

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.12 - phase: 0 /pseudo
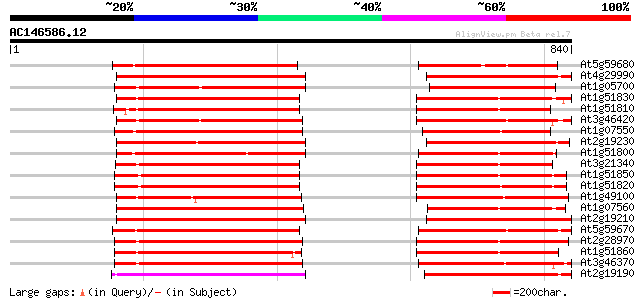
(840 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59680 serine/threonine-specific protein kinase - like 279 4e-75
At4g29990 serine/threonine-specific receptor protein kinase LRRPK 273 4e-73
At1g05700 putative light repressible receptor protein kinase 267 2e-71
At1g51830 Putative protein kinase 251 1e-66
At1g51810 Putative protein kinase 251 1e-66
At3g46420 hypothetical protein 250 3e-66
At1g07550 protein kinase like 247 2e-65
At2g19230 putative receptor-like protein kinase 247 2e-65
At1g51800 unknown protein 246 3e-65
At3g21340 serine/threonine-specific protein kinase, putative 243 4e-64
At1g51850 Putative protein kinase 241 9e-64
At1g51820 Putative protein kinase 241 2e-63
At1g49100 putative receptor-like protein kinase gb|AAD15465.1 241 2e-63
At1g07560 protein kinase, putative 241 2e-63
At2g19210 putative receptor-like protein kinase 240 2e-63
At5g59670 serine/threonine-specific protein kinase - like 238 1e-62
At2g28970 putative receptor-like protein kinase 235 9e-62
At1g51860 Putative protein kinase 233 3e-61
At3g46370 receptor-like protein kinase homolog 232 7e-61
At2g19190 putative receptor-like protein kinase 231 1e-60
>At5g59680 serine/threonine-specific protein kinase - like
Length = 882
Score = 279 bits (714), Expect = 4e-75
Identities = 139/279 (49%), Positives = 186/279 (65%), Gaps = 4/279 (1%)
Query: 155 KLTSHLQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASA 214
K S L+Y DD+YDR W S+ +W +++T+ +D N YKPP + ++TAA P NASA
Sbjct: 202 KSDSSLRYPDDIYDRQWTSFFDTEWTQINTT--SDVGNSNDYKPPKVALTTAAIPTNASA 259
Query: 215 PLQFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFS 274
PL WSS N ++QYY+Y HF+E++EL ANETREFN+ +N KL+FGPV P I S
Sbjct: 260 PLTNEWSSVNPDEQYYVYAHFSEIQELQANETREFNMLLNGKLFFGPVVPPKLAISTILS 319
Query: 275 TEPLR-RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGV 333
P + L +T STLPP+LNA+E+Y F QLET + DV + NI+ Y +
Sbjct: 320 VSPNTCEGGECNLQLIRTNRSTLPPLLNAYEVYKVIQFPQLETNETDVSAVKNIQATYEL 379
Query: 334 TR-NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYL 392
+R NWQ DPC P +MW+GLNCS D PPRIT+LNLSSSGLTG I+++I LT L+ L
Sbjct: 380 SRINWQSDPCVPQQFMWDGLNCSITDITTPPRITTLNLSSSGLTGTITAAIQNLTTLEKL 439
Query: 393 DLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSEL 431
DLSNN+L G +P+FL ++SL V+N+ N+L G +P L
Sbjct: 440 DLSNNNLTGEVPEFLSNMKSLLVINLSGNDLNGTIPQSL 478
Score = 188 bits (478), Expect = 9e-48
Identities = 99/208 (47%), Positives = 132/208 (62%), Gaps = 6/208 (2%)
Query: 613 NFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENM 672
N + +L ++NW RL IA +AA GL+YLH GC PP +HRD+K +NILLDE+
Sbjct: 658 NGDLRQHLSGKGGKPIVNWGTRLRIAAEAALGLEYLHIGCTPPMVHRDVKTTNILLDEHY 717
Query: 673 HAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELIT 732
AK+ADFGLSR+F +SH+ST AGT GY+DP ++K+D+YSFGIVLLE+IT
Sbjct: 718 KAKLADFGLSRSFPVGGESHVSTVIAGTPGYLDP-----DRLSEKSDVYSFGIVLLEMIT 772
Query: 733 GKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIE 792
+ + R +S HI QWV + GDI I+D +L G +D SAW+ +E+AMS P
Sbjct: 773 NQAVIDRNRRKS-HITQWVGSELNGGDIAKIMDLKLNGDYDSRSAWRALELAMSCADPTS 831
Query: 793 VERPDMSQILAELKECLSLDMVHRNNGR 820
RP MS ++ ELKECL + RN R
Sbjct: 832 ARRPTMSHVVIELKECLVSENSRRNMSR 859
>At4g29990 serine/threonine-specific receptor protein kinase LRRPK
Length = 876
Score = 273 bits (697), Expect = 4e-73
Identities = 139/284 (48%), Positives = 186/284 (64%), Gaps = 1/284 (0%)
Query: 160 LQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFH 219
++YKDD YDR+W Y+ + L+TSL D N ++P +IVM +A P N S PL+F+
Sbjct: 207 IRYKDDFYDRIWMPYKSPYQKTLNTSLTIDETNHNGFRPASIVMRSAIAPGNESNPLKFN 266
Query: 220 WSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEPLR 279
W+ ++ ++Y+Y+HF EV EL NETREF+I +ND + P Y D + +P+
Sbjct: 267 WAPDDPRSKFYIYMHFAEVRELQRNETREFDIYINDVILAENFRPFYLFTDTRSTVDPVG 326
Query: 280 RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTRNWQG 339
R + +I L +T STLPPI+NA EIY +F QL T Q DVD +T IK Y V +NWQG
Sbjct: 327 R-KMNEIVLQRTGVSTLPPIINAIEIYQINEFLQLPTDQQDVDAMTKIKFKYRVKKNWQG 385
Query: 340 DPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSL 399
DPC PV+ WEGL C D+N P+ +LNLSSSGLTG+I + + LT + LDLSNNSL
Sbjct: 386 DPCVPVDNSWEGLECLHSDNNTSPKSIALNLSSSGLTGQIDPAFANLTSINKLDLSNNSL 445
Query: 400 NGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
G +PDFL L +L LN+ N LTG +P++LLE+SK GSLSLR
Sbjct: 446 TGKVPDFLASLPNLTELNLEGNKLTGSIPAKLLEKSKDGSLSLR 489
Score = 213 bits (543), Expect = 3e-55
Identities = 101/217 (46%), Positives = 157/217 (71%), Gaps = 2/217 (0%)
Query: 624 ENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSR 683
++S +L+W ERL I++DAA GL+YLH GCKPP +HRD+KP+NILL+EN+ AKIADFGLSR
Sbjct: 661 KSSLILSWEERLQISLDAAQGLEYLHYGCKPPIVHRDVKPANILLNENLQAKIADFGLSR 720
Query: 684 AFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGE 743
+F + S +ST AGT GY+DP++ T N+K+D+YSFG+VLLE+ITGK A+ + E
Sbjct: 721 SFPVEGSSQVSTVVAGTIGYLDPEYYATRQMNEKSDVYSFGVVLLEVITGKPAIWHSRTE 780
Query: 744 SIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILA 803
S+H+ V ++ GDI+ I+D RL +F++ SAWK+ E+A++ S +RP MSQ++
Sbjct: 781 SVHLSDQVGSMLANGDIKGIVDQRLGDRFEVGSAWKITELALACASESSEQRPTMSQVVM 840
Query: 804 ELKECLSLDMVHRNNGRERAIVELTSLNIASDTIPLA 840
ELK+ + + +R++ ++ V + ++N+ ++ +P A
Sbjct: 841 ELKQSIFGRVNNRSDHKDP--VRMVTMNLDTEMVPRA 875
>At1g05700 putative light repressible receptor protein kinase
Length = 843
Score = 267 bits (682), Expect = 2e-71
Identities = 146/290 (50%), Positives = 193/290 (66%), Gaps = 10/290 (3%)
Query: 158 SHLQYKDDVYDRMWFSYELIDWRRLSTSL--NNDHLVQNIYKPPTIVMSTAATPVNASAP 215
S ++Y DDVYDR+W R ++TSL +D+ N Y ++VMSTA TP+N + P
Sbjct: 206 SPVRYDDDVYDRIWIPRNFGYCREINTSLPVTSDN---NSYSLSSLVMSTAMTPINTTRP 262
Query: 216 LQFHWSSNNENDQYYLYIHFNEVEELAA--NETREFNITVNDKLWFGPVTPIYRTPDLIF 273
+ +++ N +Y++Y+HF EVE+L+ N+TREF+I++N +P Y + F
Sbjct: 263 ITMTLENSDPNVRYFVYMHFAEVEDLSLKPNQTREFDISINGVTVAAGFSPKYLQTNTFF 322
Query: 274 STEPLRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGV 333
+ + SL +T STLPPI+NA EIY+A FSQ T Q+D D +T++K +Y V
Sbjct: 323 LNPESQSKIAF--SLVRTPKSTLPPIVNALEIYVANSFSQSLTNQEDGDAVTSLKTSYKV 380
Query: 334 TRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLD 393
+NW GDPC P +Y+WEGLNCS D PPRITSLNLSSSGLTG ISSS S LTM+Q LD
Sbjct: 381 KKNWHGDPCLPNDYIWEGLNCSYDS-LTPPRITSLNLSSSGLTGHISSSFSNLTMIQELD 439
Query: 394 LSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
LSNN L G +P+FL +L+ L+VLN+ N LTG VPSELLERS TGS SLR
Sbjct: 440 LSNNGLTGDIPEFLSKLKFLRVLNLENNTLTGSVPSELLERSNTGSFSLR 489
Score = 200 bits (508), Expect = 3e-51
Identities = 96/190 (50%), Positives = 133/190 (69%), Gaps = 1/190 (0%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W +RL IA+DAA GL+YLH GCKPP +HRD+K SNILL+E AK+ADFGLSR+F +
Sbjct: 653 LSWRQRLQIALDAAQGLEYLHCGCKPPIVHRDVKTSNILLNEKNRAKLADFGLSRSFHTE 712
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHIL 748
SH+ST AGT GY+DP T N+K+DIYSFG+VLLE+ITGK + + + +H+
Sbjct: 713 SRSHVSTLVAGTPGYLDPLCFETNGLNEKSDIYSFGVVLLEMITGKTVIKESQTKRVHVS 772
Query: 749 QWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKE 807
WV I+ D+ ++ID+++ FD+NS WKVVE+A+SS S +RP+M I+ L E
Sbjct: 773 DWVISILRSTNDVNNVIDSKMAKDFDVNSVWKVVELALSSVSQNVSDRPNMPHIVRGLNE 832
Query: 808 CLSLDMVHRN 817
CL + ++N
Sbjct: 833 CLQREESNKN 842
>At1g51830 Putative protein kinase
Length = 693
Score = 251 bits (641), Expect = 1e-66
Identities = 128/277 (46%), Positives = 177/277 (63%), Gaps = 4/277 (1%)
Query: 160 LQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFH 219
+++ DDVYDR W+ W +++T+LN + + IY+ P VMSTAATP+NA+A L
Sbjct: 10 VRFPDDVYDRKWYPIFQNSWTQVTTNLNVN--ISTIYELPQSVMSTAATPLNANATLNIT 67
Query: 220 WSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEPLR 279
W+ +Y YIHF E++ L AN+TREFN+T+N + GP +P + I P +
Sbjct: 68 WTIEPPTTPFYSYIHFAELQSLRANDTREFNVTLNGEYTIGPYSPKPLKTETIQDLSPEQ 127
Query: 280 -RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR-NW 337
+ L +T STLPP+LNA E + DF Q+ET +DDV I +++N YG+ R +W
Sbjct: 128 CNGGACILQLVETLKSTLPPLLNAIEAFTVIDFPQMETNEDDVTGINDVQNTYGLNRISW 187
Query: 338 QGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNN 397
QGDPC P Y W+GLNC+ D + PP I SL+LSSSGL G I+ I LT LQYLDLS+N
Sbjct: 188 QGDPCVPKQYSWDGLNCNNSDISIPPIIISLDLSSSGLNGVITQGIQNLTHLQYLDLSDN 247
Query: 398 SLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLER 434
+L G +P FL ++SL V+N+ NNLTG VP LL++
Sbjct: 248 NLTGDIPKFLADIQSLLVINLSGNNLTGSVPLSLLQK 284
Score = 203 bits (517), Expect = 3e-52
Identities = 103/238 (43%), Positives = 151/238 (63%), Gaps = 13/238 (5%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + ++ N +LNW RL I V++A GL+YLHNGCKP +HRD+K +NILL+
Sbjct: 461 YMANGDLKEHMSGTRNHFILNWGTRLKIVVESAQGLEYLHNGCKPLMVHRDIKTTNILLN 520
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E AK+ADFGLSR+F + ++H+ST AGT GY+DP++ RT +K+D+YSFG+VLLE
Sbjct: 521 EQFDAKLADFGLSRSFPIEGETHVSTAVAGTPGYLDPEYYRTNWLTEKSDVYSFGVVLLE 580
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
+IT + ++ E HI +WV ++ +GDI++I+D L G +D S WK VE+AM +
Sbjct: 581 IIT-NQPVIDPRREKPHIAEWVGEVLTKGDIKNIMDPSLNGDYDSTSVWKAVELAMCCLN 639
Query: 790 PIEVERPDMSQILAELKECLSLDMVHRNNGRERAIVEL-------TSLNIASDTIPLA 840
P RP+MSQ++ EL ECL+ + N R AI ++ SL ++ PLA
Sbjct: 640 PSSARRPNMSQVVIELNECLTSE-----NSRGGAIRDMDSEGSIEVSLTFGTEVTPLA 692
>At1g51810 Putative protein kinase
Length = 871
Score = 251 bits (641), Expect = 1e-66
Identities = 130/285 (45%), Positives = 186/285 (64%), Gaps = 10/285 (3%)
Query: 156 LTSHLQYKDDVYDRMW---FSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNA 212
L +++Y DDV+DR+W Y+ DW+ L+T+L + V N Y P VM TA TP+ A
Sbjct: 195 LKGYIEYPDDVHDRIWKQILPYQ--DWQILTTNLQIN--VSNDYDLPQRVMKTAVTPIKA 250
Query: 213 SAP-LQFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDL 271
S ++F W+ Q+YL++HF E++ L ANETREFN+ +N + F +P +
Sbjct: 251 STTTMEFPWNLEPPTSQFYLFLHFAELQSLQANETREFNVVLNGNVTFKSYSPKFLEMQT 310
Query: 272 IFSTEPLR-RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNA 330
++ST P + + L KT STLPP++NA E Y DF Q+ET D+V I NI++
Sbjct: 311 VYSTAPKQCDGGKCLLQLVKTSRSTLPPLINAMEAYTVLDFPQIETNVDEVIAIKNIQST 370
Query: 331 YGVTRN-WQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTML 389
YG+++ WQGDPC P ++W+GLNC+ DD+ PP ITSLNLSSSGLTG I +I L L
Sbjct: 371 YGLSKTTWQGDPCVPKKFLWDGLNCNNSDDSTPPIITSLNLSSSGLTGIIVLTIQNLANL 430
Query: 390 QYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLER 434
Q LDLSNN+L+G +P+FL ++SL V+N+ NNL+G+VP +L+E+
Sbjct: 431 QELDLSNNNLSGGVPEFLADMKSLLVINLSGNNLSGVVPQKLIEK 475
Score = 198 bits (504), Expect = 9e-51
Identities = 94/201 (46%), Positives = 140/201 (68%), Gaps = 1/201 (0%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + + ++ ++LNW RL IA++AA GL+YLHNGCKP +HRD+K +NILL+
Sbjct: 652 YMANGDLDEHMSGKRGGSILNWGTRLKIALEAAQGLEYLHNGCKPLMVHRDVKTTNILLN 711
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E+ K+ADFGLSR+F + ++H+ST AGT GY+DP++ RT +K+D+YSFG+VLL
Sbjct: 712 EHFDTKLADFGLSRSFPIEGETHVSTVVAGTIGYLDPEYYRTNWLTEKSDVYSFGVVLLV 771
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
+IT + ++ + E HI +WV ++ +GDI+SI D L G ++ S WK VE+AMS +
Sbjct: 772 MIT-NQPVIDQNREKRHIAEWVGGMLTKGDIKSITDPNLLGDYNSGSVWKAVELAMSCMN 830
Query: 790 PIEVERPDMSQILAELKECLS 810
P + RP MSQ++ ELKECL+
Sbjct: 831 PSSMTRPTMSQVVFELKECLA 851
>At3g46420 hypothetical protein
Length = 838
Score = 250 bits (638), Expect = 3e-66
Identities = 131/282 (46%), Positives = 181/282 (63%), Gaps = 6/282 (2%)
Query: 160 LQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFH 219
++Y DDVYDR+W SY +W+++ST+L + + + PP + TAA+P NASAPL
Sbjct: 207 IRYPDDVYDRIWGSYFEPEWKKISTTLGVNS--SSGFLPPLKALMTAASPANASAPLAIP 264
Query: 220 WSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEPL- 278
+ +D+ YL++HF+E++ L ANETREF I N KL + +P+Y I + P+
Sbjct: 265 GVLDFPSDKLYLFLHFSEIQVLKANETREFEIFWNKKLVYNAYSPVYLQTKTIRNPSPVT 324
Query: 279 -RRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR-N 336
R E + + KT+ STLPP+LNA E++ +F Q ET DV I NIK YG+TR
Sbjct: 325 CERGECI-LEMIKTERSTLPPLLNAVEVFTVVEFPQPETDASDVVAIKNIKAIYGLTRVT 383
Query: 337 WQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSN 396
WQGDPC P ++W GLNC++ + + PPRITSL+LSSSGLTG IS I LT L+ LDLSN
Sbjct: 384 WQGDPCVPQQFLWNGLNCNSMETSTPPRITSLDLSSSGLTGSISVVIQNLTHLEKLDLSN 443
Query: 397 NSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTG 438
N+L G +PDFL ++ L +N+ KNNL G +P L +R G
Sbjct: 444 NNLTGEVPDFLANMKFLVFINLSKNNLNGSIPKALRDRENKG 485
Score = 203 bits (517), Expect = 3e-52
Identities = 108/239 (45%), Positives = 152/239 (63%), Gaps = 15/239 (6%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + +L N +L+W+ RL IAVDAA GL+YLH GC+P +HRD+K +NILL
Sbjct: 606 YMSNGDLKHHLSGRNNGFVLSWSTRLQIAVDAALGLEYLHIGCRPSMVHRDVKSTNILLG 665
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E AK+ADFGLSR+F ++HIST AGT GY+DP++ RT +K+DIYSFGIVLLE
Sbjct: 666 EQFTAKMADFGLSRSFQIGDENHISTVVAGTPGYLDPEYYRTSRLAEKSDIYSFGIVLLE 725
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
+IT + A+ R + HI WV ++ RGDI IID LQG ++ S W+ +E+AMS +
Sbjct: 726 MITSQHAIDRTRVKH-HITDWVVSLISRGDITRIIDPNLQGNYNSRSVWRALELAMSCAN 784
Query: 790 PIEVERPDMSQILAELKECLSL--------DMVHRNNGRERAIVELTSLNIASDTIPLA 840
P +RP+MSQ++ +LKECL+ DM ++ +R S+N +D +P A
Sbjct: 785 PTSEKRPNMSQVVIDLKECLATENSTRSEKDMSSHSSDLDR------SMNFYTDMVPRA 837
>At1g07550 protein kinase like
Length = 864
Score = 247 bits (631), Expect = 2e-65
Identities = 127/285 (44%), Positives = 180/285 (62%), Gaps = 7/285 (2%)
Query: 158 SHLQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNI--YKPPTIVMSTAATPVNASAP 215
++++Y DV+DR+W L +W ++TS H++ +I Y PP V+ T A P NAS P
Sbjct: 204 NYIRYPQDVHDRIWVPLILPEWTHINTS---HHVIDSIDGYDPPQDVLRTGAMPANASDP 260
Query: 216 LQFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFST 275
+ W+ DQ Y YI+ E+ E+ ANETREF + VN+K+ F P P ++F+
Sbjct: 261 MTITWNLKTATDQVYGYIYIAEIMEVQANETREFEVVVNNKVHFDPFRPTRFEAQVMFNN 320
Query: 276 EPLRRAETY-QISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVT 334
PL + ++ L KT STLPP++NAFEI+ +F Q ET Q+DV + NI+ +YG+
Sbjct: 321 VPLTCEGGFCRLQLIKTPKSTLPPLMNAFEIFTGIEFPQSETNQNDVIAVKNIQASYGLN 380
Query: 335 R-NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLD 393
R +WQGDPC P ++W GL+C+ D + PPRI L+LSSSGL G I SI LT LQ LD
Sbjct: 381 RISWQGDPCVPKQFLWTGLSCNVIDVSTPPRIVKLDLSSSGLNGVIPPSIQNLTQLQELD 440
Query: 394 LSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTG 438
LS N+L G +P+FL +++ L V+N+ N L+GLVP LL+R K G
Sbjct: 441 LSQNNLTGKVPEFLAKMKYLLVINLSGNKLSGLVPQALLDRKKEG 485
Score = 183 bits (464), Expect = 4e-46
Identities = 92/191 (48%), Positives = 134/191 (69%), Gaps = 2/191 (1%)
Query: 619 YLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIAD 678
+L ++L+W RL IA+++A G++YLH GCKP +HRD+K +NILL E AKIAD
Sbjct: 643 HLSGKPGCSVLSWPIRLKIALESAIGIEYLHTGCKPKIVHRDVKSTNILLSEEFEAKIAD 702
Query: 679 FGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALV 738
FGLSR+F ++ T AGTFGY+DP++ +T + K+D+YSFG+VLLE+I+G+ ++
Sbjct: 703 FGLSRSFLIGNEAQ-PTVVAGTFGYLDPEYHKTSLLSMKSDVYSFGVVLLEIISGQD-VI 760
Query: 739 RASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDM 798
S E+ +I++W + I+E GDI SI+D L +D +SAWKVVE+AMS + ERP+M
Sbjct: 761 DLSRENCNIVEWTSFILENGDIESIVDPNLHQDYDTSSAWKVVELAMSCVNRTSKERPNM 820
Query: 799 SQILAELKECL 809
SQ++ L ECL
Sbjct: 821 SQVVHVLNECL 831
>At2g19230 putative receptor-like protein kinase
Length = 877
Score = 247 bits (630), Expect = 2e-65
Identities = 134/286 (46%), Positives = 176/286 (60%), Gaps = 5/286 (1%)
Query: 161 QYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAP-LQFH 219
+YKDD++DR W ++ L+TSL D N + PP++VMSTA P+N+S + +
Sbjct: 211 RYKDDIFDRFWMPLMFPNFLILNTSLMIDPTSSNGFLPPSVVMSTAVAPMNSSIEQIMVY 270
Query: 220 WSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKL--WFGPVTPIYRTPDLIFSTEP 277
W + N ++Y+YIHF EVE+L +NETREF++ +N + P Y D ++ P
Sbjct: 271 WEPRDPNWKFYIYIHFAEVEKLPSNETREFSVFLNKEQIDTTSVFRPSYLYTDTLYVQNP 330
Query: 278 LRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTRNW 337
+ + L + ST PPI+NA E Y +F L T Q+DVD I IK Y V +NW
Sbjct: 331 VS-GPFLEFVLRQGVKSTRPPIMNAIETYRTNEFLDLPTDQNDVDAIMKIKTKYKVKKNW 389
Query: 338 QGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNN 397
GDPCAP Y W+G+NCS NNPPRI S+NLS SGLTG+I LT LQ LDLSNN
Sbjct: 390 LGDPCAPFGYPWQGINCSYTA-NNPPRIISVNLSFSGLTGQIDPVFITLTPLQKLDLSNN 448
Query: 398 SLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
L G +PDFL L L LN+ +N LTG++P +LLERSK GSLSLR
Sbjct: 449 RLTGTVPDFLANLPDLTELNLEENKLTGILPEKLLERSKDGSLSLR 494
Score = 199 bits (507), Expect = 4e-51
Identities = 100/216 (46%), Positives = 151/216 (69%), Gaps = 2/216 (0%)
Query: 624 ENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSR 683
+NS++L+W ERL I++DAA GL+YLHNGCKPP +HRD+KP+NIL++E + AKIADFGLSR
Sbjct: 656 KNSSILSWEERLQISLDAAQGLEYLHNGCKPPIVHRDVKPTNILINEKLQAKIADFGLSR 715
Query: 684 AFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRA-SG 742
+F + DS +ST AGT GY+DP+ ++K+D+YSFG+VLLE+ITG+ + R+ +
Sbjct: 716 SFTLEGDSQVSTEVAGTIGYLDPEHYSMQQFSEKSDVYSFGVVLLEVITGQPVISRSRTE 775
Query: 743 ESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQIL 802
E+ HI V+ ++ +GDI+SI+D +L +F+ AWK+ E+A++ S R MSQ++
Sbjct: 776 ENRHISDRVSLMLSKGDIKSIVDPKLGERFNAGLAWKITEVALACASESTKTRLTMSQVV 835
Query: 803 AELKECLSLDMVHRNNGRERAIVELTSLNIASDTIP 838
AELKE L ++G + + E T +N++ P
Sbjct: 836 AELKESLCRARTSGDSG-DISFSEPTEMNVSMTVDP 870
>At1g51800 unknown protein
Length = 894
Score = 246 bits (629), Expect = 3e-65
Identities = 128/285 (44%), Positives = 184/285 (63%), Gaps = 6/285 (2%)
Query: 161 QYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFHW 220
+Y DV+DR+W Y +W ++ST N + N Y+PP I M TA+ P + A +
Sbjct: 206 RYGIDVFDRVWTPYNFGNWSQIST--NQSVNINNDYQPPEIAMVTASVPTDPDAAMNISL 263
Query: 221 SSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFS-TEPLR 279
Q+Y+++HF E++EL +N+TREFNI N+K +GP P+ T +F+ TE +
Sbjct: 264 VGVERTVQFYVFMHFAEIQELKSNDTREFNIMYNNKHIYGPFRPLNFTTSSVFTPTEVVA 323
Query: 280 RAE-TYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR-NW 337
A Y SL +T NSTLPP+LNA EIY Q ET + +VD + NIK+AYGV + +W
Sbjct: 324 DANGQYIFSLQRTGNSTLPPLLNAMEIYSVNLLPQQETDRKEVDAMMNIKSAYGVNKIDW 383
Query: 338 QGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNN 397
+GDPC P++Y W G+NC T DN P+I SL+LS+SGLTGEI IS LT L+ LDLSNN
Sbjct: 384 EGDPCVPLDYKWSGVNC-TYVDNETPKIISLDLSTSGLTGEILEFISDLTSLEVLDLSNN 442
Query: 398 SLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
SL G +P+FL + +L+++N+ N L G +P+ LL++ + GS++L
Sbjct: 443 SLTGSVPEFLANMETLKLINLSGNELNGSIPATLLDKERRGSITL 487
Score = 206 bits (525), Expect = 3e-53
Identities = 100/207 (48%), Positives = 143/207 (68%), Gaps = 2/207 (0%)
Query: 613 NFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENM 672
N + +L ++L W RL IA ++A GL+YLHNGCKP +HRD+K +NILL+E
Sbjct: 663 NGDLKEHLSGKRGPSILTWEGRLRIAAESAQGLEYLHNGCKPQIVHRDIKTTNILLNEKF 722
Query: 673 HAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELIT 732
AK+ADFGLSR+F ++H+ST AGT GY+DP++ RT +K+D++SFG+VLLEL+T
Sbjct: 723 QAKLADFGLSRSFPLGTETHVSTIVAGTPGYLDPEYYRTNWLTEKSDVFSFGVVLLELVT 782
Query: 733 GKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIE 792
+ ++ E HI +WV ++ RGDI SI+D +LQG FD N+ WKVVE AM+ +P
Sbjct: 783 -NQPVIDMKREKSHIAEWVGLMLSRGDINSIVDPKLQGDFDPNTIWKVVETAMTCLNPSS 841
Query: 793 VERPDMSQILAELKECLSLDMVHRNNG 819
RP M+Q++ +LKECL+++M RN G
Sbjct: 842 SRRPTMTQVVMDLKECLNMEMA-RNMG 867
>At3g21340 serine/threonine-specific protein kinase, putative
Length = 880
Score = 243 bits (619), Expect = 4e-64
Identities = 120/279 (43%), Positives = 183/279 (65%), Gaps = 5/279 (1%)
Query: 159 HLQYKDDVYDRMWFSY-ELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQ 217
+++Y DDV DR W+ + + +W L+T+LN + N Y PP +VM++A+TP++
Sbjct: 209 NIRYPDDVNDRKWYPFFDAKEWTELTTNLNINS--SNGYAPPEVVMASASTPISTFGTWN 266
Query: 218 FHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEP 277
F W + Q+Y+Y+HF E++ L + +TREF +T+N KL + +P + IF + P
Sbjct: 267 FSWLLPSSTTQFYVYMHFAEIQTLRSLDTREFKVTLNGKLAYERYSPKTLATETIFYSTP 326
Query: 278 LRRAE-TYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR- 335
+ + T + L+KT STLPP++NA E++ DF Q+ET DDV I +I++ YG+++
Sbjct: 327 QQCEDGTCLLELTKTPKSTLPPLMNALEVFTVIDFPQMETNPDDVAAIKSIQSTYGLSKI 386
Query: 336 NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLS 395
+WQGDPC P ++WEGLNC+ D++ PP +TSLNLSSS LTG I+ I LT LQ LDLS
Sbjct: 387 SWQGDPCVPKQFLWEGLNCNNLDNSTPPIVTSLNLSSSHLTGIIAQGIQNLTHLQELDLS 446
Query: 396 NNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLER 434
NN+L G +P+FL ++SL V+N+ NN G +P LL++
Sbjct: 447 NNNLTGGIPEFLADIKSLLVINLSGNNFNGSIPQILLQK 485
Score = 213 bits (541), Expect = 5e-55
Identities = 99/203 (48%), Positives = 145/203 (70%), Gaps = 1/203 (0%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + ++ ++LNW RL I V++A GL+YLHNGCKPP +HRD+K +NILL+
Sbjct: 648 YMANGDLREHMSGKRGGSILNWETRLKIVVESAQGLEYLHNGCKPPMVHRDVKTTNILLN 707
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E++HAK+ADFGLSR+F + ++H+ST AGT GY+DP++ RT N+K+D+YSFGIVLLE
Sbjct: 708 EHLHAKLADFGLSRSFPIEGETHVSTVVAGTPGYLDPEYYRTNWLNEKSDVYSFGIVLLE 767
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
+IT + ++ S E HI +WV ++ +GDI++I+D +L G +D S W+ VE+AMS +
Sbjct: 768 IIT-NQLVINQSREKPHIAEWVGLMLTKGDIQNIMDPKLYGDYDSGSVWRAVELAMSCLN 826
Query: 790 PIEVERPDMSQILAELKECLSLD 812
P RP MSQ++ EL ECLS +
Sbjct: 827 PSSARRPTMSQVVIELNECLSYE 849
>At1g51850 Putative protein kinase
Length = 875
Score = 241 bits (616), Expect = 9e-64
Identities = 123/281 (43%), Positives = 182/281 (63%), Gaps = 6/281 (2%)
Query: 157 TSHLQYKDDVYDRMWFSYELIDWRRLSTSLN-NDHLVQNIYKPPTIVMSTAATPVNASAP 215
++ +++ DDVYDR W+ Y W +++T+L+ N L Y+ P VM+ AATP+ A+
Sbjct: 196 STRIRFPDDVYDRKWYPYFDNSWTQVTTTLDVNTSLT---YELPQSVMAKAATPIKANDT 252
Query: 216 LQFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFST 275
L W+ ++Y Y+HF E++ L AN+ REFN+T+N +GP +P + I+
Sbjct: 253 LNITWTVEPPTTKFYSYMHFAELQTLRANDAREFNVTMNGIYTYGPYSPKPLKTETIYDK 312
Query: 276 EPLR-RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVT 334
P + + + KT STLPP+LNA E + DF Q+ET DDVD I N+++ YG++
Sbjct: 313 IPEQCDGGACLLQVVKTLKSTLPPLLNAIEAFTVIDFPQMETNGDDVDAIKNVQDTYGIS 372
Query: 335 R-NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLD 393
R +WQGDPC P ++W+GLNC+ D++ P ITSL+LSSSGLTG I+ +I LT LQ LD
Sbjct: 373 RISWQGDPCVPKLFLWDGLNCNNSDNSTSPIITSLDLSSSGLTGSITQAIQNLTNLQELD 432
Query: 394 LSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLER 434
LS+N+L G +PDFL ++SL V+N+ NNL+G VP LL++
Sbjct: 433 LSDNNLTGEIPDFLGDIKSLLVINLSGNNLSGSVPPSLLQK 473
Score = 203 bits (517), Expect = 3e-52
Identities = 100/224 (44%), Positives = 147/224 (64%), Gaps = 3/224 (1%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + ++ N LNW RL I V++A GL+YLHNGCKPP +HRD+K +NILL+
Sbjct: 643 YMANGDLKEHMSGTRNRFTLNWGTRLKIVVESAQGLEYLHNGCKPPMVHRDVKTTNILLN 702
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E+ AK+ADFGLSR+F + ++H+ST AGT GY+DP++ +T +K+D+YSFGIVLLE
Sbjct: 703 EHFQAKLADFGLSRSFPIEGETHVSTVVAGTPGYLDPEYYKTNWLTEKSDVYSFGIVLLE 762
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
LIT + ++ S E HI +WV ++ +GDI SI+D L +D S WK VE+AMS +
Sbjct: 763 LIT-NRPVIDKSREKPHIAEWVGVMLTKGDINSIMDPNLNEDYDSGSVWKAVELAMSCLN 821
Query: 790 PIEVERPDMSQILAELKECLSLDMVHRNNGRERAIVELTSLNIA 833
P RP MSQ++ EL EC++ + + G R + +S+ ++
Sbjct: 822 PSSARRPTMSQVVIELNECIASE--NSRGGASRDMDSKSSIEVS 863
>At1g51820 Putative protein kinase
Length = 883
Score = 241 bits (614), Expect = 2e-63
Identities = 121/280 (43%), Positives = 179/280 (63%), Gaps = 4/280 (1%)
Query: 157 TSHLQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPL 216
++ +++ DDVYDR W+ W +++T+L + + Y+ P VM+ AATP+ A+ L
Sbjct: 196 STRIRFPDDVYDRKWYPLFDDSWTQVTTNLKVNTSIT--YELPQSVMAKAATPIKANDTL 253
Query: 217 QFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPI-YRTPDLIFST 275
W+ Q+Y Y+H E++ L ANETREFN+T+N + FGP +PI +T ++ +
Sbjct: 254 NITWTVEPPTTQFYSYVHIAEIQALRANETREFNVTLNGEYTFGPFSPIPLKTASIVDLS 313
Query: 276 EPLRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR 335
+ + KT STLPP+LNA E + DF Q+ET ++DV I N++ YG++R
Sbjct: 314 PGQCDGGRCILQVVKTLKSTLPPLLNAIEAFTVIDFPQMETNENDVAGIKNVQGTYGLSR 373
Query: 336 -NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDL 394
+WQGDPC P +W+GLNC D + PP ITSL+LSSSGLTG I+ +I LT LQ LDL
Sbjct: 374 ISWQGDPCVPKQLLWDGLNCKNSDISTPPIITSLDLSSSGLTGIITQAIKNLTHLQILDL 433
Query: 395 SNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLER 434
S+N+L G +P+FL ++SL V+N+ NNL+G VP LL++
Sbjct: 434 SDNNLTGEVPEFLADIKSLLVINLSGNNLSGSVPPSLLQK 473
Score = 199 bits (505), Expect = 7e-51
Identities = 98/224 (43%), Positives = 148/224 (65%), Gaps = 3/224 (1%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + ++ N +LNW RL I +++A GL+YLHNGCKPP +HRD+K +NILL+
Sbjct: 651 YMANGDLKEHMSGTRNRFILNWGTRLKIVIESAQGLEYLHNGCKPPMVHRDVKTTNILLN 710
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E+ AK+ADFGLSR+F + ++H+ST AGT GY+DP++ RT +K+D+YSFGI+LLE
Sbjct: 711 EHFEAKLADFGLSRSFLIEGETHVSTVVAGTPGYLDPEYHRTNWLTEKSDVYSFGILLLE 770
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
+IT + ++ S E HI +WV ++ +GDI+SI+D L +D S WK VE+AMS +
Sbjct: 771 IITNRH-VIDQSREKPHIGEWVGVMLTKGDIQSIMDPSLNEDYDSGSVWKAVELAMSCLN 829
Query: 790 PIEVERPDMSQILAELKECLSLDMVHRNNGRERAIVELTSLNIA 833
RP MSQ++ EL ECL+ + + G R + +S+ ++
Sbjct: 830 HSSARRPTMSQVVIELNECLASE--NARGGASRDMESKSSIEVS 871
>At1g49100 putative receptor-like protein kinase gb|AAD15465.1
Length = 888
Score = 241 bits (614), Expect = 2e-63
Identities = 122/282 (43%), Positives = 182/282 (64%), Gaps = 8/282 (2%)
Query: 160 LQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFH 219
L+Y DDV DR WF + +W+ ++T+LN + N + P M++AAT VN + +F
Sbjct: 208 LRYPDDVNDRRWFPFSYKEWKIVTTTLNVN--TSNGFDLPQGAMASAATRVNDNGTWEFP 265
Query: 220 WSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEP-- 277
WS + ++++Y+HF E++ L ANETREFN+ +N K+++GP +P + D + S +P
Sbjct: 266 WSLEDSTTRFHIYLHFAELQTLLANETREFNVLLNGKVYYGPYSPKMLSIDTM-SPQPDS 324
Query: 278 --LRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR 335
+ + + L KT STLPP++NA E++ +F Q ET QD+V I I+ YG++R
Sbjct: 325 TLTCKGGSCLLQLVKTTKSTLPPLINAIELFTVVEFPQSETNQDEVIAIKKIQLTYGLSR 384
Query: 336 -NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDL 394
NWQGDPC P ++W GL CS + + PP IT LNLSSSGLTG IS SI LT LQ LDL
Sbjct: 385 INWQGDPCVPEQFLWAGLKCSNINSSTPPTITFLNLSSSGLTGIISPSIQNLTHLQELDL 444
Query: 395 SNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSK 436
SNN L G +P+FL ++SL ++N+ NN +G +P +L+++ +
Sbjct: 445 SNNDLTGDVPEFLADIKSLLIINLSGNNFSGQLPQKLIDKKR 486
Score = 207 bits (527), Expect = 2e-53
Identities = 97/227 (42%), Positives = 147/227 (64%), Gaps = 1/227 (0%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + + ++L W RL IAV+AA GL+YLH GC+PP +HRD+K +NILLD
Sbjct: 656 YMANGDLKEFFSGKRGDDVLRWETRLQIAVEAAQGLEYLHKGCRPPIVHRDVKTANILLD 715
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E+ AK+ADFGLSR+F N+ +SH+ST AGT GY+DP++ RT +K+D+YSFG+VLLE
Sbjct: 716 EHFQAKLADFGLSRSFLNEGESHVSTVVAGTIGYLDPEYYRTNWLTEKSDVYSFGVVLLE 775
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
+IT ++ + R + E HI +WV ++ +GDIR I+D L+G + +S WK VE+AM+ +
Sbjct: 776 IITNQRVIER-TREKPHIAEWVNLMITKGDIRKIVDPNLKGDYHSDSVWKFVELAMTCVN 834
Query: 790 PIEVERPDMSQILAELKECLSLDMVHRNNGRERAIVELTSLNIASDT 836
RP M+Q++ EL EC++L+ + + + + DT
Sbjct: 835 DSSATRPTMTQVVTELTECVTLENSRGGKSQNMGSTSSSEVTMTFDT 881
>At1g07560 protein kinase, putative
Length = 856
Score = 241 bits (614), Expect = 2e-63
Identities = 124/282 (43%), Positives = 177/282 (61%), Gaps = 2/282 (0%)
Query: 160 LQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFH 219
++Y +DV+DR+W + + +WR L TSL + N Y P V+ TAATP N S+PL
Sbjct: 207 VRYPEDVHDRLWSPFFMPEWRLLRTSLTVNTSDDNGYDIPEDVVVTAATPANVSSPLTIS 266
Query: 220 WSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEPLR 279
W+ +D Y Y+H E++ L N+TREFNI+ + +GPV+P +F+T P++
Sbjct: 267 WNLETPDDLVYAYLHVAEIQSLRENDTREFNISAGQDVNYGPVSPDEFLVGTLFNTSPVK 326
Query: 280 -RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR-NW 337
T + L KT STLPP+LNA E ++ +F Q ET +DV I +I+ +YG++R +W
Sbjct: 327 CEGGTCHLQLIKTPKSTLPPLLNAIEAFITVEFPQSETNANDVLAIKSIETSYGLSRISW 386
Query: 338 QGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNN 397
QGDPC P +W+GL C + + PPRI SL+LSSS LTG I I LT L+ LD SNN
Sbjct: 387 QGDPCVPQQLLWDGLTCEYTNMSTPPRIHSLDLSSSELTGIIVPEIQNLTELKKLDFSNN 446
Query: 398 SLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGS 439
+L G +P+FL +++SL V+N+ NNL+G VP LL + K GS
Sbjct: 447 NLTGGVPEFLAKMKSLLVINLSGNNLSGSVPQALLNKVKNGS 488
Score = 207 bits (527), Expect = 2e-53
Identities = 103/207 (49%), Positives = 150/207 (71%), Gaps = 6/207 (2%)
Query: 626 SNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAF 685
S++++W +RLNIAVDAA GL+YLH GCKP +HRD+K SNILLD+ + AK+ADFGLSR+F
Sbjct: 643 SSIISWVDRLNIAVDAASGLEYLHIGCKPLIVHRDVKSSNILLDDQLQAKLADFGLSRSF 702
Query: 686 DNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESI 745
+SH+ST AGTFGY+D ++ +T ++K+D+YSFG+VLLE+IT K ++ + +
Sbjct: 703 PIGDESHVSTLVAGTFGYLDHEYYQTNRLSEKSDVYSFGVVLLEIIT-NKPVIDHNRDMP 761
Query: 746 HILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
HI +WV ++ RGDI +I+D +LQG +D SAWK +E+AM+ +P ++RP+MS ++ EL
Sbjct: 762 HIAEWVKLMLTRGDISNIMDPKLQGVYDSGSAWKALELAMTCVNPSSLKRPNMSHVVHEL 821
Query: 806 KECLSLDMVHRNNGRERAIVELTSLNI 832
KECL + N R R I S++I
Sbjct: 822 KECLVSE-----NNRTRDIDTSRSMDI 843
>At2g19210 putative receptor-like protein kinase
Length = 881
Score = 240 bits (613), Expect = 2e-63
Identities = 128/285 (44%), Positives = 174/285 (60%), Gaps = 3/285 (1%)
Query: 160 LQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQFH 219
++YKDDV+DR+W + + SL D ++P VM+TA +P + S + F
Sbjct: 211 VRYKDDVFDRIWIPLRFPKYTIFNASLTIDSNNNEGFQPARFVMNTATSPEDLSQDIIFS 270
Query: 220 WSSNNENDQYYLYIHFNEVEELAANETREFNITVNDK-LWFGPVTPIYRTPDLIFSTEPL 278
W + +Y++Y+HF EV EL +NETREF + +N+K + +P Y D +F P+
Sbjct: 271 WEPKDPTWKYFVYMHFAEVVELPSNETREFKVLLNEKEINMSSFSPRYLYTDTLFVQNPV 330
Query: 279 RRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTRNWQ 338
+ + L +T STLPPI+NA E Y +F Q T Q DVD I IK+ YGV ++W
Sbjct: 331 SGPKL-EFRLQQTPRSTLPPIINAIETYRVNEFLQSPTDQQDVDAIMRIKSKYGVKKSWL 389
Query: 339 GDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNS 398
GDPCAPV Y W+ +NCS D N PRI S+NLSSSGLTGEI ++ S LT+L LDLSNNS
Sbjct: 390 GDPCAPVKYPWKDINCSYVD-NESPRIISVNLSSSGLTGEIDAAFSNLTLLHILDLSNNS 448
Query: 399 LNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
L G +PDFL L +L LN+ N L+G +P +LLERS + LR
Sbjct: 449 LTGKIPDFLGNLHNLTELNLEGNKLSGAIPVKLLERSNKKLILLR 493
Score = 197 bits (501), Expect = 2e-50
Identities = 100/219 (45%), Positives = 145/219 (65%), Gaps = 3/219 (1%)
Query: 624 ENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSR 683
E S +L+W ERL I++DAA GL+YLHNGCKPP + RD+KP+NIL++E + AKIADFGLSR
Sbjct: 663 EKSYVLSWEERLQISLDAAQGLEYLHNGCKPPIVQRDVKPANILINEKLQAKIADFGLSR 722
Query: 684 AFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRA--S 741
+ D ++ +T AGT GY+DP++ T ++K+DIYSFG+VLLE+++G+ + R+ +
Sbjct: 723 SVALDGNNQDTTAVAGTIGYLDPEYHLTQKLSEKSDIYSFGVVLLEVVSGQPVIARSRTT 782
Query: 742 GESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQI 801
E+IHI V ++ GDIR I+D +L +FD SAWK+ E+AM+ S RP MS +
Sbjct: 783 AENIHITDRVDLMLSTGDIRGIVDPKLGERFDAGSAWKITEVAMACASSSSKNRPTMSHV 842
Query: 802 LAELKECLSLDMVHRNNGRERAIVELTSLNIASDTIPLA 840
+AELKE +S +G ++ + N S P A
Sbjct: 843 VAELKESVSRARAGGGSGAS-SVTDPAMTNFDSGMFPQA 880
>At5g59670 serine/threonine-specific protein kinase - like
Length = 868
Score = 238 bits (606), Expect = 1e-62
Identities = 125/284 (44%), Positives = 177/284 (62%), Gaps = 6/284 (2%)
Query: 155 KLTSHLQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASA 214
K S L+Y DVYDR WF + +W ++ST+L + NIY+PP + AATP +ASA
Sbjct: 200 KSGSRLRYSKDVYDRSWFPRFMDEWTQISTALGV--INTNIYQPPEDALKNAATPTDASA 257
Query: 215 PLQFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWF--GPVTPIYRTPDLI 272
PL F W+S + QYY Y H+ E+++L AN+TREFNI +N + GP P +
Sbjct: 258 PLTFKWNSEKLDVQYYFYAHYAEIQDLQANDTREFNILLNGQNLSVTGPEVPDKLSIKTF 317
Query: 273 FSTEPLR-RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAY 331
S+ P+ L +TK STLPP+LNA E+Y F + ET + DV + NI +Y
Sbjct: 318 QSSSPISCNGWACNFQLIRTKRSTLPPLLNALEVYTVIQFPRSETDESDVVAMKNISASY 377
Query: 332 GVTR-NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQ 390
G++R NWQGDPC P W+ L+C+ + + PPRITSLNLSSS L G I+++I +T L+
Sbjct: 378 GLSRINWQGDPCFPQQLRWDALDCTNRNISQPPRITSLNLSSSRLNGTIAAAIQSITQLE 437
Query: 391 YLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLER 434
LDLS N+L G +P+FL +++SL V+N+ NNL G +P L ++
Sbjct: 438 TLDLSYNNLTGEVPEFLGKMKSLSVINLSGNNLNGSIPQALRKK 481
Score = 208 bits (529), Expect = 1e-53
Identities = 106/230 (46%), Positives = 155/230 (67%), Gaps = 6/230 (2%)
Query: 613 NFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENM 672
N + +L ++++NW+ RL IA++AA GL+YLH GC PP +HRD+K +NILLDEN
Sbjct: 642 NGDLKQHLSGKGGNSIINWSIRLRIALEAALGLEYLHIGCTPPMVHRDVKTANILLDENF 701
Query: 673 HAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELIT 732
AK+ADFGLSR+F + +S ST AGT GY+DP+ +G +K+D+YSFGIVLLE+IT
Sbjct: 702 KAKLADFGLSRSFQGEGESQESTTIAGTLGYLDPECYHSGRLGEKSDVYSFGIVLLEMIT 761
Query: 733 GKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIE 792
+ + + SG+S HI QWV + RGDI I+D L+ ++INSAW+ +E+AMS P
Sbjct: 762 NQPVINQTSGDS-HITQWVGFQMNRGDILEIMDPNLRKDYNINSAWRALELAMSCAYPSS 820
Query: 793 VERPDMSQILAELKECLSLDMVHRNNGRERAIVELTSLNIASDT--IPLA 840
+RP MSQ++ ELKEC++ + + R +E +N++ DT +P+A
Sbjct: 821 SKRPSMSQVIHELKECIACENTGISKNRS---LEYQEMNVSLDTTAVPMA 867
>At2g28970 putative receptor-like protein kinase
Length = 786
Score = 235 bits (599), Expect = 9e-62
Identities = 122/283 (43%), Positives = 173/283 (61%), Gaps = 4/283 (1%)
Query: 158 SHLQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQ 217
S ++Y DD YDR+W+ + + ++TSLN ++ + ++ P + +AATP NASAPL
Sbjct: 103 SEIRYDDDSYDRVWYPFFSSSFSYITTSLNINN--SDTFEIPKAALKSAATPKNASAPLI 160
Query: 218 FHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEP 277
W N + Y Y+HF E++ LAANETREF+I + +P F++ P
Sbjct: 161 ITWKPRPSNAEVYFYLHFAEIQTLAANETREFDIVFKGNFNYSAFSPTKLELLTFFTSGP 220
Query: 278 LR-RAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR- 335
++ ++ + L +T NSTLPP++NA E Y +F QLET DV+ I NIK Y +++
Sbjct: 221 VQCDSDGCNLQLVRTPNSTLPPLINALEAYTIIEFPQLETSLSDVNAIKNIKATYRLSKT 280
Query: 336 NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLS 395
+WQGDPC P WE L CS + + PP+I SLNLS+SGLTG + S LT +Q LDLS
Sbjct: 281 SWQGDPCLPQELSWENLRCSYTNSSTPPKIISLNLSASGLTGSLPSVFQNLTQIQELDLS 340
Query: 396 NNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTG 438
NNSL G +P FL ++SL +L++ NN TG VP LL+R K G
Sbjct: 341 NNSLTGLVPSFLANIKSLSLLDLSGNNFTGSVPQTLLDREKEG 383
Score = 209 bits (531), Expect = 7e-54
Identities = 100/227 (44%), Positives = 147/227 (64%), Gaps = 1/227 (0%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + +L +L+W RL +AVDAA GL+YLH GCKPP +HRD+K +NILLD
Sbjct: 554 YMPNGDLKQHLSGKRGGFVLSWESRLRVAVDAALGLEYLHTGCKPPMVHRDIKSTNILLD 613
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E AK+ADFGLSR+F + ++H+ST AGT GY+DP++ +T +K+D+YSFGIVLLE
Sbjct: 614 ERFQAKLADFGLSRSFPTENETHVSTVVAGTPGYLDPEYYQTNWLTEKSDVYSFGIVLLE 673
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
+IT + +++ S E H+++WV IV GDI +I+D L G +D+ S WK +E+AMS +
Sbjct: 674 IIT-NRPIIQQSREKPHLVEWVGFIVRTGDIGNIVDPNLHGAYDVGSVWKAIELAMSCVN 732
Query: 790 PIEVERPDMSQILAELKECLSLDMVHRNNGRERAIVELTSLNIASDT 836
RP MSQ++++LKEC+ + RE + ++ DT
Sbjct: 733 ISSARRPSMSQVVSDLKECVISENSRTGESREMNSMSSIEFSMGIDT 779
>At1g51860 Putative protein kinase
Length = 869
Score = 233 bits (594), Expect = 3e-61
Identities = 132/287 (45%), Positives = 177/287 (60%), Gaps = 11/287 (3%)
Query: 157 TSHLQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPL 216
+S ++Y +D++DR+W S+ + +ST L D N Y P VM TAA P NAS P
Sbjct: 204 SSFIRYDEDIHDRVWNSFTDDETVWISTDLPID--TSNSYDMPQSVMKTAAVPKNASEPW 261
Query: 217 QFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKL-WFGPVTPIYRTPDLIFST 275
W+ + Q Y+Y+HF EV+ L ANETREFNIT N L WF + P + IF+
Sbjct: 262 LLWWTLDENTAQSYVYMHFAEVQNLTANETREFNITYNGGLRWFSYLRPPNLSISTIFNP 321
Query: 276 EPLRRAE-TYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVT 334
+ + + + + T NSTLPP+LNA EIY D QLET +D+V + NIK YG++
Sbjct: 322 RAVSSSNGIFNFTFAMTGNSTLPPLLNALEIYTVVDILQLETNKDEVSAMMNIKETYGLS 381
Query: 335 R--NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYL 392
+ +WQGDPCAP Y WEGLNCS D RI SLNL+ S LTG I+S ISKLT+L L
Sbjct: 382 KKISWQGDPCAPQLYRWEGLNCSYPDSEG-SRIISLNLNGSELTGSITSDISKLTLLTVL 440
Query: 393 DLSNNSLNGPLPDFLIQLRSLQVLNVGKN---NLTGLVPSELLERSK 436
DLSNN L+G +P F +++SL+++N+ N NLT +P L +RSK
Sbjct: 441 DLSNNDLSGDIPTFFAEMKSLKLINLSGNPNLNLTA-IPDSLQQRSK 486
Score = 203 bits (517), Expect = 3e-52
Identities = 94/212 (44%), Positives = 143/212 (67%), Gaps = 1/212 (0%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + + N+L W R+ IAV+AA GL+YLHNGC+PP +HRD+K +NILL+
Sbjct: 637 YMANGDLRENMSGKRGGNVLTWENRMQIAVEAAQGLEYLHNGCRPPMVHRDVKTTNILLN 696
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
E AK+ADFGLSR+F D + H+ST AGT GY+DP++ RT ++K+D+YSFG+VLLE
Sbjct: 697 ERCGAKLADFGLSRSFPIDGECHVSTVVAGTPGYLDPEYYRTNWLSEKSDVYSFGVVLLE 756
Query: 730 LITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTS 789
++T + ++ + E HI WV ++ +GDI+SI+D +L G +D N AWK+VE+A++ +
Sbjct: 757 IVT-NQPVIDKTRERPHINDWVGFMLTKGDIKSIVDPKLMGDYDTNGAWKIVELALACVN 815
Query: 790 PIEVERPDMSQILAELKECLSLDMVHRNNGRE 821
P RP M+ ++ EL +C++L+ R E
Sbjct: 816 PSSNRRPTMAHVVMELNDCVALENARRQGSEE 847
>At3g46370 receptor-like protein kinase homolog
Length = 793
Score = 232 bits (591), Expect = 7e-61
Identities = 124/284 (43%), Positives = 175/284 (60%), Gaps = 4/284 (1%)
Query: 157 TSHLQYKDDVYDRMWFSYELIDWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPL 216
T ++Y +D YDRMW + +W+++ST+L + N Y P V+ TAA PVN SA L
Sbjct: 116 TEVIRYPNDFYDRMWVPHFETEWKQISTNLKVNS--SNGYLLPQDVLMTAAIPVNTSARL 173
Query: 217 QFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTE 276
F + +D+ YLY HF+EV+ L AN++REF+I N + + P Y +++
Sbjct: 174 SFTENLEFPHDELYLYFHFSEVQVLQANQSREFSILWNGMVIYPDFIPDYLGAATVYNPS 233
Query: 277 P-LRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTR 335
P L + L +T+ STLPP+LNA E++ +F Q ET DDV IT IK+ + + R
Sbjct: 234 PSLCEVGKCLLELERTQKSTLPPLLNAIEVFTVMNFPQSETNDDDVIAITKIKDTHRLNR 293
Query: 336 -NWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDL 394
+WQGDPC P + W GL+C + + PPRI SLNLSSSGLTG I++ I LT LQ LDL
Sbjct: 294 TSWQGDPCVPQLFSWAGLSCIDTNVSTPPRIISLNLSSSGLTGNIATGIQNLTKLQKLDL 353
Query: 395 SNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTG 438
SNN+L G +P+FL ++SL +++ KN L G +P LL+R K G
Sbjct: 354 SNNNLTGVVPEFLANMKSLLFIDLRKNKLNGSIPKTLLDRKKKG 397
Score = 191 bits (485), Expect = 1e-48
Identities = 98/231 (42%), Positives = 150/231 (64%), Gaps = 6/231 (2%)
Query: 613 NFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENM 672
N + +L + + +L W+ RL IAVDAA GL+YLH GC+P +HRD+K +NILLD+ +
Sbjct: 565 NGDLKDHLSGKKGNAVLKWSTRLRIAVDAALGLEYLHYGCRPSIVHRDVKSTNILLDDQL 624
Query: 673 HAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELIT 732
AKIADFGLSR+F +S ST AGT GY+DP++ RT + +D+YSFGI+LLE+IT
Sbjct: 625 MAKIADFGLSRSFKLGEESQASTVVAGTLGYLDPEYYRTCRLAEMSDVYSFGILLLEIIT 684
Query: 733 GKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIE 792
+ + A E HI +WV +++ GD+ I+D L G+++ S W+ +E+AMS +P
Sbjct: 685 NQNVIDHAR-EKAHITEWVGLVLKGGDVTRIVDPNLDGEYNSRSVWRALELAMSCANPSS 743
Query: 793 VERPDMSQILAELKECLSLD---MVHRNNGRERAIVELTSLNIASDTIPLA 840
RP MSQ++ +LKECL+ + + +N+ +EL+S + ++ +P A
Sbjct: 744 EHRPIMSQVVIDLKECLNTENSMKIKKNDTDNDGSLELSSSD--TEAVPCA 792
>At2g19190 putative receptor-like protein kinase
Length = 876
Score = 231 bits (589), Expect = 1e-60
Identities = 126/292 (43%), Positives = 173/292 (59%), Gaps = 3/292 (1%)
Query: 153 TMKLTSHLQYKDDVYDRMWFSYELI-DWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVN 211
T KL S +YKDD+YDR+W + +++ L+TSL D + N Y+P + VMSTA T N
Sbjct: 205 TGKLPS--RYKDDIYDRIWTPRIVSSEYKILNTSLTVDQFLNNGYQPASTVMSTAETARN 262
Query: 212 ASAPLQFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDL 271
S L + + N ++Y+Y+HF E+E L +N+TREF+I +N+ + Y D
Sbjct: 263 ESLYLTLSFRPPDPNAKFYVYMHFAEIEVLKSNQTREFSIWLNEDVISPSFKLRYLLTDT 322
Query: 272 IFSTEPLRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAY 331
+ +P+ L LPPI+NA E+Y +F Q+ T DVD + IK Y
Sbjct: 323 FVTPDPVSGITINFSLLQPPGEFVLPPIINALEVYQVNEFLQIPTHPQDVDAMRKIKATY 382
Query: 332 GVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQY 391
V +NWQGDPC PV+Y WEG++C D+ PR+ SLN+S S L G+I + S LT ++
Sbjct: 383 RVKKNWQGDPCVPVDYSWEGIDCIQSDNTTNPRVVSLNISFSELRGQIDPAFSNLTSIRK 442
Query: 392 LDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
LDLS N+L G +P FL L +L LNV N LTG+VP L ERSK GSLSLR
Sbjct: 443 LDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIVPQRLHERSKNGSLSLR 494
Score = 207 bits (528), Expect = 2e-53
Identities = 100/219 (45%), Positives = 152/219 (68%), Gaps = 2/219 (0%)
Query: 622 AVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGL 681
A + S +L+W ERL I++DAA GL+YLHNGCKPP +HRD+KP+NILL+E + AK+ADFGL
Sbjct: 659 AGKRSFILSWEERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGL 718
Query: 682 SRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRAS 741
SR+F + IST AG+ GY+DP++ T N+K+D+YS G+VLLE+ITG+ A+ +
Sbjct: 719 SRSFSVEGSGQISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSK 778
Query: 742 GESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQI 801
E +HI V I+ GDIR I+D RL+ ++D+ SAWK+ EIA++ T +RP MSQ+
Sbjct: 779 TEKVHISDHVRSILANGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQV 838
Query: 802 LAELKECLSLDMVHRNNGRERAIVELTSLNIASDTIPLA 840
+ ELK+ + + + N + ++ ++N+ ++ +P A
Sbjct: 839 VMELKQIVYGIVTDQENYDDS--TKMLTVNLDTEMVPRA 875
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.343 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,224,471
Number of Sequences: 26719
Number of extensions: 704786
Number of successful extensions: 9558
Number of sequences better than 10.0: 1168
Number of HSP's better than 10.0 without gapping: 962
Number of HSP's successfully gapped in prelim test: 206
Number of HSP's that attempted gapping in prelim test: 3294
Number of HSP's gapped (non-prelim): 4286
length of query: 840
length of database: 11,318,596
effective HSP length: 108
effective length of query: 732
effective length of database: 8,432,944
effective search space: 6172915008
effective search space used: 6172915008
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146586.12


