIHAEVQDFVQNTLATMLRTTFRKKKDLSRILSDMRTLSADWMANT-RPEHEMPSS 539
Query: 535 QHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVVSGGNLRRPGGLFGNSGSEVPVNDLK 594
QHG +ES+ N FYPR VAPTAAQVHCLQFLIYEVVSGGNLRRPGG FGN+GSE+P+ L
Sbjct: 540 QHGNDESRGNFFYPRPVAPTAAQVHCLQFLIYEVVSGGNLRRPGGFFGNNGSEIPLCKLT 599
Query: 595 QLETFVMLAVTVATLTDLGFLWFREFYLESSRVIQFPIECSLPWMLVDCVLESPNSGLLE 654
F M A ++ LTDLGFLWFREFYLESSRVIQFPIECSLPWML+D +LE+ NSGLLE
Sbjct: 600 PQLQFTMAAASIGILTDLGFLWFREFYLESSRVIQFPIECSLPWMLIDYILEAQNSGLLE 659
Query: 655 SVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDIFVARLCETIFTYYKSWAASEL 714
SVL+PFDIYNDSAQQALV+L+QRFLYDEIEAEVDH FDIFV+RL E+IFTYYKSW+ASEL
Sbjct: 660 SVLLPFDIYNDSAQQALVVLRQRFLYDEIEAEVDHGFDIFVSRLSESIFTYYKSWSASEL 719
Query: 715 LDPTFLFASENAEKYAVQPMRLNMLLKMTRVK------ELEKLLDVLKHSHELLSIDLSV 768
LDP+FLFA +N EK+++QP+R L KMT+VK ELEKL+D+LKHSHELLS DLS+
Sbjct: 720 LDPSFLFALDNGEKFSIQPVRFTALFKMTKVKDLCAVVELEKLIDILKHSHELLSQDLSI 779
Query: 769 DSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSSKTVPVQKP 828
D FSLMLNEMQENISLVSFSSRLA+QIWSEMQSDFLPNFILCNTTQRF+RSSK P QKP
Sbjct: 780 DPFSLMLNEMQENISLVSFSSRLATQIWSEMQSDFLPNFILCNTTQRFVRSSKVPPTQKP 839
Query: 829 SIPSAKPSFYCGTQ 842
S+PSAKPSFYCGTQ
Sbjct: 840 SVPSAKPSFYCGTQ 853
Score = 97.8 bits (242), Expect = 3e-20
Identities = 45/64 (70%), Positives = 51/64 (79%)
Query: 1053 GYLEESAQVQSSSPERLGDSVAWGGCTIIYLLGQQLHFELFDFSYQILNIAEVEAASVVQ 1112
GYLEE QS+ E LGDS+AWGGCTIIYLLGQQLHFELFDFSYQ+LN++EVE S
Sbjct: 1101 GYLEEITAPQSAQHEVLGDSIAWGGCTIIYLLGQQLHFELFDFSYQVLNVSEVETVSASH 1160
Query: 1113 TQKN 1116
T +N
Sbjct: 1161 THRN 1164
>At5g40520 putative protein
Length = 693
Score = 33.9 bits (76), Expect = 0.55
Identities = 43/178 (24%), Positives = 76/178 (42%), Gaps = 26/178 (14%)
Query: 59 KALNQLSSL-TQEGKEMASVLYTYRSCVKALPQLPDSMK-QSQADLYLETYQVLDLEMSR 116
K LN+L SL TQ + M L R +PQL +S + Q ++DL + Q + +S
Sbjct: 197 KVLNELDSLSTQTLRAMKRKLKGSRM----IPQLKNSRRGQRRSDLINQVRQASEKMLSE 252
Query: 117 LREIQRWQASASSKL-----------------ATDMQRFSRPERRINGPTISHLWSMLKL 159
L + Q + L ATD RFS + + + +W + K+
Sbjct: 253 LSAGDKLQEQLAKALSVVDLSLKLSPGYKTAAATDFFRFSPETKNLQNEIVKAVWLLRKV 312
Query: 160 -FDVLVQLDHLKNAKASIPNDF--SWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTR 214
F L +L + +A + ND S ++T + + D D++ + L + +++R
Sbjct: 313 RFRELKRLHLCLDPEAEVSNDSLRSAVRKTLIEYLFECSDLDTIPKSLMEALSLVNSR 370
>At1g15540 hypothetical protein
Length = 320
Score = 32.3 bits (72), Expect = 1.6
Identities = 29/91 (31%), Positives = 39/91 (41%), Gaps = 5/91 (5%)
Query: 697 RLCETIFTYYKSWAASELLDPTFLFASENAEKYAVQ----PMRLNMLLKMTRVKELEKLL 752
R CET+ Y K A E L LF S N EKY + L LLK R+ E
Sbjct: 127 RFCETVHAYAKMQAELEQLVIRMLFESYNVEKYTEKYIGGTRYLLRLLKYRRLPNGEPNR 186
Query: 753 DVLKHSHE-LLSIDLSVDSFSLMLNEMQENI 782
+ H+ + +SI LML +E++
Sbjct: 187 KFISHTDKSFISILHQNHITGLMLKSEKEDV 217
>At1g22260 hypothetical protein
Length = 851
Score = 32.0 bits (71), Expect = 2.1
Identities = 34/148 (22%), Positives = 65/148 (42%), Gaps = 7/148 (4%)
Query: 17 LEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKALNQLSSLTQEGKEMAS 76
LE+ + E Q ++ TER T S IE D +L S + LN + +L + +++
Sbjct: 181 LEELKLEKQQKEMFYQTERCGTASLIEKKDAVITKLEASAAERKLN-IENLNSQLEKVHL 239
Query: 77 VLYTYRSCVKALPQLPDSMK------QSQADLYLETYQVLDLEMSRLREIQRWQASASSK 130
L T VK L + + ++ Q AD E + E+ +L E+ ++ + ++
Sbjct: 240 ELTTKEDEVKDLVSIQEKLEKEKTSVQLSADNCFEKLVSSEQEVKKLDELVQYLVAELTE 299
Query: 131 LATDMQRFSRPERRINGPTISHLWSMLK 158
L F +++G +H+ + K
Sbjct: 300 LDKKNLTFKEKFDKLSGLYDTHIMLLQK 327
>At5g26770 unknown protein
Length = 335
Score = 31.6 bits (70), Expect = 2.7
Identities = 11/44 (25%), Positives = 26/44 (59%)
Query: 502 KKKDLSRILSDMRTLSADWMANTNKSESELQSSQHGGEESKANI 545
K ++++++ D++ +SA W T + ES+L+ + +E K +
Sbjct: 218 KDEEIAKLKDDVKLMSAHWKLKTKELESQLERQRRADQELKKKV 261
>At4g04790 hypothetical protein
Length = 731
Score = 30.8 bits (68), Expect = 4.7
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 8/63 (12%)
Query: 427 DASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQRCDTLVADALWETIHA--EV 484
D PS + YEK+V+Y+ + E +++V + G M+ AD L +HA EV
Sbjct: 292 DVMPSSTSYEKLVKYSCDSNEVVTALDVVEKMGEAGLMIS------ADILHSLLHAIDEV 345
Query: 485 QDF 487
+F
Sbjct: 346 LEF 348
>At5g56160 putative protein
Length = 592
Score = 30.0 bits (66), Expect = 8.0
Identities = 19/75 (25%), Positives = 34/75 (45%), Gaps = 2/75 (2%)
Query: 88 LPQLPDSMKQSQADLYLETYQVLDLEMSRL--REIQRWQASASSKLATDMQRFSRPERRI 145
+P L D S D ++ T + ++ S+L + + A+ S + + RF +I
Sbjct: 405 VPPLSDKASTSDGDKFITTVESIESAQSQLLDADTENTFANTSVREGGQILRFGALREKI 464
Query: 146 NGPTISHLWSMLKLF 160
N I HL +L +F
Sbjct: 465 NSENIFHLVKILLVF 479
>At3g56040 unknown protein
Length = 883
Score = 30.0 bits (66), Expect = 8.0
Identities = 27/82 (32%), Positives = 36/82 (42%), Gaps = 2/82 (2%)
Query: 494 SMLRTTFRKKKDLSRILSDMRTLSADWMANTNKSESELQSSQHGGEESKANIFYPRAVAP 553
S + T K D+S L AD+ ANTN +L S++ G S A P V
Sbjct: 519 SCIEYTEFDKFDISNRSPSSNGLQADFPANTNILYVDLHSAELIGSSSNAKSL-PNMVLN 577
Query: 554 TAAQVHCL-QFLIYEVVSGGNL 574
T ++ L Q+ Y V GG L
Sbjct: 578 TKKRIEYLDQYGDYHSVMGGRL 599
>At3g21000 unknown protein
Length = 405
Score = 30.0 bits (66), Expect = 8.0
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Query: 158 KLFDVLVQLDHLKNAKASIPNDFSWYKRTFTQVSGQWQDTDSMREELDDL 207
K ++L +L K K+ D+ K FT +SG + DSM EEL D+
Sbjct: 143 KALEILERLGRAKLEKS----DYEICKNVFTTLSGSFDGLDSMLEELIDV 188
>At2g25460 SYNC1 protein
Length = 423
Score = 30.0 bits (66), Expect = 8.0
Identities = 20/64 (31%), Positives = 34/64 (52%), Gaps = 1/64 (1%)
Query: 445 AEERKALVELVSCIKSVGSMMQRCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKK 504
A ++++ VE I+S GS++ + TLV + + + E DF+Q S + + KKK
Sbjct: 125 ASKQESTVERKLPIRSKGSVLSKEATLVVNVTFSEVRTEPDDFMQLGQIS-VDSAIPKKK 183
Query: 505 DLSR 508
SR
Sbjct: 184 STSR 187
>At1g12380 hypothetical protein
Length = 793
Score = 30.0 bits (66), Expect = 8.0
Identities = 25/82 (30%), Positives = 48/82 (58%), Gaps = 13/82 (15%)
Query: 679 LYDEIEAEV---DHCFDI---FVARLCETIF--TYYKSWAASELLDPTFLFASENAEKYA 730
L+DE+ A+V D F++ V ++ E F +Y+ +WAA+ +LDP +L +++ KY
Sbjct: 563 LWDELRAKVKDWDSKFNVGEGHVEKVVERRFKKSYHPAWAAAFILDPLYLI-RDSSGKY- 620
Query: 731 VQPMRLNMLLKMTRVKELEKLL 752
+ P + L + K+++KL+
Sbjct: 621 LPPFK---CLSPEQEKDVDKLI 639
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.331 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,067,396
Number of Sequences: 26719
Number of extensions: 967003
Number of successful extensions: 2986
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 2979
Number of HSP's gapped (non-prelim): 12
length of query: 1157
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1047
effective length of database: 8,379,506
effective search space: 8773342782
effective search space used: 8773342782
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146587.10


