

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.11 - phase: 0 /pseudo
(291 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
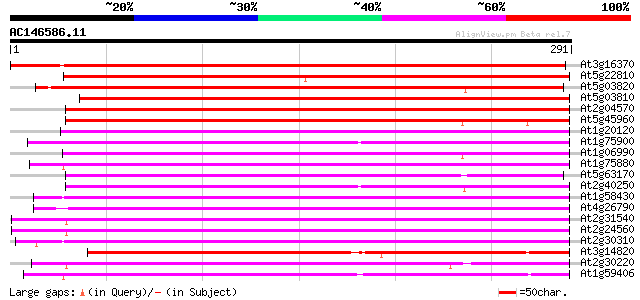
Score E
Sequences producing significant alignments: (bits) Value
At3g16370 putative APG protein 418 e-117
At5g22810 GDSL-motif lipase/hydrolase-like protein 316 1e-86
At5g03820 putative protein 311 3e-85
At5g03810 putative protein 300 6e-82
At2g04570 putative GDSL-motif lipase/hydrolase 242 2e-64
At5g45960 GDSL-motif lipase/hydrolase-like protein 229 2e-60
At1g20120 anter-specific proline-rich like protein (APG precursor) 221 4e-58
At1g75900 family II lipase EXL3 (EXL3) 219 2e-57
At1g06990 unknown protein 219 2e-57
At1g75880 family II extracellular lipase 1 like protein (EXL1) 214 3e-56
At5g63170 lipase/acylhydrolase-like protein 211 3e-55
At2g40250 putative GDSL-motif lipase/hydrolase 211 3e-55
At1g58430 unknown protein 211 5e-55
At4g26790 putative APG protein 208 2e-54
At2g31540 putative GDSL-motif lipase/hydrolase 207 7e-54
At2g24560 putative GDSL-motif lipase/hydrolase 206 2e-53
At2g30310 putative GDSL-motif lipase/hydrolase 205 3e-53
At3g14820 hypothetical protein 204 3e-53
At2g30220 putative GDSL-motif lipase/hydrolase 204 6e-53
At1g59406 proline-rich protein, putative 203 8e-53
>At3g16370 putative APG protein
Length = 353
Score = 418 bits (1074), Expect = e-117
Identities = 207/288 (71%), Positives = 240/288 (82%), Gaps = 2/288 (0%)
Query: 1 MMNMNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPY 60
M S L+ L+ + + SFAQ LVPAIMTFGDS VDVGNN+YLPTLF+A+YPPY
Sbjct: 1 MDRCTSSFLLLTLVSTLSILQISFAQ--LVPAIMTFGDSVVDVGNNNYLPTLFRADYPPY 58
Query: 61 GRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYD 120
GRDF N + TGRFCNGKLATD TAETLGFT + PAYLSP+ASGKNLL+GANFASAASGYD
Sbjct: 59 GRDFANHKATGRFCNGKLATDITAETLGFTKYPPAYLSPEASGKNLLIGANFASAASGYD 118
Query: 121 EKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTN 180
+KAA LNHAIPL QQ+EYFKEY+ KL ++AGSKKA SIIK ++ +LSAGSSDFVQNYY N
Sbjct: 119 DKAALLNHAIPLYQQVEYFKEYKSKLIKIAGSKKADSIIKGAICLLSAGSSDFVQNYYVN 178
Query: 181 PWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGC 240
P + + TVD Y S+L+D+F+ FIK VY +GARKIGVTSLPP GCLPAARTLFG+HE GC
Sbjct: 179 PLLYKVYTVDAYGSFLIDNFSTFIKQVYAVGARKIGVTSLPPTGCLPAARTLFGFHEKGC 238
Query: 241 VARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPS 288
V+R+NTDAQ FNKK+++AAS LQKQ LKIV+FDIY PLYDLVQNPS
Sbjct: 239 VSRLNTDAQNFNKKLNAAASKLQKQYSDLKIVVFDIYSPLYDLVQNPS 286
>At5g22810 GDSL-motif lipase/hydrolase-like protein
Length = 337
Score = 316 bits (809), Expect = 1e-86
Identities = 153/265 (57%), Positives = 201/265 (75%), Gaps = 3/265 (1%)
Query: 29 LVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLG 88
LVPAI FGDS VDVGNN+ + T+ KAN+PPYGRDFT PTGRFCNGKLATDFTAE LG
Sbjct: 9 LVPAIFIFGDSVVDVGNNNDIYTIVKANFPPYGRDFTTHTPTGRFCNGKLATDFTAENLG 68
Query: 89 FTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQ 148
F S+ AYLS +A GKNLL+GANFASAASGY + A L AI L QQLE++K+Y ++ +
Sbjct: 69 FKSYPQAYLSKKAKGKNLLIGANFASAASGYYDGTAKLYSAISLPQQLEHYKDYISRIQE 128
Query: 149 VAGS---KKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIK 205
+A S A++II + +Y++SAGSSDF+QNYY NP + + + D++S L+ S+++FI+
Sbjct: 129 IATSNNNSNASAIISNGIYIVSAGSSDFIQNYYINPLLYRDQSPDEFSDLLILSYSSFIQ 188
Query: 206 GVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQ 265
+Y LGAR+IGVT+LPPLGCLPAA T+ G HE GC ++N DA FN K+++ + +L++
Sbjct: 189 NLYSLGARRIGVTTLPPLGCLPAAITVVGPHEGGCSEKLNNDAISFNNKLNTTSQDLKRN 248
Query: 266 LPGLKIVIFDIYKPLYDLVQNPSNF 290
L GL +V+FDIY+PLYDL PS F
Sbjct: 249 LIGLNLVVFDIYQPLYDLATRPSEF 273
>At5g03820 putative protein
Length = 354
Score = 311 bits (796), Expect = 3e-85
Identities = 153/276 (55%), Positives = 199/276 (71%), Gaps = 3/276 (1%)
Query: 14 IVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRF 73
+++CF G + LVPA++ GDS VD GNN+ L TL KAN+PPYGRDF TGRF
Sbjct: 13 VIACFYA-GVGTGEPLVPALIIMGDSVVDAGNNNRLNTLIKANFPPYGRDFLAHNATGRF 71
Query: 74 CNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLS 133
NGKLATDFTAE+LGFTS+ YLS +A+G NLL GANFAS ASGYD+ A +AI L+
Sbjct: 72 SNGKLATDFTAESLGFTSYPVPYLSQEANGTNLLTGANFASGASGYDDGTAIFYNAITLN 131
Query: 134 QQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYS 193
QQL+ +KEYQ K+ + GS++A I ++++LS GSSDF+Q+YY NP +N+ T DQYS
Sbjct: 132 QQLKNYKEYQNKVTNIVGSERANKIFSGAIHLLSTGSSDFLQSYYINPILNRIFTPDQYS 191
Query: 194 SYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGY--HENGCVARINTDAQGF 251
L+ ++ F++ +Y LGARKIGVT+LPPLGCLPAA TLFG + N CV R+N DA F
Sbjct: 192 DRLMKPYSTFVQNLYDLGARKIGVTTLPPLGCLPAAITLFGETGNNNTCVERLNQDAVSF 251
Query: 252 NKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNP 287
N K+++ + NL LPGLK+V+FDIY PL ++ NP
Sbjct: 252 NTKLNNTSMNLTNNLPGLKLVVFDIYNPLLNMAMNP 287
>At5g03810 putative protein
Length = 320
Score = 300 bits (768), Expect = 6e-82
Identities = 147/255 (57%), Positives = 188/255 (73%), Gaps = 1/255 (0%)
Query: 37 GDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAY 96
GDS VD GNN++ TL KAN+PPYGRDF TGRF NGKLATDFTAE LGFTS+ AY
Sbjct: 2 GDSVVDAGNNNHRITLVKANFPPYGRDFVAHSATGRFSNGKLATDFTAENLGFTSYPVAY 61
Query: 97 LSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAA 156
LS +A+ NLL GANFAS ASG+D+ A +AI LSQQL+ +KEYQ K+ + G ++A
Sbjct: 62 LSQEANETNLLTGANFASGASGFDDATAIFYNAITLSQQLKNYKEYQNKVTNIVGKERAN 121
Query: 157 SIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIG 216
I ++++LS GSSDF+Q+YY NP +N+ T DQYS +LL S++ F++ +YGLGAR+IG
Sbjct: 122 EIFSGAIHLLSTGSSDFLQSYYINPILNRIFTPDQYSDHLLRSYSTFVQNLYGLGARRIG 181
Query: 217 VTSLPPLGCLPAARTLF-GYHENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFD 275
VT+LPPLGCLPAA TLF G N CV R+N DA FN K+++ + NL LPGLK+V+FD
Sbjct: 182 VTTLPPLGCLPAAITLFGGVGNNMCVERLNQDAVSFNTKLNNTSINLTNNLPGLKLVVFD 241
Query: 276 IYKPLYDLVQNPSNF 290
IY PL ++V NP +
Sbjct: 242 IYNPLLNMVINPVEY 256
>At2g04570 putative GDSL-motif lipase/hydrolase
Length = 350
Score = 242 bits (618), Expect = 2e-64
Identities = 115/261 (44%), Positives = 162/261 (62%)
Query: 30 VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
+PAI+ FGDS+VD GNN+Y+PT+ ++N+ PYGRDF +PTGRFCNGK+ATDF +E LG
Sbjct: 26 IPAIIVFGDSSVDAGNNNYIPTVARSNFEPYGRDFVGGKPTGRFCNGKIATDFMSEALGL 85
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQV 149
PAYL P + + G FASAA+GYD + + +PL +QLEY+KEYQ KL
Sbjct: 86 KPIIPAYLDPSYNISDFATGVTFASAATGYDNATSDVLSVLPLWKQLEYYKEYQTKLKAY 145
Query: 150 AGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYG 209
G + I+ SLY++S G++DF++NY+ P + +V Y +L F+K ++G
Sbjct: 146 QGKDRGTETIESSLYLISIGTNDFLENYFAFPGRSSQYSVSLYQDFLAGIAKEFVKKLHG 205
Query: 210 LGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQLPGL 269
LGARKI + LPP+GC+P R CV R N A FN K+ L K+LPG
Sbjct: 206 LGARKISLGGLPPMGCMPLERATNIGTGGECVGRYNDIAVQFNSKLDKMVEKLSKELPGS 265
Query: 270 KIVIFDIYKPLYDLVQNPSNF 290
+V + Y+P +++NPS+F
Sbjct: 266 NLVFSNPYEPFMRIIKNPSSF 286
>At5g45960 GDSL-motif lipase/hydrolase-like protein
Length = 375
Score = 229 bits (583), Expect = 2e-60
Identities = 113/266 (42%), Positives = 162/266 (60%), Gaps = 5/266 (1%)
Query: 30 VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
V AI+ FGDS VD GNN+Y+ T+FK N+PPYG DF NK PTGRFCNG+L TDF A +G
Sbjct: 45 VSAILVFGDSTVDPGNNNYIDTVFKCNFPPYGLDFRNKTPTGRFCNGRLVTDFIASYIGV 104
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQV 149
P YL P L+ G +FASA SGYD T+ + I + QLEYF+EY+ KL
Sbjct: 105 KENVPPYLDPNLGINELISGVSFASAGSGYDPLTPTITNVIDIPTQLEYFREYKRKLEGK 164
Query: 150 AGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYG 209
G ++ I+++++ +SAG++DFV NY+T P + T++ Y +++ + FI+G++
Sbjct: 165 MGKQEMEKHIEEAMFCVSAGTNDFVINYFTIPIRRKTFTIEAYQQFVISNLKQFIQGLWK 224
Query: 210 LGARKIGVTSLPPLGCLPAARTLF---GYHENGCVARINTDAQGFNKKVSSAASNLQKQL 266
GARKI V LPP+GCLP TLF C+ R +T A +N + + +Q L
Sbjct: 225 EGARKITVAGLPPIGCLPIVITLFSGEALTNRRCIDRFSTVATNYNFLLQKQLALMQVGL 284
Query: 267 P--GLKIVIFDIYKPLYDLVQNPSNF 290
G KI D+Y P+Y+++++P F
Sbjct: 285 AHLGSKIFYLDVYNPVYEVIRDPRKF 310
>At1g20120 anter-specific proline-rich like protein (APG precursor)
Length = 402
Score = 221 bits (563), Expect = 4e-58
Identities = 111/264 (42%), Positives = 154/264 (58%)
Query: 27 DTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAET 86
+T PAI FGDS +D GNNDY+ TL KAN+ PYG +F +K PTGRFCNGK+ +DF A+
Sbjct: 73 NTTFPAIFAFGDSILDTGNNDYILTLIKANFLPYGMNFPDKVPTGRFCNGKIPSDFIADY 132
Query: 87 LGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKL 146
+G PAYL P + ++LL G +FAS SGYD + AIP+S+QL YF+EY K+
Sbjct: 133 IGVKPVVPAYLRPGLTQEDLLTGVSFASGGSGYDPLTPIVVSAIPMSKQLTYFQEYIEKV 192
Query: 147 AQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKG 206
G +KA II L ++ AGS D YY +D Y+S++ S +F
Sbjct: 193 KGFVGKEKAEHIISKGLAIVVAGSDDLANTYYGEHLEEFLYDIDTYTSFMASSAASFAMQ 252
Query: 207 VYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQL 266
+Y GA+KIG + P+GC+P RT G + C +N AQ FN K+S++ + L K +
Sbjct: 253 LYESGAKKIGFIGVSPIGCIPIQRTTRGGLKRKCADELNFAAQLFNSKLSTSLNELAKTM 312
Query: 267 PGLKIVIFDIYKPLYDLVQNPSNF 290
+V DIY D++QNP +
Sbjct: 313 KNTTLVYIDIYSSFNDMIQNPKKY 336
>At1g75900 family II lipase EXL3 (EXL3)
Length = 364
Score = 219 bits (557), Expect = 2e-57
Identities = 111/281 (39%), Positives = 162/281 (57%), Gaps = 1/281 (0%)
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L +L ++ +T ++PA++ FGDS VD G N+ + T+ K ++ PYG +F +
Sbjct: 21 LSVLFLTETITAVKLPPKLIIPAVIAFGDSIVDTGMNNNVKTVVKCDFLPYGINFQSGVA 80
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHA 129
TGRFC+G++ D AE LG S PAYL P K+LL G +FAS SGYD L
Sbjct: 81 TGRFCDGRVPADLLAEELGIKSIVPAYLDPNLKSKDLLTGVSFASGGSGYDPITPKLVAV 140
Query: 130 IPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITV 189
I L QL YF+EY K+ + G + I+ +SL++L AGS D YYT V
Sbjct: 141 ISLEDQLSYFEEYIEKVKNIVGEARKDFIVANSLFLLVAGSDDIANTYYTLR-ARPEYDV 199
Query: 190 DQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQ 249
D Y++ + DS + F+ +YG G R++ V PP+GC+P+ RTL G C N A+
Sbjct: 200 DSYTTLMSDSASEFVTKLYGYGVRRVAVFGAPPIGCVPSQRTLGGGILRDCADNYNEAAK 259
Query: 250 GFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
FN K+S +L+K LPG+K + +IY PL+D++QNP+N+
Sbjct: 260 LFNSKLSPKLDSLRKTLPGIKPIYINIYDPLFDIIQNPANY 300
>At1g06990 unknown protein
Length = 360
Score = 219 bits (557), Expect = 2e-57
Identities = 104/265 (39%), Positives = 159/265 (59%), Gaps = 2/265 (0%)
Query: 28 TLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETL 87
++ PAI+ FGDS +D GNN+Y+ T +AN+PPYG +F TGRF NGKL DF A +
Sbjct: 33 SMFPAILVFGDSTIDTGNNNYIKTYIRANFPPYGCNFPGHNATGRFSNGKLIPDFIASLM 92
Query: 88 GFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLA 147
G P +L P S +++ G FASA SGYD + + +Q + + Y +L+
Sbjct: 93 GIKDTVPPFLDPHLSDSDIITGVCFASAGSGYDNLTDRATSTLSVDKQADMLRSYVERLS 152
Query: 148 QVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGV 207
Q+ G +KAASI+ ++L ++S+G++DF N Y P Q + VD Y S++L + NF++ +
Sbjct: 153 QIVGDEKAASIVSEALVIVSSGTNDFNLNLYDTPSRRQKLGVDGYQSFILSNVHNFVQEL 212
Query: 208 YGLGARKIGVTSLPPLGCLPAARTLF--GYHENGCVARINTDAQGFNKKVSSAASNLQKQ 265
Y +G RKI V LPP+GCLP T+ +E C+ + N+D+Q FN+K+ ++ + +Q
Sbjct: 213 YDIGCRKIMVLGLPPVGCLPIQMTMAMQKQNERRCIDKQNSDSQEFNQKLKNSLTEMQSN 272
Query: 266 LPGLKIVIFDIYKPLYDLVQNPSNF 290
L G I DIY L+D+ NP +
Sbjct: 273 LTGSVIFYGDIYGALFDMATNPQRY 297
>At1g75880 family II extracellular lipase 1 like protein (EXL1)
Length = 374
Score = 214 bits (546), Expect = 3e-56
Identities = 116/283 (40%), Positives = 163/283 (56%), Gaps = 3/283 (1%)
Query: 11 VLLIVSCFLTCGSFAQ---DTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNK 67
VL+++S T + + +T VPA++ FGDS VD GNND + T + +Y PYG DF
Sbjct: 28 VLVLLSTTSTTNALVKIPKNTTVPAVIVFGDSIVDAGNNDDMITEARCDYAPYGIDFDGG 87
Query: 68 QPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLN 127
TGRF NGK+ D AE LG PAY +P + LL G FAS +GY +
Sbjct: 88 VATGRFSNGKVPGDIVAEELGIKPNIPAYRNPNLKPEELLTGVTFASGGAGYVPLTTKIA 147
Query: 128 HAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAI 187
IPL QQL YF+EY KL Q+ G K+ IIK+SL+V+ GS+D +++T P +
Sbjct: 148 GGIPLPQQLIYFEEYIEKLKQMVGEKRTKFIIKNSLFVVICGSNDIANDFFTLPPVRLHY 207
Query: 188 TVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTD 247
TV +++ + D+ +F + +YG GAR+I V PP+GC+P+ RT+ G CVAR N
Sbjct: 208 TVASFTALMADNARSFAQTLYGYGARRILVFGAPPIGCVPSQRTVAGGPTRDCVARFNDA 267
Query: 248 AQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
A+ FN K+S+ L + L I+ DIY PL DL+ NP +
Sbjct: 268 AKLFNTKLSANIDVLSRTLQDPTIIYIDIYSPLLDLILNPHQY 310
>At5g63170 lipase/acylhydrolase-like protein
Length = 338
Score = 211 bits (538), Expect = 3e-55
Identities = 109/259 (42%), Positives = 153/259 (58%), Gaps = 4/259 (1%)
Query: 30 VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
+PA++ FGDS +D GNN+YL TL K N+ PYGRDF ++ TGRF NG++ TD AE LG
Sbjct: 26 IPAVIAFGDSILDTGNNNYLMTLTKVNFYPYGRDFVTRRATGRFGNGRIPTDLIAEGLGI 85
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQV 149
+ PAY SP ++L G +FAS SG D A + I + QL FK Y KL +
Sbjct: 86 KNIVPAYRSPFLEPNDILTGVSFASGGSGLDPMTARIQGVIWVPDQLNDFKAYIAKLNSI 145
Query: 150 AG-SKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVY 208
G +K SII ++++V+SAG++D Y+TNP N T+ Y+ ++ +FIK +Y
Sbjct: 146 TGDEEKTRSIISNAVFVISAGNNDIAITYFTNPIRNTRYTIFSYTDLMVSWTQSFIKELY 205
Query: 209 GLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQLPG 268
LGARK + PLGCLP A G C+ N A+ FN+K++ +NL LPG
Sbjct: 206 NLGARKFAIMGTLPLGCLPGASNALG---GLCLEPANAVARLFNRKLADEVNNLNSMLPG 262
Query: 269 LKIVIFDIYKPLYDLVQNP 287
+ + D+Y PL +LV+NP
Sbjct: 263 SRSIYVDMYNPLLELVKNP 281
>At2g40250 putative GDSL-motif lipase/hydrolase
Length = 361
Score = 211 bits (538), Expect = 3e-55
Identities = 103/267 (38%), Positives = 159/267 (58%), Gaps = 7/267 (2%)
Query: 30 VPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGF 89
+ A+ FGDS VD GNN+Y+PTLF++N+PPYG+ F +K TGRF +GKLATDF +LG
Sbjct: 34 ITALYAFGDSTVDSGNNNYIPTLFQSNHPPYGKSFPSKLSTGRFSDGKLATDFIVSSLGL 93
Query: 90 TSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQV 149
PAYL+P +LL G +FASA G D++ A + I + +Q YF+E GK+ +
Sbjct: 94 KPTLPAYLNPSVKPVDLLTGVSFASAGGGLDDRTAKSSLTITMDKQWSYFEEALGKMKSL 153
Query: 150 AGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYG 209
G + +IK++++V+SAG++D + N Y + + I+V Y LL F++ +Y
Sbjct: 154 VGDSETNRVIKNAVFVISAGTNDMIFNVYDHV-LGSLISVSDYQDSLLTKVEVFVQRLYE 212
Query: 210 LGARKIGVTSLPPLGCLPAARTLFG------YHENGCVARINTDAQGFNKKVSSAASNLQ 263
GAR+I + LPP+GCLP TL +H C N D++ +N+K+ L
Sbjct: 213 AGARRITIAGLPPIGCLPVQVTLTSINTPRIFHHRICTEHQNDDSRVYNQKLQKLIFGLS 272
Query: 264 KQLPGLKIVIFDIYKPLYDLVQNPSNF 290
++ G K++ DIY PL D++++P +
Sbjct: 273 QRFRGSKVLYLDIYSPLIDMIKHPRKY 299
>At1g58430 unknown protein
Length = 360
Score = 211 bits (536), Expect = 5e-55
Identities = 112/281 (39%), Positives = 160/281 (56%), Gaps = 4/281 (1%)
Query: 13 LIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDY-LPTLFKANYPPYGRDFTNKQPTG 71
L+ SC + + Q L PAI+ FGDS VD GNN+Y T+F+A + PYG D N P G
Sbjct: 17 LLGSCNASAKAKTQP-LFPAILIFGDSTVDTGNNNYPSQTIFRAKHVPYGIDLPNHSPNG 75
Query: 72 RFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIP 131
RF NGK+ +D A L F P +L P + + ++ G FASA +GYD++ + AI
Sbjct: 76 RFSNGKIFSDIIATKLNIKQFVPPFLQPNLTDQEIVTGVCFASAGAGYDDQTSLTTQAIR 135
Query: 132 LSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNP-WINQAITVD 190
+S+Q FK Y +L + G KKA II ++L V+SAG +DF+ NYY P W ++
Sbjct: 136 VSEQPNMFKSYIARLKSIVGDKKAMKIINNALVVVSAGPNDFILNYYEVPSWRRMYPSIS 195
Query: 191 QYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTL-FGYHENGCVARINTDAQ 249
Y ++L NF+K +Y LG RKI V LPP+GCLP T F C+ + N D+
Sbjct: 196 DYQDFVLSRLNNFVKELYSLGCRKILVGGLPPMGCLPIQMTAQFRNVLRFCLEQENRDSV 255
Query: 250 GFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
+N+K+ Q L G KI+ D+Y P+ +++QNPS +
Sbjct: 256 LYNQKLQKLLPQTQASLTGSKILYSDVYDPMMEMLQNPSKY 296
>At4g26790 putative APG protein
Length = 351
Score = 208 bits (530), Expect = 2e-54
Identities = 104/278 (37%), Positives = 166/278 (59%), Gaps = 6/278 (2%)
Query: 13 LIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGR 72
L+V TC F PA++ FGDS VD GNN+ + T+ K+N+ PYGRD+ + + TGR
Sbjct: 16 LLVKIPETCAKF------PALIVFGDSTVDSGNNNQISTVLKSNFQPYGRDYFDGKATGR 69
Query: 73 FCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPL 132
F NG++A DF +E LG + PAYL P + + G FASA +G D + + +PL
Sbjct: 70 FSNGRIAPDFISEGLGLKNAVPAYLDPAYNIADFATGVCFASAGTGLDNATSAVLSVMPL 129
Query: 133 SQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQY 192
+++EY+KEYQ +L G +KA II +SLY++S G++DF++NYY P + +V++Y
Sbjct: 130 WKEVEYYKEYQTRLRSYLGEEKANEIISESLYLISIGTNDFLENYYLLPRKLRKYSVNEY 189
Query: 193 SSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFN 252
+L+ +F+ +Y LGARK+ ++ L P GCLP RT ++ + C+ N A+ FN
Sbjct: 190 QYFLIGIAADFVTDIYRLGARKMSLSGLSPFGCLPLERTTQLFYGSKCIEEYNIVARDFN 249
Query: 253 KKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
K+ L + L G+++V + Y + +++ +P F
Sbjct: 250 IKMEEKVFQLNRDLNGIQLVFSNPYDLVSEIIYHPEAF 287
>At2g31540 putative GDSL-motif lipase/hydrolase
Length = 360
Score = 207 bits (526), Expect = 7e-54
Identities = 114/296 (38%), Positives = 166/296 (55%), Gaps = 7/296 (2%)
Query: 2 MNMNSKETLVLLIVSCFLT-CGSFAQDT---LVPAIMTFGDSAVDVGNNDY-LPTLFKAN 56
M+ + TL L I + L C + A T L PAI+ FGDS VD GNN+Y LPT+F+A
Sbjct: 1 MSTSKAITLTLFIATTLLAPCNAAANATTKPLFPAILIFGDSTVDTGNNNYPLPTIFRAE 60
Query: 57 YPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAA 116
+ PYG D + + GRF NGKL +D A L F P +L P S +++L G FASA
Sbjct: 61 HFPYGMDLPDGKANGRFSNGKLISDIIATKLNIKEFIPPFLQPNLSDQDILTGVCFASAG 120
Query: 117 SGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQN 176
+GYD+ + AI +S+Q FK Y +L + G KKA II ++ V+SAG +DF+ N
Sbjct: 121 AGYDDLTSLSTQAIRVSEQPNMFKSYIARLKGIVGDKKAMEIINNAFVVVSAGPNDFILN 180
Query: 177 YYTNPWIN-QAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTL-FG 234
YY P + + Y ++L NF++ +Y LG R + V LPP+GCLP T F
Sbjct: 181 YYEIPSRRLEYPFISGYQDFILKRLENFVRELYSLGVRNVLVGGLPPMGCLPIHMTAKFR 240
Query: 235 YHENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
C+ N D+ +N+K+ + ++ LPG K + D+Y P+ +++QNPS +
Sbjct: 241 NIFRFCLEHHNKDSVLYNEKLQNLLPQIEASLPGSKFLYADVYNPMMEMIQNPSKY 296
>At2g24560 putative GDSL-motif lipase/hydrolase
Length = 360
Score = 206 bits (523), Expect = 2e-53
Identities = 112/295 (37%), Positives = 164/295 (54%), Gaps = 6/295 (2%)
Query: 2 MNMNSKETLVLLIVSCFLTCGSFAQDT---LVPAIMTFGDSAVDVGNNDY-LPTLFKANY 57
M+ + T L I + +C + T L PAI+ FGDS VD GNN+Y T+FKA +
Sbjct: 1 MSTSKTITFTLFIAALLSSCDAATNATSQPLFPAILIFGDSTVDTGNNNYHSQTIFKAKH 60
Query: 58 PPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAAS 117
PYG D N + +GRF NGK+ +D A L F P +L P S + ++ G FASA +
Sbjct: 61 LPYGIDLPNHKASGRFTNGKIFSDIIATKLNIKQFVPPFLQPNLSDQEIVTGVCFASAGA 120
Query: 118 GYDEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNY 177
GYD+ + AI + Q + FK Y +L + G KKA IIK++L V+SAG +DF+ NY
Sbjct: 121 GYDDHTSLSTQAIRVLDQQKMFKNYIARLKSIVGDKKAMEIIKNALVVISAGPNDFILNY 180
Query: 178 YTNPWINQAIT-VDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTL-FGY 235
Y P + Y ++L NF++ +Y LG RKI V LPP+GCLP T F
Sbjct: 181 YDIPSRRLEFPHISGYQDFVLQRLDNFVRELYSLGCRKIMVGGLPPMGCLPIQMTAKFRN 240
Query: 236 HENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
C+ + N D+ +N+K+ + ++ L G KI+ ++Y P+ D++QNPS +
Sbjct: 241 ALRFCLEQENRDSVLYNQKLQNLLPQIEASLTGSKILYSNVYDPMMDMMQNPSKY 295
>At2g30310 putative GDSL-motif lipase/hydrolase
Length = 359
Score = 205 bits (521), Expect = 3e-53
Identities = 112/296 (37%), Positives = 168/296 (55%), Gaps = 10/296 (3%)
Query: 4 MNSKETLVL------LIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDY-LPTLFKAN 56
M++ +T+V L+VSC + + Q L PAI+ FGDS VD GNN+Y T+FKA
Sbjct: 1 MSTSKTIVFGLFVATLLVSCNVAANATTQP-LFPAILIFGDSTVDTGNNNYHSQTIFKAK 59
Query: 57 YPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAA 116
+ PYG D + GR+ NGK+ +D A L P +L P S ++++ G +FASA
Sbjct: 60 HLPYGVDLPGHEANGRYSNGKVISDVIASKLNIKELVPPFLQPNISHQDIVTGVSFASAG 119
Query: 117 SGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQN 176
+GYD++++ + AIP+SQQ FK Y +L + G KKA II ++L V+SAG +DF+ N
Sbjct: 120 AGYDDRSSLSSKAIPVSQQPSMFKNYIARLKGIVGDKKAMEIINNALVVISAGPNDFILN 179
Query: 177 YYTNPWIN-QAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTL-FG 234
+Y P + T+ Y ++L F++ +Y LG R I V LPP+GCLP T
Sbjct: 180 FYDIPTRRLEYPTIHGYQEFILKRLDGFVRELYSLGCRNIVVGGLPPMGCLPIQMTAKMR 239
Query: 235 YHENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
CV + N D+ +N+K+ +Q LPG + ++Y PL D++QNPS +
Sbjct: 240 NILRFCVEQENKDSVLYNQKLVKKLPEIQASLPGSNFLYANVYDPLMDMIQNPSKY 295
>At3g14820 hypothetical protein
Length = 311
Score = 204 bits (520), Expect = 3e-53
Identities = 104/252 (41%), Positives = 158/252 (62%), Gaps = 8/252 (3%)
Query: 41 VDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQ 100
+D GNN+ +PTL K+N+PPYGRDF PTGRF +GK+ +D AE+LG P YL
Sbjct: 1 MDTGNNNDIPTLLKSNFPPYGRDFPGAIPTGRFSDGKVPSDIIAESLGIAKTLPPYLGSN 60
Query: 101 ASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIK 160
+LL G FAS SGYD +TL + +S QL+YF+EY K+ Q G +K I++
Sbjct: 61 LKPHDLLKGVIFASGGSGYDPLTSTLLSVVSMSDQLKYFQEYLAKIKQHFGEEKVKFILE 120
Query: 161 DSLYVLSAGSSDFVQNYYTNPWINQAITVDQ--YSSYLLDSFTNFIKGVYGLGARKIGVT 218
S++++ + S+D + Y W+ +++ D+ Y+ YL++ + FIK + LGA+ IG+
Sbjct: 121 KSVFLVVSSSNDLAETY----WV-RSVEYDRNSYAEYLVELASEFIKELSELGAKNIGLF 175
Query: 219 SLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYK 278
S P+GCLPA RTLFG E C ++N A FN K+SS+ L+K+LP +++ D+Y
Sbjct: 176 SGVPVGCLPAQRTLFGGFERKCYEKLNNMALHFNSKLSSSLDTLKKELPS-RLIFIDVYD 234
Query: 279 PLYDLVQNPSNF 290
L D+++NP+N+
Sbjct: 235 TLLDIIKNPTNY 246
>At2g30220 putative GDSL-motif lipase/hydrolase
Length = 358
Score = 204 bits (518), Expect = 6e-53
Identities = 110/288 (38%), Positives = 164/288 (56%), Gaps = 13/288 (4%)
Query: 12 LLIVSCFLTCGSFAQDT--LVPAIMTFGDSAVDVGNNDYLP-TLFKANYPPYGRDFTNKQ 68
L + + ++C + A T L PAI+ FGDS D GNN+Y +FKAN+ PYG D +
Sbjct: 11 LFVATLLVSCNADANTTQPLFPAILIFGDSTADTGNNNYYSQAVFKANHLPYGVDLPGHE 70
Query: 69 PTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNH 128
GRF NGKL +D + L F P +L P S ++++ G FASA +GYD++ + +
Sbjct: 71 ANGRFSNGKLISDVISTKLNIKEFVPPFLQPNISDQDIVTGVCFASAGAGYDDETSLSSK 130
Query: 129 AIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWIN-QAI 187
AIP+SQQ FK Y +L + G KKA II ++L V+SAG +DF+ N+Y P +
Sbjct: 131 AIPVSQQPSMFKNYIARLKGIVGDKKAMEIINNALVVISAGPNDFILNFYDIPIRRLEYP 190
Query: 188 TVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLP-----AARTLFGYHENGCVA 242
T+ Y ++L F++ +Y LG R I V LPP+GCLP RT+ G CV
Sbjct: 191 TIYGYQDFVLKRLDGFVRELYSLGCRNILVGGLPPMGCLPIQLTAKLRTILGI----CVE 246
Query: 243 RINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
+ N D+ +N+K+ +Q LPG K + ++Y P+ D+++NPS +
Sbjct: 247 QENKDSILYNQKLVKKLPEIQASLPGSKFLYANVYDPVMDMIRNPSKY 294
>At1g59406 proline-rich protein, putative
Length = 349
Score = 203 bits (517), Expect = 8e-53
Identities = 106/286 (37%), Positives = 168/286 (58%), Gaps = 7/286 (2%)
Query: 8 ETLVLLIVSCFLTCGSFAQ--DTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFT 65
+ L+ +V F+ + Q +T +PA++ FGDS +D GNN+ LPTL K N+PPYG+D+
Sbjct: 4 QILLFALVLIFVEANAATQGKNTTIPALIVFGDSIMDTGNNNNLPTLLKCNFPPYGKDYP 63
Query: 66 NKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAAT 125
TGRF +G++ +D AE LG PAY++P ++LL G FAS +GYD A
Sbjct: 64 GGFATGRFSDGRVPSDLIAEKLGLAKTLPAYMNPYLKPEDLLKGVTFASGGTGYDPLTAK 123
Query: 126 LNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQ 185
+ I + QL FKEY K+ + G +KA I++ S +++ + S+D Y
Sbjct: 124 IMSVISVWDQLINFKEYISKIKRHFGEEKAKDILEHSFFLVVSSSNDLAHTYLAQ---TH 180
Query: 186 AITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLF-GYHENGCVARI 244
Y+++L DS +F++ ++ LGARKIGV S P+GC+P RT+F G+ GC +
Sbjct: 181 RYDRTSYANFLADSAVHFVRELHKLGARKIGVFSAVPVGCVPLQRTVFGGFFTRGCNQPL 240
Query: 245 NTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
N A+ FN ++S A +L K+L G+ I+ ++Y L+D++Q+P +
Sbjct: 241 NNMAKQFNARLSPALDSLDKELDGV-ILYINVYDTLFDMIQHPKKY 285
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,474,786
Number of Sequences: 26719
Number of extensions: 271813
Number of successful extensions: 973
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 110
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 593
Number of HSP's gapped (non-prelim): 123
length of query: 291
length of database: 11,318,596
effective HSP length: 98
effective length of query: 193
effective length of database: 8,700,134
effective search space: 1679125862
effective search space used: 1679125862
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146586.11


