

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.14 - phase: 0
(891 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
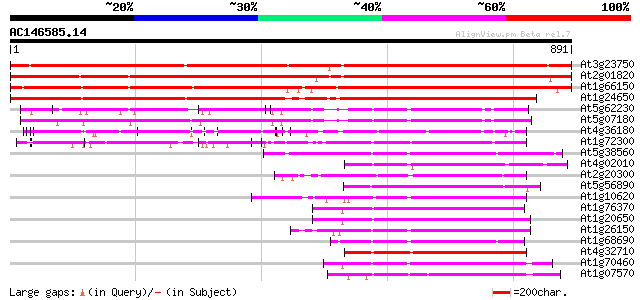
Score E
Sequences producing significant alignments: (bits) Value
At3g23750 receptor-like kinase, putative 993 0.0
At2g01820 putative receptor-like protein kinase 858 0.0
At1g66150 putative receptor protein kinase (TMK1) 840 0.0
At1g24650 putative protein kinase 738 0.0
At5g62230 unknown protein 281 9e-76
At5g07180 receptor-like protein kinase 278 7e-75
At4g36180 receptor protein kinase like protein 275 1e-73
At1g72300 leucine-rich receptor-like protein kinase, putative 272 7e-73
At5g38560 putative protein 255 7e-68
At4g02010 putative NAK-like ser/thr protein kinase 240 2e-63
At2g20300 protein kinase like protein 240 2e-63
At5g56890 unknown protein 239 4e-63
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 235 7e-62
At1g76370 putative protein kinase 234 1e-61
At1g20650 unknown protein 233 3e-61
At1g26150 Pto kinase interactor, putative 232 6e-61
At1g68690 protein kinase, putative 231 1e-60
At4g32710 putative protein kinase 231 1e-60
At1g70460 putative protein kinase 231 1e-60
At1g07570 protein kinase APK1A 231 1e-60
>At3g23750 receptor-like kinase, putative
Length = 928
Score = 993 bits (2566), Expect = 0.0
Identities = 523/909 (57%), Positives = 650/909 (70%), Gaps = 28/909 (3%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLT 60
M +L +S P P+ WSS T FC+WSGV+C+ RVT+I+L+D+ L G + +++L++L
Sbjct: 30 MLALAKSFNPPPSDWSSTTDFCKWSGVRCTG-GRVTTISLADKSLTGFIAPEISTLSELK 88
Query: 61 TLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPW 120
++ +Q N LSG +PS A LSSL ++ + NNF V GAF+GL SLQ LSL +N N++ W
Sbjct: 89 SVSIQRNKLSGTIPSFAKLSSLQEIYMDENNFVGVETGAFAGLTSLQILSLSDNNNITTW 148
Query: 121 TFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQ 180
+FP+EL S++L +I ++ I G LPDIF S +SL L L+YNN++G LP SL S IQ
Sbjct: 149 SFPSELVDSTSLTTIYLDNTNIAGVLPDIFDSLASLQNLRLSYNNITGVLPPSLGKSSIQ 208
Query: 181 SFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLTGV 240
+ WINN G++G+I V+S+MT L+Q WLH N F GPIPDLS+ ++ DLQLRDN LTG+
Sbjct: 209 NLWINNQDLGMSGTIEVLSSMTSLSQAWLHKNHFFGPIPDLSKSENLFDLQLRDNDLTGI 268
Query: 241 VPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDL 300
VP +L++++ L+N++L NN+ QGP+P+F +VK D HN FC A C +VM L
Sbjct: 269 VPPTLLTLASLKNISLDNNKFQGPLPLFSPEVKVTID---HNVFCTTKAGQSCSPQVMTL 325
Query: 301 LHIVGGFGYPIQFAKSWTGNDPCKDWLCVIC--GGGKITKLNFAKQGLQGTISPAFANLT 358
L + GG GYP A+SW G+D C W V C G + LN K G G ISPA ANLT
Sbjct: 326 LAVAGGLGYPSMLAESWQGDDACSGWAYVSCDSAGKNVVTLNLGKHGFTGFISPAIANLT 385
Query: 359 DLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKF-ITDGNVWLGK 417
L +LYLNGN+LTG IP+ L ++ L+ +DVSNN+L GE+PKF VKF GN LG
Sbjct: 386 SLKSLYLNGNDLTGVIPKELTFMTSLQLIDVSNNNLRGEIPKFPATVKFSYKPGNALLGT 445
Query: 418 NHGGGAP---GSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGL 474
N G G+ G A GG SG G S KV +I+ +++ V+ F+ + +K ++
Sbjct: 446 NGGDGSSPGTGGASGGPGGSSGGGGS--KVGVIVGVIVAVLVFLAILGFVVYKFVMKRKY 503
Query: 475 RRFARVGNPENGEGNVKLDLASVSNGYGGASS--------ELQSQSSGDHSDLHGFDGGN 526
RF R + G+ V +++ +G GG ++ L S SSGD+SD +GG+
Sbjct: 504 GRFNRTDPEKVGKILVSDAVSNGGSGNGGYANGHGANNFNALNSPSSGDNSDRFLLEGGS 563
Query: 527 GGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNE 586
TI + VLRQVTN+FS+DNILGRGGFG+VY GEL DGTK AVKRM A G+KG++E
Sbjct: 564 ---VTIPMEVLRQVTNNFSEDNILGRGGFGVVYAGELHDGTKTAVKRMECAAMGNKGMSE 620
Query: 587 FQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQ 646
FQAEI VLTKVRHRHLVALLGYC+NGNERLLVYE+MPQG L QHLFE E GY+PLTWKQ
Sbjct: 621 FQAEIAVLTKVRHRHLVALLGYCVNGNERLLVYEYMPQGNLGQHLFEWSELGYSPLTWKQ 680
Query: 647 RLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVE 706
R+ IALDV RGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG YSVE
Sbjct: 681 RVSIALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE 740
Query: 707 TKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRR 766
T+LAGTFGYLAPEYAATGRVTTKVDVYAFGVVLME++TGRKALDDS+PDE SHLVTWFRR
Sbjct: 741 TRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMEILTGRKALDDSLPDERSHLVTWFRR 800
Query: 767 VLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQ 826
+L NKENIPKA+DQTL+ DEETM SIY+VAELAGHCT R P QRPD+GHAVNVL PLV++
Sbjct: 801 ILINKENIPKALDQTLEADEETMESIYRVAELAGHCTAREPQQRPDMGHAVNVLGPLVEK 860
Query: 827 WEPTTHTDESTCADDNQMSLTQALQRWQANEGT--STFFNG--MTSQTQSSSTSKPPVFA 882
W+P+ +E + D MSL QALQRWQ NEGT ST F+G SQTQSS K F
Sbjct: 861 WKPSCQEEEESFGIDVNMSLPQALQRWQ-NEGTSSSTMFHGDFSYSQTQSSIPPKASGFP 919
Query: 883 DSLHSPDCR 891
++ S D R
Sbjct: 920 NTFDSADGR 928
>At2g01820 putative receptor-like protein kinase
Length = 943
Score = 858 bits (2218), Expect = 0.0
Identities = 474/919 (51%), Positives = 596/919 (64%), Gaps = 36/919 (3%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLT 60
MQSL+ SL + + SN + C+W V+C NRVT I L + + GTLP NL SL++L
Sbjct: 33 MQSLKSSLNLTSDVDWSNPNPCKWQSVQCDGSNRVTKIQLKQKGIRGTLPTNLQSLSELV 92
Query: 61 TLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPW 120
L L N +SGP+P L+ LS L +NL N F+SV FSG++SLQ + L EN PW
Sbjct: 93 ILELFLNRISGPIPDLSGLSRLQTLNLHDNLFTSVPKNLFSGMSSLQEMYL-ENNPFDPW 151
Query: 121 TFPTELTQSSNLNSIDINQAKINGTLPDIFGSFS--SLNTLHLAYNNLSGGLPNSLAGSG 178
P + ++++L ++ ++ I G +PD FGS S SL L L+ N L G LP S AG+
Sbjct: 152 VIPDTVKEATSLQNLTLSNCSIIGKIPDFFGSQSLPSLTNLKLSQNGLEGELPMSFAGTS 211
Query: 179 IQSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLT 238
IQS ++N L GSI+V+ NMT L +V L N+F+GPIPDLS S++ +R+NQLT
Sbjct: 212 IQSLFLNGQK--LNGSISVLGNMTSLVEVSLQGNQFSGPIPDLSGLVSLRVFNVRENQLT 269
Query: 239 GVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVM 298
GVVP SLVS+S L V L NN LQGP P+FGK V + + + N+FC N A CD RV
Sbjct: 270 GVVPQSLVSLSSLTTVNLTNNYLQGPTPLFGKSVGVDIVN-NMNSFCTNVAGEACDPRVD 328
Query: 299 DLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLT 358
L+ + FGYP++ A+SW GN+PC +W+ + C GG IT +N KQ L GTISP+ A LT
Sbjct: 329 TLVSVAESFGYPVKLAESWKGNNPCVNWVGITCSGGNITVVNMRKQDLSGTISPSLAKLT 388
Query: 359 DLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKN 418
L + L N L+G IP L TLS+L LDVSNND G PKF V +T+GN +GKN
Sbjct: 389 SLETINLADNKLSGHIPDELTTLSKLRLLDVSNNDFYGIPPKFRDTVTLVTEGNANMGKN 448
Query: 419 ---HGGGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWK--CYSRKG 473
APG++PG P+G G+ K + I++ VVG VVG C K
Sbjct: 449 GPNKTSDAPGASPGSKPSGGSDGSETSKKSSNVKIIVPVVGGVVGALCLVGLGVCLYAKK 508
Query: 474 LRRFARVGNPENG----------EGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFD 523
+R ARV +P + ++KL +A+ S GG S S S SD+H +
Sbjct: 509 RKRPARVQSPSSNMVIHPHHSGDNDDIKLTVAASSLNSGGGSDSY-SHSGSAASDIHVVE 567
Query: 524 GGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKG 583
GN ISI VLR VTN+FS++NILGRGGFG VYKGEL DGTKIAVKRM S KG
Sbjct: 568 AGN---LVISIQVLRNVTNNFSEENILGRGGFGTVYKGELHDGTKIAVKRMESSVVSDKG 624
Query: 584 LNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLT 643
L EF++EI VLTK+RHRHLVALLGYC++GNERLLVYE+MPQGTL+QHLF +E G PL
Sbjct: 625 LTEFKSEITVLTKMRHRHLVALLGYCLDGNERLLVYEYMPQGTLSQHLFHWKEEGRKPLD 684
Query: 644 WKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNY 703
W +RL IALDV RGVEYLH+LA QSFIHRDLKPSNILLGDDMRAKV+DFGLV+ APDG Y
Sbjct: 685 WTRRLAIALDVARGVEYLHTLAHQSFIHRDLKPSNILLGDDMRAKVSDFGLVRLAPDGKY 744
Query: 704 SVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTW 763
S+ET++AGTFGYLAPEYA TGRVTTKVD+++ GV+LMELITGRKALD++ P++S HLVTW
Sbjct: 745 SIETRVAGTFGYLAPEYAVTGRVTTKVDIFSLGVILMELITGRKALDETQPEDSVHLVTW 804
Query: 764 FRRVLTNKEN--IPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLC 821
FRRV +K+ AID + D++T+ SI KV ELAGHC R PYQRPD+ H VNVL
Sbjct: 805 FRRVAASKDENAFKNAIDPNISLDDDTVASIEKVWELAGHCCAREPYQRPDMAHIVNVLS 864
Query: 822 PLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANE---------GTSTFFNGMTSQTQS 872
L QW+PT + D M L Q L++WQA E G+S+ G TQ+
Sbjct: 865 SLTVQWKPTETDPDDVYGIDYDMPLPQVLKKWQAFEGLSQTADDSGSSSSAYGSKDNTQT 924
Query: 873 SSTSKPPVFADSLHSPDCR 891
S ++P FADS S D R
Sbjct: 925 SIPTRPSGFADSFTSVDGR 943
>At1g66150 putative receptor protein kinase (TMK1)
Length = 942
Score = 840 bits (2171), Expect = 0.0
Identities = 476/922 (51%), Positives = 591/922 (63%), Gaps = 42/922 (4%)
Query: 1 MQSLRQSLKP-SPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQL 59
M SL++SL P S GWS C+W+ + C+ RVT I + L GTL +L +L++L
Sbjct: 32 MLSLKKSLNPPSSFGWSDPDP-CKWTHIVCTGTKRVTRIQIGHSGLQGTLSPDLRNLSEL 90
Query: 60 TTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
L LQ N +SGP+PSL+ L+SL + L +NNF S+ F GL SLQ++ + N
Sbjct: 91 ERLELQWNNISGPVPSLSGLASLQVLMLSNNNFDSIPSDVFQGLTSLQSVEIDNN-PFKS 149
Query: 120 WTFPTELTQSSNLNSIDINQAKINGTLPDIFG--SFSSLNTLHLAYNNLSGGLPNSLAGS 177
W P L +S L + N A ++G+LP G F L+ LHLA+NNL G LP SLAGS
Sbjct: 150 WEIPESLRNASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELPMSLAGS 209
Query: 178 GIQSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQL 237
+QS W+N LTG ITV+ NMT L +VWLH NKF+GP+PD S ++ L LRDN
Sbjct: 210 QVQSLWLNGQK--LTGDITVLQNMTGLKEVWLHSNKFSGPLPDFSGLKELESLSLRDNSF 267
Query: 238 TGVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARV 297
TG VP SL+S+ L+ V L NN LQGPVPVF V + D S N+FC ++ CD RV
Sbjct: 268 TGPVPASLLSLESLKVVNLTNNHLQGPVPVFKSSVSVDLDKDS-NSFCLSSPG-ECDPRV 325
Query: 298 MDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANL 357
LL I F YP + A+SW GNDPC +W+ + C G IT ++ K L GTISP F +
Sbjct: 326 KSLLLIASSFDYPPRLAESWKGNDPCTNWIGIACSNGNITVISLEKMELTGTISPEFGAI 385
Query: 358 TDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGK 417
L + L NNLTG IPQ L TL L+TLDVS+N L G+VP F V T+GN +GK
Sbjct: 386 KSLQRIILGINNLTGMIPQELTTLPNLKTLDVSSNKLFGKVPGFRSNVVVNTNGNPDIGK 445
Query: 418 NHGG-GAPGSAPGGSPAGSGKGAS-----MKKVWIIIIIVLIVVG-----FVVGGAWFSW 466
+ +PGS+ +GSG MK I IIV V+G F++G F
Sbjct: 446 DKSSLSSPGSSSPSGGSGSGINGDKDRRGMKSSTFIGIIVGSVLGGLLSIFLIGLLVF-- 503
Query: 467 KCYSRKGLRRF-------ARVGNPEN-GEGN--VKLDLASVSNGYGGASSELQSQSSGDH 516
C+ +K +RF A V +P + G N VK+ +A S GG S + +
Sbjct: 504 -CWYKKRQKRFSGSESSNAVVVHPRHSGSDNESVKITVAGSSVSVGGISDTYTLPGTSEV 562
Query: 517 SDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMIS 576
D GN ISI VLR VTN+FS DNILG GGFG+VYKGEL DGTKIAVKRM +
Sbjct: 563 GD--NIQMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMEN 620
Query: 577 VAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECRE 636
KG EF++EI VLTKVRHRHLV LLGYC++GNE+LLVYE+MPQGTL++HLFE E
Sbjct: 621 GVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSE 680
Query: 637 HGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVK 696
G PL WKQRL +ALDV RGVEYLH LA QSFIHRDLKPSNILLGDDMRAKVADFGLV+
Sbjct: 681 EGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVR 740
Query: 697 NAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDE 756
AP+G S+ET++AGTFGYLAPEYA TGRVTTKVDVY+FGV+LMELITGRK+LD+S P+E
Sbjct: 741 LAPEGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEE 800
Query: 757 SSHLVTWFRRVLTNKE-NIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGH 815
S HLV+WF+R+ NKE + KAID T+D DEET+ S++ VAELAGHC R PYQRPD+GH
Sbjct: 801 SIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGH 860
Query: 816 AVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMTS------Q 869
AVN+L LV+ W+P+ E D MSL QAL++WQA EG S + +S
Sbjct: 861 AVNILSSLVELWKPSDQNPEDIYGIDLDMSLPQALKKWQAYEGRSDLESSTSSLLPSLDN 920
Query: 870 TQSSSTSKPPVFADSLHSPDCR 891
TQ S ++P FA+S S D R
Sbjct: 921 TQMSIPTRPYGFAESFTSVDGR 942
>At1g24650 putative protein kinase
Length = 886
Score = 738 bits (1906), Expect = 0.0
Identities = 414/843 (49%), Positives = 532/843 (62%), Gaps = 37/843 (4%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSG-VKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQL 59
M +LR SLK S N S + C+WS +KC + NRVT+I + D+ ++G LP +L LT L
Sbjct: 27 MIALRDSLKLSGNPNWSGSDPCKWSMFIKCDASNRVTAIQIGDRGISGKLPPDLGKLTSL 86
Query: 60 TTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
T + N L+GP+PSLA L SL V N+F+SV FSGL+SLQ +SL N
Sbjct: 87 TKFEVMRNRLTGPIPSLAGLKSLVTVYANDNDFTSVPEDFFSGLSSLQHVSLDNN-PFDS 145
Query: 120 WTFPTELTQSSNLNSIDINQAKINGTLPDIF---GSFSSLNTLHLAYNNLSGGLPNSLAG 176
W P L +++L ++G +PD FSSL TL L+YN+L P + +
Sbjct: 146 WVIPPSLENATSLVDFSAVNCNLSGKIPDYLFEGKDFSSLTTLKLSYNSLVCEFPMNFSD 205
Query: 177 SGIQSFWINNNL--PGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRD 234
S +Q +N L GSI+ + MT LT V L N F+GP+PD S S+K +R+
Sbjct: 206 SRVQVLMLNGQKGREKLHGSISFLQKMTSLTNVTLQGNSFSGPLPDFSGLVSLKSFNVRE 265
Query: 235 NQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVF-GKDVKYNSDDISHNNFCNNNASVPC 293
NQL+G+VP SL + L +V L NN LQGP P F D+K + + + N+FC + C
Sbjct: 266 NQLSGLVPSSLFELQSLSDVALGNNLLQGPTPNFTAPDIKPDLNGL--NSFCLDTPGTSC 323
Query: 294 DARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPA 353
D RV LL IV FGYP+ FA+ W GNDPC W+ + C G IT +NF GL GTISP
Sbjct: 324 DPRVNTLLSIVEAFGYPVNFAEKWKGNDPCSGWVGITCTGTDITVINFKNLGLNGTISPR 383
Query: 354 FANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNV 413
FA+ L + L+ NNL G+IPQ LA LS L+TLDVS N L GEVP+F+ + T GN
Sbjct: 384 FADFASLRVINLSQNNLNGTIPQELAKLSNLKTLDVSKNRLCGEVPRFNTTI-VNTTGNF 442
Query: 414 WLGKNHGGGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKG 473
N G S+ G GS G I++ L+++G + + K
Sbjct: 443 EDCPNGNAGKKASSNAGKIVGSVIG---------ILLALLLIGVAI--------FFLVKK 485
Query: 474 LRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATIS 533
++ ++ +P+ + ++ N G S +S SG+ + L G GN IS
Sbjct: 486 KMQYHKM-HPQQQSSDQDAFKITIENLCTGVS---ESGFSGNDAHL-----GEAGNIVIS 536
Query: 534 IHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGV 593
I VLR T +F + NILGRGGFGIVYKGEL DGTKIAVKRM S KGL+EF++EI V
Sbjct: 537 IQVLRDATYNFDEKNILGRGGFGIVYKGELHDGTKIAVKRMESSIISGKGLDEFKSEIAV 596
Query: 594 LTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALD 653
LT+VRHR+LV L GYC+ GNERLLVY++MPQGTL++H+F +E G PL W +RLIIALD
Sbjct: 597 LTRVRHRNLVVLHGYCLEGNERLLVYQYMPQGTLSRHIFYWKEEGLRPLEWTRRLIIALD 656
Query: 654 VGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTF 713
V RGVEYLH+LA QSFIHRDLKPSNILLGDDM AKVADFGLV+ AP+G S+ETK+AGTF
Sbjct: 657 VARGVEYLHTLAHQSFIHRDLKPSNILLGDDMHAKVADFGLVRLAPEGTQSIETKIAGTF 716
Query: 714 GYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKEN 773
GYLAPEYA TGRVTTKVDVY+FGV+LMEL+TGRKALD + +E HL TWFRR+ NK +
Sbjct: 717 GYLAPEYAVTGRVTTKVDVYSFGVILMELLTGRKALDVARSEEEVHLATWFRRMFINKGS 776
Query: 774 IPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHT 833
PKAID+ ++ +EET+ SI VAELA C++R P RPD+ H VNVL LV QW+PT +
Sbjct: 777 FPKAIDEAMEVNEETLRSINIVAELANQCSSREPRDRPDMNHVVNVLVSLVVQWKPTERS 836
Query: 834 DES 836
+S
Sbjct: 837 SDS 839
>At5g62230 unknown protein
Length = 966
Score = 281 bits (720), Expect = 9e-76
Identities = 240/910 (26%), Positives = 394/910 (42%), Gaps = 149/910 (16%)
Query: 18 NTSFCQWSGVKCSSDN-------------------------RVTSINLSDQKLAGTLPDN 52
N+ C W GV C + + + SI+L KLAG +PD
Sbjct: 55 NSDLCSWRGVFCDNVSYSVVSLNLSSLNLGGEISPAIGDLRNLQSIDLQGNKLAGQIPDE 114
Query: 53 LNSLTQLTTLYLQNNALSGPLP-SLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSL 111
+ + L L L N L G +P S++ L L +NL +N + P + + +L+ L L
Sbjct: 115 IGNCASLVYLDLSENLLYGDIPFSISKLKQLETLNLKNNQLTGPVPATLTQIPNLKRLDL 174
Query: 112 ------GENINLSPW----------------TFPTELTQSSNLNSIDINQAKINGTLPDI 149
GE L W T +++ Q + L D+ + GT+P+
Sbjct: 175 AGNHLTGEISRLLYWNEVLQYLGLRGNMLTGTLSSDMCQLTGLWYFDVRGNNLTGTIPES 234
Query: 150 FGSFSSLNTLHLAYNNLSGGLPNSL-----AGSGIQSFWINNNLPGLTGSIT-------- 196
G+ +S L ++YN ++G +P ++ A +Q + +P + G +
Sbjct: 235 IGNCTSFQILDISYNQITGEIPYNIGFLQVATLSLQGNRLTGRIPEVIGLMQALAVLDLS 294
Query: 197 ----------VISNMTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSL 245
++ N++ +++LH N TGPIP +L + + LQL DN+L G +P L
Sbjct: 295 DNELVGPIPPILGNLSFTGKLYLHGNMLTGPIPSELGNMSRLSYLQLNDNKLVGTIPPEL 354
Query: 246 VSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVG 305
+ L + L NN+L GP+P + H N + S+P R + L +
Sbjct: 355 GKLEQLFELNLANNRLVGPIPSNISSCAALNQFNVHGNLLSG--SIPLAFRNLGSLTYLN 412
Query: 306 ------------GFGYPIQFAK-SWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISP 352
G+ I K +GN+ + + LN ++ L G +
Sbjct: 413 LSSNNFKGKIPVELGHIINLDKLDLSGNNFSGSIPLTLGDLEHLLILNLSRNHLSGQLPA 472
Query: 353 AFANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGN 412
F NL + + ++ N L+G IP L L L +L ++NN L G++P + + N
Sbjct: 473 EFGNLRSIQMIDVSFNLLSGVIPTELGQLQNLNSLILNNNKLHGKIPDQLTNCFTLVNLN 532
Query: 413 VWLG---------KNHGGGAPGSAPG-----GSPAGSGKGASMK-KVWIIIIIVLIVVGF 457
V KN AP S G G+ GS G K +V+ ++ IV+G
Sbjct: 533 VSFNNLSGIVPPMKNFSRFAPASFVGNPYLCGNWVGSICGPLPKSRVFSRGALICIVLGV 592
Query: 458 VVGGAWFSWKCYSRKGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHS 517
+ Y ++ + G+ + EG KL + +
Sbjct: 593 ITLLCMIFLAVYKSMQQKKILQ-GSSKQAEGLTKLVILHMDMA----------------- 634
Query: 518 DLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISV 577
+H FD + +VT + ++ I+G G VYK L IA+KR+ +
Sbjct: 635 -IHTFDD------------IMRVTENLNEKFIIGYGASSTVYKCALKSSRPIAIKRLYN- 680
Query: 578 AKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREH 637
+ L EF+ E+ + +RHR++V+L GY ++ LL Y++M G+L L H
Sbjct: 681 -QYPHNLREFETELETIGSIRHRNIVSLHGYALSPTGNLLFYDYMENGSLWDLL-----H 734
Query: 638 G---YTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGL 694
G L W+ RL IA+ +G+ YLH IHRD+K SNILL ++ A ++DFG+
Sbjct: 735 GSLKKVKLDWETRLKIAVGAAQGLAYLHHDCTPRIIHRDIKSSNILLDENFEAHLSDFGI 794
Query: 695 VKNAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVP 754
K+ P T + GT GY+ PEYA T R+ K D+Y+FG+VL+EL+TG+KA+D+
Sbjct: 795 AKSIPASKTHASTYVLGTIGYIDPEYARTSRINEKSDIYSFGIVLLELLTGKKAVDN--- 851
Query: 755 DESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIG 814
+ + H + + + + +A+D + + I K +LA CT R+P +RP +
Sbjct: 852 EANLHQLILSK---ADDNTVMEAVDPEVTVTCMDLGHIRKTFQLALLCTKRNPLERPTML 908
Query: 815 HAVNVLCPLV 824
VL LV
Sbjct: 909 EVSRVLLSLV 918
Score = 97.8 bits (242), Expect = 2e-20
Identities = 103/355 (29%), Positives = 157/355 (44%), Gaps = 33/355 (9%)
Query: 68 ALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLG-ENINLSPWTFPTEL 126
A+ G +L N+ L D + N+ G F S +SL ++NL P +
Sbjct: 35 AIKGSFSNLVNM--LLDWDDVHNSDLCSWRGVFCDNVSYSVVSLNLSSLNLGGEISPA-I 91
Query: 127 TQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAG-SGIQSFWIN 185
NL SID+ K+ G +PD G+ +SL L L+ N L G +P S++ +++ +
Sbjct: 92 GDLRNLQSIDLQGNKLAGQIPDEIGNCASLVYLDLSENLLYGDIPFSISKLKQLETLNLK 151
Query: 186 NNLPGLTGSI-TVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKD-LQLRDNQLTGVVPD 243
NN LTG + ++ + L ++ L N TG I L N + L LR N LTG +
Sbjct: 152 NNQ--LTGPVPATLTQIPNLKRLDLAGNHLTGEISRLLYWNEVLQYLGLRGNMLTGTLSS 209
Query: 244 SLVSMSGLQNVTLRNNQLQGPVPV-FGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLH 302
+ ++GL +R N L G +P G + DIS+N
Sbjct: 210 DMCQLTGLWYFDVRGNNLTGTIPESIGNCTSFQILDISYNQ------------------- 250
Query: 303 IVGGFGYPIQFAK----SWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLT 358
I G Y I F + S GN VI + L+ + L G I P NL+
Sbjct: 251 ITGEIPYNIGFLQVATLSLQGNRLTGRIPEVIGLMQALAVLDLSDNELVGPIPPILGNLS 310
Query: 359 DLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNV 413
LYL+GN LTG IP L +S+L L +++N L G +P K++ + + N+
Sbjct: 311 FTGKLYLHGNMLTGPIPSELGNMSRLSYLQLNDNKLVGTIPPELGKLEQLFELNL 365
Score = 57.4 bits (137), Expect = 4e-08
Identities = 36/111 (32%), Positives = 50/111 (44%), Gaps = 4/111 (3%)
Query: 300 LLHIVGGFGYPIQFAKSWTG--NDPCKDWLCVICGGGK--ITKLNFAKQGLQGTISPAFA 355
L+ I G F + W N W V C + LN + L G ISPA
Sbjct: 33 LMAIKGSFSNLVNMLLDWDDVHNSDLCSWRGVFCDNVSYSVVSLNLSSLNLGGEISPAIG 92
Query: 356 NLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVK 406
+L +L ++ L GN L G IP + + L LD+S N L G++P K+K
Sbjct: 93 DLRNLQSIDLQGNKLAGQIPDEIGNCASLVYLDLSENLLYGDIPFSISKLK 143
>At5g07180 receptor-like protein kinase
Length = 932
Score = 278 bits (712), Expect = 7e-75
Identities = 241/916 (26%), Positives = 394/916 (42%), Gaps = 150/916 (16%)
Query: 18 NTSFCQWSGVKCSSDN-RVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLP-- 74
N FC W GV C + + V S+NLS+ L G + L L L ++ LQ N L G +P
Sbjct: 22 NHDFCSWRGVFCDNVSLNVVSLNLSNLNLGGEISSALGDLMNLQSIDLQGNKLGGQIPDE 81
Query: 75 -----------------------SLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSL 111
S++ L L +NL +N + P + + +L+TL L
Sbjct: 82 IGNCVSLAYVDFSTNLLFGDIPFSISKLKQLEFLNLKNNQLTGPIPATLTQIPNLKTLDL 141
Query: 112 GENI----------------------NLSPWTFPTELTQSSNLNSIDINQAKINGTLPDI 149
N N+ T ++ Q + L D+ + GT+P+
Sbjct: 142 ARNQLTGEIPRLLYWNEVLQYLGLRGNMLTGTLSPDMCQLTGLWYFDVRGNNLTGTIPES 201
Query: 150 FGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNN-LPG------------------ 190
G+ +S L ++YN ++G +P ++ + + + N L G
Sbjct: 202 IGNCTSFEILDVSYNQITGVIPYNIGFLQVATLSLQGNKLTGRIPEVIGLMQALAVLDLS 261
Query: 191 ---LTGSIT-VISNMTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSL 245
LTG I ++ N++ +++LH NK TG IP +L + + LQL DN+L G +P L
Sbjct: 262 DNELTGPIPPILGNLSFTGKLYLHGNKLTGQIPPELGNMSRLSYLQLNDNELVGKIPPEL 321
Query: 246 VSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLL---- 301
+ L + L NN L G +P + H NF + +VP + R + L
Sbjct: 322 GKLEQLFELNLANNNLVGLIPSNISSCAALNQFNVHGNFLSG--AVPLEFRNLGSLTYLN 379
Query: 302 --------HIVGGFGYPIQF-AKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISP 352
I G+ I +GN+ + + LN ++ L GT+
Sbjct: 380 LSSNSFKGKIPAELGHIINLDTLDLSGNNFSGSIPLTLGDLEHLLILNLSRNHLNGTLPA 439
Query: 353 AFANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGN 412
F NL + + ++ N L G IP L L + +L ++NN + G++P + + N
Sbjct: 440 EFGNLRSIQIIDVSFNFLAGVIPTELGQLQNINSLILNNNKIHGKIPDQLTNCFSLANLN 499
Query: 413 VWLG---------KNHGGGAPGSAPG-----GSPAGSGKGASMKK--VWIIIIIVLIVVG 456
+ KN +P S G G+ GS G S+ K V+ + ++ +V+G
Sbjct: 500 ISFNNLSGIIPPMKNFTRFSPASFFGNPFLCGNWVGSICGPSLPKSQVFTRVAVICMVLG 559
Query: 457 FVVGGAWFSWKCYSRKGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDH 516
F+ Y K + + G+ + EG+ KL + +
Sbjct: 560 FITLICMIFIAVYKSKQQKPVLK-GSSKQPEGSTKLVILHMDMA---------------- 602
Query: 517 SDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMIS 576
+H FD + +VT + + I+G G VYK IA+KR+ +
Sbjct: 603 --IHTFDD------------IMRVTENLDEKYIIGYGASSTVYKCTSKTSRPIAIKRIYN 648
Query: 577 VAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECRE 636
+ EF+ E+ + +RHR++V+L GY ++ LL Y++M G+L L
Sbjct: 649 --QYPSNFREFETELETIGSIRHRNIVSLHGYALSPFGNLLFYDYMENGSLWDLL----- 701
Query: 637 HG---YTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFG 693
HG L W+ RL IA+ +G+ YLH IHRD+K SNILL + A+++DFG
Sbjct: 702 HGPGKKVKLDWETRLKIAVGAAQGLAYLHHDCTPRIIHRDIKSSNILLDGNFEARLSDFG 761
Query: 694 LVKNAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSV 753
+ K+ P T + GT GY+ PEYA T R+ K D+Y+FG+VL+EL+TG+KA+D+
Sbjct: 762 IAKSIPATKTYASTYVLGTIGYIDPEYARTSRLNEKSDIYSFGIVLLELLTGKKAVDN-- 819
Query: 754 PDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDI 813
+ + H + + + + +A+D + I K +LA CT R+P +RP +
Sbjct: 820 -EANLHQMILSK---ADDNTVMEAVDAEVSVTCMDSGHIKKTFQLALLCTKRNPLERPTM 875
Query: 814 GHAVNVLCPLVQQWEP 829
VL LV P
Sbjct: 876 QEVSRVLLSLVPSPPP 891
>At4g36180 receptor protein kinase like protein
Length = 1136
Score = 275 bits (702), Expect = 1e-73
Identities = 223/807 (27%), Positives = 399/807 (48%), Gaps = 69/807 (8%)
Query: 34 RVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPS-LANLSSLTDVNLGSNNF 92
R+ + L++ L G +P + L L + N+L G +P L + +L ++LG N+F
Sbjct: 357 RLEELKLANNSLTGEIPVEIKQCGSLDVLDFEGNSLKGQIPEFLGYMKALKVLSLGRNSF 416
Query: 93 SSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIFGS 152
S P + L L+ L+LGEN NL+ +FP EL ++L+ +D++ + +G +P +
Sbjct: 417 SGYVPSSMVNLQQLERLNLGEN-NLNG-SFPVELMALTSLSELDLSGNRFSGAVPVSISN 474
Query: 153 FSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLPGLTGSITV-ISNMTLLTQVWLHV 211
S+L+ L+L+ N SG +P S+ G+ + ++ + ++G + V +S + + + L
Sbjct: 475 LSNLSFLNLSGNGFSGEIPASV-GNLFKLTALDLSKQNMSGEVPVELSGLPNVQVIALQG 533
Query: 212 NKFTGPIPD-LSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPV-FG 269
N F+G +P+ S S++ + L N +G +P + + L +++L +N + G +P G
Sbjct: 534 NNFSGVVPEGFSSLVSLRYVNLSSNSFSGEIPQTFGFLRLLVSLSLSDNHISGSIPPEIG 593
Query: 270 KDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCV 329
++ N + +P D + L ++ + +G P +
Sbjct: 594 NCSALEVLELRSNRLMGH---IPADLSRLPRLKVLD------LGQNNLSGEIPPE----- 639
Query: 330 ICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQ-LETLD 388
I + L+ L G I +F+ L++LT + L+ NNLTG IP +LA +S L +
Sbjct: 640 ISQSSSLNSLSLDHNHLSGVIPGSFSGLSNLTKMDLSVNNLTGEIPASLALISSNLVYFN 699
Query: 389 VSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAPGSAPGGSPAGSGKGASMKKVWIII 448
VS+N+L GE+P + I + + + G G P + S GK KK +I+
Sbjct: 700 VSSNNLKGEIP--ASLGSRINNTSEFSGNTELCGKPLNRRCESSTAEGK---KKKRKMIL 754
Query: 449 IIVLIVVGFVVGGAWFSWKCYS----RKGLRRFARVGNPENGEGNVKLDLASVSNGYGGA 504
+IV+ +G + + + Y+ RK L++ + G + G S G
Sbjct: 755 MIVMAAIGAFLLSLFCCFYVYTLLKWRKKLKQQSTTGEKKRSPGRT-------SAGSRVR 807
Query: 505 SSELQSQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELP 564
SS +S + L F N I++ + T F ++N+L R +G+++K
Sbjct: 808 SSTSRSSTENGEPKLVMF------NNKITLAETIEATRQFDEENVLSRTRYGLLFKANYN 861
Query: 565 DGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNE-RLLVYEHMP 623
DG ++++R+ + + ++ L F+ E VL KV+HR++ L GY + RLLVY++MP
Sbjct: 862 DGMVLSIRRLPNGSLLNENL--FKKEAEVLGKVKHRNITVLRGYYAGPPDLRLLVYDYMP 919
Query: 624 QGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGD 683
G L+ L E L W R +IAL + RG+ +LH Q + +H D+KP N+L
Sbjct: 920 NGNLSTLLQEASHQDGHVLNWPMRHLIALGIARGLGFLH---QSNMVHGDIKPQNVLFDA 976
Query: 684 DMRAKVADFGLVK---NAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLM 740
D A ++DFGL + +P + +V GT GY++PE +G +T + D+Y+FG+VL+
Sbjct: 977 DFEAHISDFGLDRLTIRSPSRS-AVTANTIGTLGYVSPEATLSGEITRESDIYSFGIVLL 1035
Query: 741 ELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQ---TLDPD----EETMLSIY 793
E++TG++ + + + +V W ++ L + + + ++ LDP+ EE +L I
Sbjct: 1036 EILTGKRPV---MFTQDEDIVKWVKKQL-QRGQVTELLEPGLLELDPESSEWEEFLLGI- 1090
Query: 794 KVAELAGHCTTRSPYQRPDIGHAVNVL 820
KV L CT P RP + V +L
Sbjct: 1091 KVGLL---CTATDPLDRPTMSDVVFML 1114
Score = 131 bits (330), Expect = 1e-30
Identities = 114/409 (27%), Positives = 180/409 (43%), Gaps = 56/409 (13%)
Query: 22 CQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLP-SLANLS 80
C W GV C++ +RVT I L +L+G + D ++ L L L L++N+ +G +P SLA +
Sbjct: 58 CDWRGVGCTN-HRVTEIRLPRLQLSGRISDRISGLRMLRKLSLRSNSFNGTIPTSLAYCT 116
Query: 81 SLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSIDINQA 140
L V L N+ S P A L SL+ ++ N LS P L S+L +DI+
Sbjct: 117 RLLSVFLQYNSLSGKLPPAMRNLTSLEVFNVAGN-RLSG-EIPVGLP--SSLQFLDISSN 172
Query: 141 KINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLPGLTGSITVISN 200
+G +P + + L L+L+YN L+G +P SL N
Sbjct: 173 TFSGQIPSGLANLTQLQLLNLSYNQLTGEIPASL------------------------GN 208
Query: 201 MTLLTQVWLHVNKFTGPIPD-LSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNN 259
+ L +WL N G +P +S C+S+ L +N++ GV+P + ++ L+ ++L NN
Sbjct: 209 LQSLQYLWLDFNLLQGTLPSAISNCSSLVHLSASENEIGGVIPAAYGALPKLEVLSLSNN 268
Query: 260 QLQGPVPV-FGKDVKYNSDDISHNNFCN-----NNASVPCDARVMDLLHIVGGFGYPIQF 313
G VP + + N F + A+ +V+DL +P+
Sbjct: 269 NFSGTVPFSLFCNTSLTIVQLGFNAFSDIVRPETTANCRTGLQVLDLQENRISGRFPL-- 326
Query: 314 AKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGS 373
WL I + L+ + G I P NL L L L N+LTG
Sbjct: 327 ------------WLTNIL---SLKNLDVSGNLFSGEIPPDIGNLKRLEELKLANNSLTGE 371
Query: 374 IPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGG 422
IP + L+ LD N L G++P+F +K + + LG+N G
Sbjct: 372 IPVEIKQCGSLDVLDFEGNSLKGQIPEFLGYMKALK--VLSLGRNSFSG 418
Score = 126 bits (317), Expect = 5e-29
Identities = 114/417 (27%), Positives = 188/417 (44%), Gaps = 51/417 (12%)
Query: 38 INLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPS-LANLSSLTDVNLGSNNFSSVT 96
+NLS +L G +P +L +L L L+L N L G LPS ++N SSL ++ N V
Sbjct: 191 LNLSYNQLTGEIPASLGNLQSLQYLWLDFNLLQGTLPSAISNCSSLVHLSASENEIGGVI 250
Query: 97 PGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSI--------------------- 135
P A+ L L+ LSL N N S T P L +++L +
Sbjct: 251 PAAYGALPKLEVLSLSNN-NFSG-TVPFSLFCNTSLTIVQLGFNAFSDIVRPETTANCRT 308
Query: 136 -----DINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAG-SGIQSFWINNNLP 189
D+ + +I+G P + SL L ++ N SG +P + ++ + NN
Sbjct: 309 GLQVLDLQENRISGRFPLWLTNILSLKNLDVSGNLFSGEIPPDIGNLKRLEELKLANN-- 366
Query: 190 GLTGSITV-ISNMTLLTQVWLHVNKFTGPIPD-LSQCNSIKDLQLRDNQLTGVVPDSLVS 247
LTG I V I L + N G IP+ L ++K L L N +G VP S+V+
Sbjct: 367 SLTGEIPVEIKQCGSLDVLDFEGNSLKGQIPEFLGYMKALKVLSLGRNSFSGYVPSSMVN 426
Query: 248 MSGLQNVTLRNNQLQGPVPVFGKDVKYNSD-DISHNNFCNNNASVPCDARVMDLLHIVGG 306
+ L+ + L N L G PV + S+ D+S N F + +VP + L +
Sbjct: 427 LQQLERLNLGENNLNGSFPVELMALTSLSELDLSGNRF---SGAVPVSISNLSNLSFLNL 483
Query: 307 FGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLN 366
G ++G P + K+T L+ +KQ + G + + L ++ + L
Sbjct: 484 SG------NGFSGEIPAS-----VGNLFKLTALDLSKQNMSGEVPVELSGLPNVQVIALQ 532
Query: 367 GNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGA 423
GNN +G +P+ ++L L +++S+N SGE+P+ ++ + ++ L NH G+
Sbjct: 533 GNNFSGVVPEGFSSLVSLRYVNLSSNSFSGEIPQTFGFLRLLV--SLSLSDNHISGS 587
Score = 98.6 bits (244), Expect = 1e-20
Identities = 79/268 (29%), Positives = 127/268 (46%), Gaps = 39/268 (14%)
Query: 27 VKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLP-SLANLSSLTDV 85
V S+ + ++ +NLS +G +P ++ +L +LT L L +SG +P L+ L ++ +
Sbjct: 470 VSISNLSNLSFLNLSGNGFSGEIPASVGNLFKLTALDLSKQNMSGEVPVELSGLPNVQVI 529
Query: 86 NLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSN----LNSIDINQAK 141
L NNFS V P FS L SL+ +NLS +F E+ Q+ L S+ ++
Sbjct: 530 ALQGNNFSGVVPEGFSSLVSLR------YVNLSSNSFSGEIPQTFGFLRLLVSLSLSDNH 583
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLPGLTGSITVISNM 201
I+G++P G+ S+L L L N L G +P L S +
Sbjct: 584 ISGSIPPEIGNCSALEVLELRSNRLMGHIPADL------------------------SRL 619
Query: 202 TLLTQVWLHVNKFTGPI-PDLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQ 260
L + L N +G I P++SQ +S+ L L N L+GV+P S +S L + L N
Sbjct: 620 PRLKVLDLGQNNLSGEIPPEISQSSSLNSLSLDHNHLSGVIPGSFSGLSNLTKMDLSVNN 679
Query: 261 LQGPVPVFGKDVKYNSDDISHNNFCNNN 288
L G +P + S ++ + N +NN
Sbjct: 680 LTGEIPA---SLALISSNLVYFNVSSNN 704
Score = 53.5 bits (127), Expect = 5e-07
Identities = 59/230 (25%), Positives = 86/230 (36%), Gaps = 42/230 (18%)
Query: 204 LTQVWLHVNKFTGPIPD-LSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQ 262
+T++ L + +G I D +S ++ L LR N G +P SL + L +V L+ N L
Sbjct: 70 VTEIRLPRLQLSGRISDRISGLRMLRKLSLRSNSFNGTIPTSLAYCTRLLSVFLQYNSLS 129
Query: 263 GPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTGNDP 322
G +P N + N N S I G +QF
Sbjct: 130 GKLP----PAMRNLTSLEVFNVAGNRLSG----------EIPVGLPSSLQF--------- 166
Query: 323 CKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLS 382
L+ + G I ANLT L L L+ N LTG IP +L L
Sbjct: 167 ----------------LDISSNTFSGQIPSGLANLTQLQLLNLSYNQLTGEIPASLGNLQ 210
Query: 383 QLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAPGSAPGGSP 432
L+ L + N L G +P + ++ +N GG +A G P
Sbjct: 211 SLQYLWLDFNLLQGTLPSAISNCSSLV--HLSASENEIGGVIPAAYGALP 258
Score = 52.4 bits (124), Expect = 1e-06
Identities = 37/126 (29%), Positives = 60/126 (47%), Gaps = 7/126 (5%)
Query: 310 PIQFAKSWTGNDPCK--DWLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNG 367
P+ SW + P DW V C ++T++ + L G IS + L L L L
Sbjct: 42 PLGALTSWDPSTPAAPCDWRGVGCTNHRVTEIRLPRLQLSGRISDRISGLRMLRKLSLRS 101
Query: 368 NNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAPGSA 427
N+ G+IP +LA ++L ++ + N LSG++P P ++ +T V+ G G
Sbjct: 102 NSFNGTIPTSLAYCTRLLSVFLQYNSLSGKLP---PAMRNLTSLEVF--NVAGNRLSGEI 156
Query: 428 PGGSPA 433
P G P+
Sbjct: 157 PVGLPS 162
Score = 50.8 bits (120), Expect = 3e-06
Identities = 37/130 (28%), Positives = 60/130 (45%), Gaps = 13/130 (10%)
Query: 330 ICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDV 389
I G + KL+ GTI + A T L +++L N+L+G +P + L+ LE +V
Sbjct: 88 ISGLRMLRKLSLRSNSFNGTIPTSLAYCTRLLSVFLQYNSLSGKLPPAMRNLTSLEVFNV 147
Query: 390 SNNDLSGEVPKFSP-KVKFI-TDGNVWLGKNHGGGAP-----------GSAPGGSPAGSG 436
+ N LSGE+P P ++F+ N + G+ G A G PA G
Sbjct: 148 AGNRLSGEIPVGLPSSLQFLDISSNTFSGQIPSGLANLTQLQLLNLSYNQLTGEIPASLG 207
Query: 437 KGASMKKVWI 446
S++ +W+
Sbjct: 208 NLQSLQYLWL 217
Score = 41.6 bits (96), Expect = 0.002
Identities = 34/100 (34%), Positives = 50/100 (50%), Gaps = 14/100 (14%)
Query: 30 SSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLP-SLANLSS-LTDVNL 87
S + + S++L L+G +P + + L+ LT + L N L+G +P SLA +SS L N+
Sbjct: 641 SQSSSLNSLSLDHNHLSGVIPGSFSGLSNLTKMDLSVNNLTGEIPASLALISSNLVYFNV 700
Query: 88 GSNNFSSVTPGAFSGLNSLQTLSLGENI-NLSPWTFPTEL 126
SNN P SLG I N S ++ TEL
Sbjct: 701 SSNNLKGEIPA-----------SLGSRINNTSEFSGNTEL 729
>At1g72300 leucine-rich receptor-like protein kinase, putative
Length = 1095
Score = 272 bits (695), Expect = 7e-73
Identities = 244/815 (29%), Positives = 386/815 (46%), Gaps = 86/815 (10%)
Query: 33 NRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSL--ANLSSLTDVNLGSN 90
++++S+ L L G++P +L + T+L L L+ N L G L ++ + SL+ ++LG+N
Sbjct: 319 SKLSSLQLHVNNLMGSIPVSLANCTKLVKLNLRVNQLGGTLSAIDFSRFQSLSILDLGNN 378
Query: 91 NFSSVTPGAFSGLNSLQTLSLGENI---NLSPWTFPTELTQSSNLNSIDINQAKINGTLP 147
+F+ P + + N +SP EL S D + G L
Sbjct: 379 SFTGEFPSTVYSCKMMTAMRFAGNKLTGQISPQVL--ELESLSFFTFSDNKMTNLTGALS 436
Query: 148 DIFGSFSSLNTLHLAYNNLSGGLPNS---LAGSGIQSFWINN-NLPGLTGSITV-ISNMT 202
+ G L+TL +A N +P++ L G S I LTG I + +
Sbjct: 437 ILQGC-KKLSTLIMAKNFYDETVPSNKDFLRSDGFPSLQIFGIGACRLTGEIPAWLIKLQ 495
Query: 203 LLTQVWLHVNKFTGPIPD-LSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNV----TLR 257
+ + L +N+F G IP L + L L DN LTG +P L + L +
Sbjct: 496 RVEVMDLSMNRFVGTIPGWLGTLPDLFYLDLSDNFLTGELPKELFQLRALMSQKAYDATE 555
Query: 258 NNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAK-- 315
N L+ PV V N ++++ N N +S+P P + K
Sbjct: 556 RNYLELPVFV-------NPNNVTTNQQYNQLSSLP-----------------PTIYIKRN 591
Query: 316 SWTGNDPCKDWLCVICGGGKITK-LNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSI 374
+ TG P V G K+ L G+I +NLT+L L L+ NNL+G I
Sbjct: 592 NLTGTIP------VEVGQLKVLHILELLGNNFSGSIPDELSNLTNLERLDLSNNNLSGRI 645
Query: 375 PQNLATLSQLETLDVSNNDLSGEVPKFS-----PKVKFITDGNVWLGKNHGGGAPGSA-- 427
P +L L L +V+NN LSG +P + PK F +GN L GG S
Sbjct: 646 PWSLTGLHFLSYFNVANNTLSGPIPTGTQFDTFPKANF--EGNPLL---CGGVLLTSCDP 700
Query: 428 PGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPENGE 487
S GKG + + ++ +++ + G + + S++ + NP + E
Sbjct: 701 TQHSTTKMGKG-KVNRTLVLGLVLGLFFGVSLILVLLALLVLSKRRV-------NPGDSE 752
Query: 488 GNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDD 547
N +L++ S G+ SE+ S D S + F ++I L + T++FS
Sbjct: 753 -NAELEINS-----NGSYSEVPPGSDKDISLVLLFGNSRYEVKDLTIFELLKATDNFSQA 806
Query: 548 NILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGL--NEFQAEIGVLTKVRHRHLVAL 605
NI+G GGFG+VYK L +GTK+AVK++ G G+ EF+AE+ VL++ +H +LVAL
Sbjct: 807 NIIGCGGFGLVYKATLDNGTKLAVKKL----TGDYGMMEKEFKAEVEVLSRAKHENLVAL 862
Query: 606 LGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLA 665
GYC++ + R+L+Y M G+L L E E G L W +RL I G+ Y+H +
Sbjct: 863 QGYCVHDSARILIYSFMENGSLDYWLHENPE-GPAQLDWPKRLNIMRGASSGLAYMHQIC 921
Query: 666 QQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYLAPEYAATGR 725
+ +HRD+K SNILL + +A VADFGL + V T+L GT GY+ PEY
Sbjct: 922 EPHIVHRDIKSSNILLDGNFKAYVADFGLSRLILPYRTHVTTELVGTLGYIPPEYGQAWV 981
Query: 726 VTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPD 785
T + DVY+FGVV++EL+TG++ ++ P S LV W + ++ P+ + TL +
Sbjct: 982 ATLRGDVYSFGVVMLELLTGKRPMEVFRPKMSRELVAWVHTM--KRDGKPEEVFDTLLRE 1039
Query: 786 EETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
++ +V ++A C ++P +RP+I V+ L
Sbjct: 1040 SGNEEAMLRVLDIACMCVNQNPMKRPNIQQVVDWL 1074
Score = 131 bits (330), Expect = 1e-30
Identities = 133/473 (28%), Positives = 201/473 (42%), Gaps = 86/473 (18%)
Query: 11 SPNGWSSNTSFCQWSGVKC--SSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNA 68
SP W+S+ C W G+ C S +NRVTSI LS + L+G LP ++ L +L+ L L +N
Sbjct: 68 SPLHWNSSIDCCSWEGISCDKSPENRVTSIILSSRGLSGNLPSSVLDLQRLSRLDLSHNR 127
Query: 69 LSGPLPS--LANLSSLTDVNLGSNNFSSVTP------GAFSGLNSLQTLSLGENI----- 115
LSGPLP L+ L L ++L N+F P +G+ +QT+ L N+
Sbjct: 128 LSGPLPPGFLSALDQLLVLDLSYNSFKGELPLQQSFGNGSNGIFPIQTVDLSSNLLEGEI 187
Query: 116 ----------------NLSPWTFPTEL-----TQSSNLNSIDINQAKINGTLPDIFGSFS 154
N+S +F + T S L +D + +G L S
Sbjct: 188 LSSSVFLQGAFNLTSFNVSNNSFTGSIPSFMCTASPQLTKLDFSYNDFSGDLSQELSRCS 247
Query: 155 SLNTLHLAYNNLSGGLPNSLAG--SGIQSFWINNNLPG-LTGSITVISNMTLLTQVWLHV 211
L+ L +NNLSG +P + Q F N L G + IT ++ +TLL L+
Sbjct: 248 RLSVLRAGFNNLSGEIPKEIYNLPELEQLFLPVNRLSGKIDNGITRLTKLTLLE---LYS 304
Query: 212 NKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPV--F 268
N G IP D+ + + + LQL N L G +P SL + + L + LR NQL G + F
Sbjct: 305 NHIEGEIPKDIGKLSKLSSLQLHVNNLMGSIPVSLANCTKLVKLNLRVNQLGGTLSAIDF 364
Query: 269 GKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVG----GFGYP----IQFAKSWTGN 320
+ + D+ +N+F S ++M + G G P ++ +T +
Sbjct: 365 SRFQSLSILDLGNNSFTGEFPSTVYSCKMMTAMRFAGNKLTGQISPQVLELESLSFFTFS 424
Query: 321 D----PCKDWLCVICGGGKITKLNFAKQ-----------------------------GLQ 347
D L ++ G K++ L AK L
Sbjct: 425 DNKMTNLTGALSILQGCKKLSTLIMAKNFYDETVPSNKDFLRSDGFPSLQIFGIGACRLT 484
Query: 348 GTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPK 400
G I L + + L+ N G+IP L TL L LD+S+N L+GE+PK
Sbjct: 485 GEIPAWLIKLQRVEVMDLSMNRFVGTIPGWLGTLPDLFYLDLSDNFLTGELPK 537
Score = 90.1 bits (222), Expect = 5e-18
Identities = 98/358 (27%), Positives = 154/358 (42%), Gaps = 20/358 (5%)
Query: 35 VTSINLSDQKLAGTLPDNLNSLT-QLTTLYLQNNALSGPLPS-LANLSSLTDVNLGSNNF 92
+TS N+S+ G++P + + + QLT L N SG L L+ S L+ + G NN
Sbjct: 200 LTSFNVSNNSFTGSIPSFMCTASPQLTKLDFSYNDFSGDLSQELSRCSRLSVLRAGFNNL 259
Query: 93 SSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIFGS 152
S P L L+ L L +N +T+ + L +++ I G +P G
Sbjct: 260 SGEIPKEIYNLPELEQLFLP--VNRLSGKIDNGITRLTKLTLLELYSNHIEGEIPKDIGK 317
Query: 153 FSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLPGLTGSITVI--SNMTLLTQVWLH 210
S L++L L NNL G +P SLA + +N + L G+++ I S L+ + L
Sbjct: 318 LSKLSSLQLHVNNLMGSIPVSLANC-TKLVKLNLRVNQLGGTLSAIDFSRFQSLSILDLG 376
Query: 211 VNKFTGPIPD-LSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQ---LQGPVP 266
N FTG P + C + ++ N+LTG + ++ + L T +N+ L G +
Sbjct: 377 NNSFTGEFPSTVYSCKMMTAMRFAGNKLTGQISPQVLELESLSFFTFSDNKMTNLTGALS 436
Query: 267 VFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDW 326
+ + K S I NF + D D + FG A TG P W
Sbjct: 437 IL-QGCKKLSTLIMAKNFYDETVPSNKDFLRSDGFPSLQIFGIG---ACRLTGEIPA--W 490
Query: 327 LCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQL 384
L + ++ ++ + GTI L DL L L+ N LTG +P+ L L L
Sbjct: 491 LIKL---QRVEVMDLSMNRFVGTIPGWLGTLPDLFYLDLSDNFLTGELPKELFQLRAL 545
Score = 48.5 bits (114), Expect = 2e-05
Identities = 31/104 (29%), Positives = 49/104 (46%), Gaps = 5/104 (4%)
Query: 300 LLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGG---KITKLNFAKQGLQGTISPAFAN 356
LL G P+ W + C W + C ++T + + +GL G + + +
Sbjct: 56 LLWFSGNVSSPVS-PLHWNSSIDCCSWEGISCDKSPENRVTSIILSSRGLSGNLPSSVLD 114
Query: 357 LTDLTALYLNGNNLTGSIPQN-LATLSQLETLDVSNNDLSGEVP 399
L L+ L L+ N L+G +P L+ L QL LD+S N GE+P
Sbjct: 115 LQRLSRLDLSHNRLSGPLPPGFLSALDQLLVLDLSYNSFKGELP 158
Score = 47.4 bits (111), Expect = 4e-05
Identities = 32/110 (29%), Positives = 53/110 (48%), Gaps = 8/110 (7%)
Query: 11 SPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALS 70
+PN ++N + Q S + +I + L GT+P + L L L L N S
Sbjct: 566 NPNNVTTNQQYNQLSSLP-------PTIYIKRNNLTGTIPVEVGQLKVLHILELLGNNFS 618
Query: 71 GPLPS-LANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
G +P L+NL++L ++L +NN S P + +GL+ L ++ N P
Sbjct: 619 GSIPDELSNLTNLERLDLSNNNLSGRIPWSLTGLHFLSYFNVANNTLSGP 668
>At5g38560 putative protein
Length = 681
Score = 255 bits (652), Expect = 7e-68
Identities = 179/488 (36%), Positives = 255/488 (51%), Gaps = 29/488 (5%)
Query: 403 PKVKFITDGNVWLGKNHGGGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGA 462
P+ K I N P S+PG S G+G V I +I+ L+ + V G
Sbjct: 203 PREKPIAKPTGPASNNGNNTLPSSSPGKSEVGTGG-----IVAIGVIVGLVFLSLFVMGV 257
Query: 463 WFSWKCYSRKGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGF 522
WF+ K RK F P + + + + N A +++S S D+
Sbjct: 258 WFTRK-RKRKDPGTFVGYTMPPSAYSSPQGSDVVLFNSRSSAPPKMRSHSGSDYMYASS- 315
Query: 523 DGGNGGN--ATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKG 580
D G N + S L QVT+ FS+ N+LG GGFG VYKG L DG ++AVK++ G
Sbjct: 316 DSGMVSNQRSWFSYDELSQVTSGFSEKNLLGEGGFGCVYKGVLSDGREVAVKQL--KIGG 373
Query: 581 SKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYT 640
S+G EF+AE+ ++++V HRHLV L+GYCI+ RLLVY+++P TL HL G
Sbjct: 374 SQGEREFKAEVEIISRVHHRHLVTLVGYCISEQHRLLVYDYVPNNTLHYHL---HAPGRP 430
Query: 641 PLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP- 699
+TW+ R+ +A RG+ YLH IHRD+K SNILL + A VADFGL K A
Sbjct: 431 VMTWETRVRVAAGAARGIAYLHEDCHPRIIHRDIKSSNILLDNSFEALVADFGLAKIAQE 490
Query: 700 -DGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESS 758
D N V T++ GTFGY+APEYA +G+++ K DVY++GV+L+ELITGRK +D S P
Sbjct: 491 LDLNTHVSTRVMGTFGYMAPEYATSGKLSEKADVYSYGVILLELITGRKPVDTSQPLGDE 550
Query: 759 HLVTWFRRVLTNK-ENIPKAIDQTLDPDEETML---SIYKVAELAGHCTTRSPYQRPDIG 814
LV W R +L EN + D+ +DP ++++ E A C S +RP +
Sbjct: 551 SLVEWARPLLGQAIEN--EEFDELVDPRLGKNFIPGEMFRMVEAAAACVRHSAAKRPKMS 608
Query: 815 HAVNVLCPLVQQWEPTT--HTDESTCADDNQMSL-TQALQR--WQANEGTSTFFNGMTSQ 869
V L L + + T +S D Q S + QR + + + +S FF+ SQ
Sbjct: 609 QVVRALDTLEEATDITNGMRPGQSQVFDSRQQSAQIRMFQRMAFGSQDYSSDFFD--RSQ 666
Query: 870 TQSSSTSK 877
+ SS S+
Sbjct: 667 SHSSWGSR 674
>At4g02010 putative NAK-like ser/thr protein kinase
Length = 725
Score = 240 bits (613), Expect = 2e-63
Identities = 147/371 (39%), Positives = 216/371 (57%), Gaps = 32/371 (8%)
Query: 532 ISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEI 591
+S L++ T++F +ILG GGFG VY+G L DGT +A+K++ S G +G EFQ EI
Sbjct: 368 LSYEELKEATSNFESASILGEGGFGKVYRGILADGTAVAIKKLTS--GGPQGDKEFQVEI 425
Query: 592 GVLTKVRHRHLVALLGYCIN--GNERLLVYEHMPQGTLTQHLFECREHG----YTPLTWK 645
+L+++ HR+LV L+GY + ++ LL YE +P G+L L HG PL W
Sbjct: 426 DMLSRLHHRNLVKLVGYYSSRDSSQHLLCYELVPNGSLEAWL-----HGPLGLNCPLDWD 480
Query: 646 QRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS- 704
R+ IALD RG+ YLH +Q S IHRD K SNILL ++ AKVADFGL K AP+G +
Sbjct: 481 TRMKIALDAARGLAYLHEDSQPSVIHRDFKASNILLENNFNAKVADFGLAKQAPEGRGNH 540
Query: 705 VETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWF 764
+ T++ GTFGY+APEYA TG + K DVY++GVVL+EL+TGRK +D S P +LVTW
Sbjct: 541 LSTRVMGTFGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRKPVDMSQPSGQENLVTWT 600
Query: 765 RRVLTNKENIPKAIDQTLD---PDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAV---N 818
R VL +K+ + + +D L+ P E+ +V +A C QRP +G V
Sbjct: 601 RPVLRDKDRLEELVDSRLEGKYPKED----FIRVCTIAAACVAPEASQRPTMGEVVQSLK 656
Query: 819 VLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMTSQTQS---SST 875
++ +V+ +P +T + Q S T +++ +S F +G S + +
Sbjct: 657 MVQRVVEYQDPVLNTSNKARPNRRQSSAT-----FESEVTSSMFSSGPYSGLSAFDHENI 711
Query: 876 SKPPVFADSLH 886
++ VF++ LH
Sbjct: 712 TRTTVFSEDLH 722
>At2g20300 protein kinase like protein
Length = 744
Score = 240 bits (613), Expect = 2e-63
Identities = 152/414 (36%), Positives = 215/414 (51%), Gaps = 39/414 (9%)
Query: 421 GGAPGSAPGGSP--------AGSGKGASMKKVWII----IIIVLIVVGFVVGGAWFSWKC 468
G G APGG P A +G + + II +++L++VG + WK
Sbjct: 228 GDVTGDAPGGLPIPINATTFANKSQGIGFRTIAIIALSGFVLILVLVGAI--SIIVKWK- 284
Query: 469 YSRKGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQS--QSSGDHSDLHGFDGGN 526
++G N G LA N GA S S +SSG S +
Sbjct: 285 ----------KIGKSSNAVGPA---LAPSINKRPGAGSMFSSSARSSGSDSLMSSMATCA 331
Query: 527 GGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNE 586
T ++ L + T+ FS +LG GGFG VY+G + DGT++AVK + + E
Sbjct: 332 LSVKTFTLSELEKATDRFSAKRVLGEGGFGRVYQGSMEDGTEVAVKLLTRDNQNRD--RE 389
Query: 587 FQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQ 646
F AE+ +L+++ HR+LV L+G CI G R L+YE + G++ HL E L W
Sbjct: 390 FIAEVEMLSRLHHRNLVKLIGICIEGRTRCLIYELVHNGSVESHLHE------GTLDWDA 443
Query: 647 RLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVE 706
RL IAL RG+ YLH + IHRD K SN+LL DD KV+DFGL + A +G+ +
Sbjct: 444 RLKIALGAARGLAYLHEDSNPRVIHRDFKASNVLLEDDFTPKVSDFGLAREATEGSQHIS 503
Query: 707 TKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRR 766
T++ GTFGY+APEYA TG + K DVY++GVVL+EL+TGR+ +D S P +LVTW R
Sbjct: 504 TRVMGTFGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRRPVDMSQPSGEENLVTWARP 563
Query: 767 VLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
+L N+E + + +D L + KVA +A C + RP +G V L
Sbjct: 564 LLANREGLEQLVDPAL-AGTYNFDDMAKVAAIASMCVHQEVSHRPFMGEVVQAL 616
>At5g56890 unknown protein
Length = 1113
Score = 239 bits (611), Expect = 4e-63
Identities = 137/321 (42%), Positives = 185/321 (56%), Gaps = 12/321 (3%)
Query: 531 TISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAE 590
T + + + TN+F + +LG GGFG VY+G DGTK+AVK + +G EF AE
Sbjct: 710 TFTASEIMKATNNFDESRVLGEGGFGRVYEGVFDDGTKVAVK--VLKRDDQQGSREFLAE 767
Query: 591 IGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLII 650
+ +L+++ HR+LV L+G CI R LVYE +P G++ HL + +PL W RL I
Sbjct: 768 VEMLSRLHHRNLVNLIGICIEDRNRSLVYELIPNGSVESHLHGI-DKASSPLDWDARLKI 826
Query: 651 ALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG--NYSVETK 708
AL RG+ YLH + IHRD K SNILL +D KV+DFGL +NA D N + T+
Sbjct: 827 ALGAARGLAYLHEDSSPRVIHRDFKSSNILLENDFTPKVSDFGLARNALDDEDNRHISTR 886
Query: 709 LAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVL 768
+ GTFGY+APEYA TG + K DVY++GVVL+EL+TGRK +D S P +LV+W R L
Sbjct: 887 VMGTFGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRKPVDMSQPPGQENLVSWTRPFL 946
Query: 769 TNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL------CP 822
T+ E + IDQ+L P E + SI KVA +A C RP +G V L C
Sbjct: 947 TSAEGLAAIIDQSLGP-EISFDSIAKVAAIASMCVQPEVSHRPFMGEVVQALKLVSNECD 1005
Query: 823 LVQQWEPTTHTDESTCADDNQ 843
++ T + DD Q
Sbjct: 1006 EAKELNSLTSISKDDFRDDTQ 1026
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 235 bits (600), Expect = 7e-62
Identities = 166/463 (35%), Positives = 239/463 (50%), Gaps = 45/463 (9%)
Query: 384 LETLDVSNNDLSGEVPKFS--PKVKFITDGNVWLGKNHGGGAPGSAPGGSPAGSGKGASM 441
L+ LD S VP S P ++ N G ++ A + GG+ S +
Sbjct: 200 LQPLDSPLGGESNRVPSSSSSPSPPSLSGSNNHSGGSNRHNANSNGDGGTSQQSNESNYT 259
Query: 442 KKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPENGEGNVKLDLASVS--- 498
+K I I I ++V + G +F +RR + G+ + N L A+VS
Sbjct: 260 EKTVIGIGIAGVLVILFIAGVFF---------VRRKQKKGS-SSPRSNQYLPPANVSVNT 309
Query: 499 NGY-------GGASSELQSQSSGDHSDLHGFDGGNGGN-----ATISIHV----LRQVTN 542
G+ G +S Q+ SS D + L G G T IH L Q+T
Sbjct: 310 EGFIHYRQKPGNGNSSAQN-SSPDTNSLGNPKHGRGTPDSAVIGTSKIHFTYEELSQITE 368
Query: 543 DFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHL 602
F ++G GGFG VYKG L +G +A+K++ SV+ ++G EF+AE+ ++++V HRHL
Sbjct: 369 GFCKSFVVGEGGFGCVYKGILFEGKPVAIKQLKSVS--AEGYREFKAEVEIISRVHHRHL 426
Query: 603 VALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYT--PLTWKQRLIIALDVGRGVEY 660
V+L+GYCI+ R L+YE +P TL HL HG L W +R+ IA+ +G+ Y
Sbjct: 427 VSLVGYCISEQHRFLIYEFVPNNTLDYHL-----HGKNLPVLEWSRRVRIAIGAAKGLAY 481
Query: 661 LHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYLAPEY 720
LH IHRD+K SNILL D+ A+VADFGL + + T++ GTFGYLAPEY
Sbjct: 482 LHEDCHPKIIHRDIKSSNILLDDEFEAQVADFGLARLNDTAQSHISTRVMGTFGYLAPEY 541
Query: 721 AATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVL---TNKENIPKA 777
A++G++T + DV++FGVVL+ELITGRK +D S P LV W R L K +I +
Sbjct: 542 ASSGKLTDRSDVFSFGVVLLELITGRKPVDTSQPLGEESLVEWARPRLIEAIEKGDISEV 601
Query: 778 IDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
+D L+ D +YK+ E A C S +RP + V L
Sbjct: 602 VDPRLEND-YVESEVYKMIETAASCVRHSALKRPRMVQVVRAL 643
>At1g76370 putative protein kinase
Length = 381
Score = 234 bits (598), Expect = 1e-61
Identities = 133/342 (38%), Positives = 193/342 (55%), Gaps = 10/342 (2%)
Query: 482 NPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGN-----GGNATISIHV 536
N + + + +D S Y ++++ + +G S + N GG + +
Sbjct: 8 NTQTNDMRINIDTLSDLTDYASVATKIDPRGTGSKSGILVNGKVNSPKPGGGARSFTFKE 67
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTK 596
L T +F + NI+G+GGFG VYKG L G +A+K++ G +G EF E+ +L+
Sbjct: 68 LAAATKNFREGNIIGKGGFGSVYKGRLDSGQVVAIKQLNP--DGHQGNQEFIVEVCMLSV 125
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGR 656
H +LV L+GYC +G +RLLVYE+MP G+L HLF+ E TPL+W R+ IA+ R
Sbjct: 126 FHHPNLVTLIGYCTSGAQRLLVYEYMPMGSLEDHLFDL-EPDQTPLSWYTRMKIAVGAAR 184
Query: 657 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS-VETKLAGTFGY 715
G+EYLH S I+RDLK +NILL + K++DFGL K P GN + V T++ GT+GY
Sbjct: 185 GIEYLHCKISPSVIYRDLKSANILLDKEFSVKLSDFGLAKVGPVGNRTHVSTRVMGTYGY 244
Query: 716 LAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIP 775
APEYA +GR+T K D+Y+FGVVL+ELI+GRKA+D S P+ +LV W R L + +
Sbjct: 245 CAPEYAMSGRLTIKSDIYSFGVVLLELISGRKAIDLSKPNGEQYLVAWARPYLKDPKKFG 304
Query: 776 KAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAV 817
+D L Y ++ + C RP IG V
Sbjct: 305 LLVDPLLRGKFSKRCLNYAIS-ITEMCLNDEANHRPKIGDVV 345
>At1g20650 unknown protein
Length = 381
Score = 233 bits (595), Expect = 3e-61
Identities = 140/353 (39%), Positives = 197/353 (55%), Gaps = 12/353 (3%)
Query: 482 NPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGN-------GGNATISI 534
NP + V +D A ++ Y SS S ++G S G GG + +
Sbjct: 9 NPRTKDIRVDIDNARCNSRYQTDSSVHGSDTTGTESISGILVNGKVNSPIPGGGARSFTF 68
Query: 535 HVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVL 594
L T +F + N+LG GGFG VYKG L G +A+K++ G +G EF E+ +L
Sbjct: 69 KELAAATRNFREVNLLGEGGFGRVYKGRLDSGQVVAIKQLNP--DGLQGNREFIVEVLML 126
Query: 595 TKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDV 654
+ + H +LV L+GYC +G++RLLVYE+MP G+L HLF+ E PL+W R+ IA+
Sbjct: 127 SLLHHPNLVTLIGYCTSGDQRLLVYEYMPMGSLEDHLFDL-ESNQEPLSWNTRMKIAVGA 185
Query: 655 GRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS-VETKLAGTF 713
RG+EYLH A I+RDLK +NILL + K++DFGL K P G+ + V T++ GT+
Sbjct: 186 ARGIEYLHCTANPPVIYRDLKSANILLDKEFSPKLSDFGLAKLGPVGDRTHVSTRVMGTY 245
Query: 714 GYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKEN 773
GY APEYA +G++T K D+Y FGVVL+ELITGRKA+D +LVTW R L +++
Sbjct: 246 GYCAPEYAMSGKLTVKSDIYCFGVVLLELITGRKAIDLGQKQGEQNLVTWSRPYLKDQKK 305
Query: 774 IPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQ 826
+D +L Y +A +A C + RP IG V L L Q
Sbjct: 306 FGHLVDPSLRGKYPRRCLNYAIAIIA-MCLNEEAHYRPFIGDIVVALEYLAAQ 357
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 232 bits (592), Expect = 6e-61
Identities = 147/393 (37%), Positives = 217/393 (54%), Gaps = 35/393 (8%)
Query: 447 IIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPENGEGNVKLDLASVSNGYGGASS 506
+ +++L ++G VV C +K +R + +G G + S+ S+
Sbjct: 337 VALVLLTLIGVVV--------CCLKKRKKRLSTIG------GGYVMPTPMESSSPRSDSA 382
Query: 507 ELQSQSS----GDHSDLHGF-----DGGNGGNATI-SIHVLRQVTNDFSDDNILGRGGFG 556
L++QSS G+ S + GG G + + S L TN FSD+N+LG GGFG
Sbjct: 383 LLKTQSSAPLVGNRSSNRTYLSQSEPGGFGQSRELFSYEELVIATNGFSDENLLGEGGFG 442
Query: 557 IVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERL 616
VYKG LPD +AVK++ G +G EF+AE+ +++V HR+L++++GYCI+ N RL
Sbjct: 443 RVYKGVLPDERVVAVKQL--KIGGGQGDREFKAEVDTISRVHHRNLLSMVGYCISENRRL 500
Query: 617 LVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKP 676
L+Y+++P L HL HG L W R+ IA RG+ YLH IHRD+K
Sbjct: 501 LIYDYVPNNNLYFHL-----HGTPGLDWATRVKIAAGAARGLAYLHEDCHPRIIHRDIKS 555
Query: 677 SNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFG 736
SNILL ++ A V+DFGL K A D N + T++ GTFGY+APEYA++G++T K DV++FG
Sbjct: 556 SNILLENNFHALVSDFGLAKLALDCNTHITTRVMGTFGYMAPEYASSGKLTEKSDVFSFG 615
Query: 737 VVLMELITGRKALDDSVPDESSHLVTWFRRVLTN---KENIPKAIDQTLDPDEETMLSIY 793
VVL+ELITGRK +D S P LV W R +L+N E D L + + ++
Sbjct: 616 VVLLELITGRKPVDASQPLGDESLVEWARPLLSNATETEEFTALADPKLGRN-YVGVEMF 674
Query: 794 KVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQ 826
++ E A C S +RP + V L ++
Sbjct: 675 RMIEAAAACIRHSATKRPRMSQIVRAFDSLAEE 707
>At1g68690 protein kinase, putative
Length = 708
Score = 231 bits (590), Expect = 1e-60
Identities = 131/312 (41%), Positives = 186/312 (58%), Gaps = 14/312 (4%)
Query: 510 SQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKI 569
S+ SG + G G A S L + TN FS +N+LG GGFG VYKG LPDG +
Sbjct: 345 SKRSGSYQSQSG--GLGNSKALFSYEELVKATNGFSQENLLGEGGFGCVYKGILPDGRVV 402
Query: 570 AVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQ 629
AVK++ G +G EF+AE+ L+++ HRHLV+++G+CI+G+ RLL+Y+++ L
Sbjct: 403 AVKQL--KIGGGQGDREFKAEVETLSRIHHRHLVSIVGHCISGDRRLLIYDYVSNNDLYF 460
Query: 630 HLFECREHGY-TPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAK 688
HL HG + L W R+ IA RG+ YLH IHRD+K SNILL D+ A+
Sbjct: 461 HL-----HGEKSVLDWATRVKIAAGAARGLAYLHEDCHPRIIHRDIKSSNILLEDNFDAR 515
Query: 689 VADFGLVKNAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKA 748
V+DFGL + A D N + T++ GTFGY+APEYA++G++T K DV++FGVVL+ELITGRK
Sbjct: 516 VSDFGLARLALDCNTHITTRVIGTFGYMAPEYASSGKLTEKSDVFSFGVVLLELITGRKP 575
Query: 749 LDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDP---DEETMLSIYKVAELAGHCTTR 805
+D S P LV W R ++++ + D DP ++++ E AG C
Sbjct: 576 VDTSQPLGDESLVEWARPLISHAIE-TEEFDSLADPKLGGNYVESEMFRMIEAAGACVRH 634
Query: 806 SPYQRPDIGHAV 817
+RP +G V
Sbjct: 635 LATKRPRMGQIV 646
>At4g32710 putative protein kinase
Length = 731
Score = 231 bits (589), Expect = 1e-60
Identities = 130/295 (44%), Positives = 186/295 (62%), Gaps = 14/295 (4%)
Query: 533 SIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGS-KGLNEFQAEI 591
S L + T FS++N+LG GGFG V+KG L +GT++AVK++ GS +G EFQAE+
Sbjct: 378 SYEELSKATGGFSEENLLGEGGFGYVHKGVLKNGTEVAVKQL---KIGSYQGEREFQAEV 434
Query: 592 GVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIA 651
+++V H+HLV+L+GYC+NG++RLLVYE +P+ TL HL E R + L W+ RL IA
Sbjct: 435 DTISRVHHKHLVSLVGYCVNGDKRLLVYEFVPKDTLEFHLHENRG---SVLEWEMRLRIA 491
Query: 652 LDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS---VETK 708
+ +G+ YLH + IHRD+K +NILL AKV+DFGL K D N S + T+
Sbjct: 492 VGAAKGLAYLHEDCSPTIIHRDIKAANILLDSKFEAKVSDFGLAKFFSDTNSSFTHISTR 551
Query: 709 LAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVL 768
+ GTFGY+APEYA++G+VT K DVY+FGVVL+ELITGR ++ + LV W R +L
Sbjct: 552 VVGTFGYMAPEYASSGKVTDKSDVYSFGVVLLELITGRPSIFAKDSSTNQSLVDWARPLL 611
Query: 769 T---NKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
T + E+ +D L+ + +T + +A A C +S + RP + V L
Sbjct: 612 TKAISGESFDFLVDSRLEKNYDT-TQMANMAACAAACIRQSAWLRPRMSQVVRAL 665
>At1g70460 putative protein kinase
Length = 710
Score = 231 bits (589), Expect = 1e-60
Identities = 142/379 (37%), Positives = 201/379 (52%), Gaps = 30/379 (7%)
Query: 499 NGYGGASSELQSQSSGDHSDLHGFDGGN------------GGNATISIHVLRQVTNDFSD 546
+G GG +S+ QS S G G G + L +T FS
Sbjct: 296 SGPGGYNSQQQSNSGNSFGSQRGGGGYTRSGSAPDSAVMGSGQTHFTYEELTDITEGFSK 355
Query: 547 DNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALL 606
NILG GGFG VYKG+L DG +AVK++ V G +G EF+AE+ ++++V HRHLV+L+
Sbjct: 356 HNILGEGGFGCVYKGKLNDGKLVAVKQL-KVGSG-QGDREFKAEVEIISRVHHRHLVSLV 413
Query: 607 GYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQ 666
GYCI +ERLL+YE++P TL HL G L W +R+ IA+ +G+ YLH
Sbjct: 414 GYCIADSERLLIYEYVPNQTLEHHL---HGKGRPVLEWARRVRIAIGSAKGLAYLHEDCH 470
Query: 667 QSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYLAPEYAATGRV 726
IHRD+K +NILL D+ A+VADFGL K V T++ GTFGYLAPEYA +G++
Sbjct: 471 PKIIHRDIKSANILLDDEFEAQVADFGLAKLNDSTQTHVSTRVMGTFGYLAPEYAQSGKL 530
Query: 727 TTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVL---TNKENIPKAIDQTLD 783
T + DV++FGVVL+ELITGRK +D P LV W R +L + + +D+ L+
Sbjct: 531 TDRSDVFSFGVVLLELITGRKPVDQYQPLGEESLVEWARPLLHKAIETGDFSELVDRRLE 590
Query: 784 PDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTDESTCADDNQ 843
++++ E A C S +RP + V L + D ++ N+
Sbjct: 591 -KHYVENEVFRMIETAAACVRHSGPKRPRMVQVVRAL---------DSEGDMGDISNGNK 640
Query: 844 MSLTQALQRWQANEGTSTF 862
+ + A Q N T F
Sbjct: 641 VGQSSAYDSGQYNNDTMKF 659
>At1g07570 protein kinase APK1A
Length = 410
Score = 231 bits (589), Expect = 1e-60
Identities = 152/406 (37%), Positives = 215/406 (52%), Gaps = 50/406 (12%)
Query: 505 SSELQSQSSGDHSDLHGFDGGNGGNATISIHV-----------------------LRQVT 541
S++++++SSG + D G+ G+ S+ V L+ T
Sbjct: 6 SAQVKAESSGASTKYDAKDIGSLGSKASSVSVRPSPRTEGEILQSPNLKSFSFAELKSAT 65
Query: 542 NDFSDDNILGRGGFGIVYKGELPD----------GTKIAVKRMISVAKGSKGLNEFQAEI 591
+F D++LG GGFG V+KG + + G IAVK++ G +G E+ AE+
Sbjct: 66 RNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQ--DGWQGHQEWLAEV 123
Query: 592 GVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIA 651
L + HRHLV L+GYC+ RLLVYE MP+G+L HLF R + PL+WK RL +A
Sbjct: 124 NYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFR-RGLYFQPLSWKLRLKVA 182
Query: 652 LDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS-VETKLA 710
L +G+ +LHS ++ I+RD K SNILL + AK++DFGL K+ P G+ S V T++
Sbjct: 183 LGAAKGLAFLHS-SETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTRVM 241
Query: 711 GTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTN 770
GT GY APEY ATG +TTK DVY+FGVVL+EL++GR+A+D + P +LV W + L N
Sbjct: 242 GTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYLVN 301
Query: 771 KENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPT 830
K I + ID L D+ +M KVA L+ C T RP++ V+ L
Sbjct: 302 KRKIFRVIDNRLQ-DQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHL---------- 350
Query: 831 THTDESTCADDNQMSLTQALQRWQANEGTSTFFN-GMTSQTQSSST 875
H A M T R +++ S N G QT ST
Sbjct: 351 EHIQSLNAAIGGNMDKTDRRMRRRSDSVVSKKVNAGFARQTAVGST 396
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,452,884
Number of Sequences: 26719
Number of extensions: 1014075
Number of successful extensions: 18810
Number of sequences better than 10.0: 1208
Number of HSP's better than 10.0 without gapping: 997
Number of HSP's successfully gapped in prelim test: 213
Number of HSP's that attempted gapping in prelim test: 3634
Number of HSP's gapped (non-prelim): 4875
length of query: 891
length of database: 11,318,596
effective HSP length: 108
effective length of query: 783
effective length of database: 8,432,944
effective search space: 6602995152
effective search space used: 6602995152
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146585.14


