

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.12 + phase: 0 /pseudo
(942 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
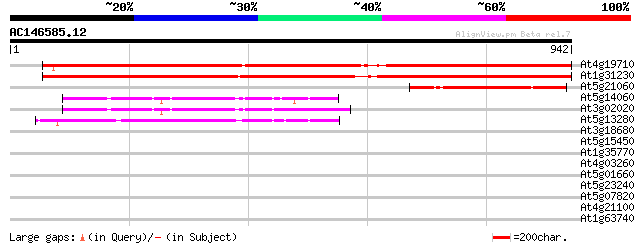
At4g19710 aspartate kinase-homoserine dehydrogenase - like protein 1308 0.0
At1g31230 aspartate kinase-homoserine dehydrogenase like protein 1298 0.0
At5g21060 homoserine dehydrogenase-like protein 190 3e-48
At5g14060 lysine-sensitive aspartate kinase (gb|AAB63104.1) 182 8e-46
At3g02020 putative aspartate kinase 181 1e-45
At5g13280 aspartate kinase 176 7e-44
At3g18680 uridylate kinase like protein 42 0.002
At5g15450 clpB heat shock protein-like 33 0.58
At1g35770 hypothetical protein 33 0.58
At4g03260 protein phosphatase regulatory subunit like 32 1.3
At5g01660 putative protein 31 3.7
At5g23240 unknown protein 30 4.9
At5g07820 putative protein 30 4.9
At4g21100 UV-damaged DNA-binding protein- like 30 4.9
At1g63740 disease resistance like protein 30 8.3
>At4g19710 aspartate kinase-homoserine dehydrogenase - like protein
Length = 916
Score = 1308 bits (3384), Expect = 0.0
Identities = 671/892 (75%), Positives = 763/892 (85%), Gaps = 34/892 (3%)
Query: 55 RRESPSSGICASLTDVS----VNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIV 110
R E P + A+ T VS VN+ V++ ++ KG+ WSVHKFGGTC+G+SQRI+NV +++
Sbjct: 54 RCELPDFHLSATATTVSGVSTVNL-VDQVQIPKGEMWSVHKFGGTCVGNSQRIRNVAEVI 112
Query: 111 LNDDSERKLVVVSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETL 170
+ND+SERKLVVVSAMSKVTDMMYDLI KAQSRD+SY+S+L+AVLEKH TA D+LDG+ L
Sbjct: 113 INDNSERKLVVVSAMSKVTDMMYDLIRKAQSRDDSYLSALEAVLEKHRLTARDLLDGDDL 172
Query: 171 AIFLSKLHEDISNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCKW 230
A FLS LH DISNLKAMLRAIYIAGH +ESF+DFV GHGELWSAQMLS V+RK G++CKW
Sbjct: 173 ASFLSHLHNDISNLKAMLRAIYIAGHASESFSDFVAGHGELWSAQMLSYVVRKTGLECKW 232
Query: 231 MDTREVLIVNPTSSNQVDPDYLESERRLEKWYSLNPCKVIIATGFIASTPENIPTTLKRD 290
MDTR+VLIVNPTSSNQVDPD+ ESE+RL+KW+SLNP K+IIATGFIASTP+NIPTTLKRD
Sbjct: 233 MDTRDVLIVNPTSSNQVDPDFGESEKRLDKWFSLNPSKIIIATGFIASTPQNIPTTLKRD 292
Query: 291 GSDFSAAIMGSLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVL 350
GSDFSAAIMG+L RARQVTIWTDVDGVYSADPRKV+EAVIL+TLSYQEAWEMSYFGANVL
Sbjct: 293 GSDFSAAIMGALLRARQVTIWTDVDGVYSADPRKVNEAVILQTLSYQEAWEMSYFGANVL 352
Query: 351 HPRTIIPVMRYGIPILIRNIFNLSAPGTKICHPVVSDYEDKSNLQNYVKGFATIDNLALV 410
HPRTIIPVMRY IPI+IRNIFNLSAPGT IC P DY+ K L VKGFATIDNLAL+
Sbjct: 353 HPRTIIPVMRYNIPIVIRNIFNLSAPGTIICQPPEDDYDLK--LTTPVKGFATIDNLALI 410
Query: 411 NVEGTGMAGVPGTASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRF 470
NVEGTGMAGVPGTAS IF VKDVGANVIMISQASSEHSVCFAVPEKEV AV+EAL+SRF
Sbjct: 411 NVEGTGMAGVPGTASDIFGCVKDVGANVIMISQASSEHSVCFAVPEKEVNAVSEALRSRF 470
Query: 471 RQALDNGRLSQVAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYN 530
+AL GRLSQ+ VIPNCSILAAVGQKMASTPGVS TLF+ALAKANINVRAI+QGCSEYN
Sbjct: 471 SEALQAGRLSQIEVIPNCSILAAVGQKMASTPGVSCTLFSALAKANINVRAISQGCSEYN 530
Query: 531 ITVVIKREDSIKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQAKHSHLYISYL 590
+TVVIKREDS+KALRAVHSRF+LSRTT+AMGI+GPGLIG+TLLDQLRDQA +
Sbjct: 531 VTVVIKREDSVKALRAVHSRFFLSRTTLAMGIVGPGLIGATLLDQLRDQAAVLKQEFNID 590
Query: 591 LHTRRLLEICGKLMQQFIQCRLQF*KKNLTLICV*WA*LAQSQCFSVMSKWKELREERGE 650
L R+L I G KK L S +S+W+EL E+G
Sbjct: 591 L---RVLGITGS-------------KKMLL-----------SDIGIDLSRWRELLNEKGT 623
Query: 651 VANLEKFAQHVHGNNFIPNTALVDCTADSIIAGHYYEWLCKGIHVITPNKKANSGPLEQY 710
A+L+KF Q VHGN+FIPN+ +VDCTADS IA YY+WL KGIHVITPNKKANSGPL+QY
Sbjct: 624 EADLDKFTQQVHGNHFIPNSVVVDCTADSAIASRYYDWLRKGIHVITPNKKANSGPLDQY 683
Query: 711 LRLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKD 770
L+LR LQR+SYTHYFYEATVGAGLPI+STLRGLLETGDKIL+IEGI SGTLSY+FNNF
Sbjct: 684 LKLRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRIEGICSGTLSYLFNNFVG 743
Query: 771 GRAFSEVVGEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPEPL 830
R+FSEVV EAK AG+TEPDPRDDLSGTDVARKVIILARESGLKL+L+++PI SLVPEPL
Sbjct: 744 DRSFSEVVTEAKNAGFTEPDPRDDLSGTDVARKVIILARESGLKLDLADLPIRSLVPEPL 803
Query: 831 RACASAQEFMQQLPKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDHPF 890
+ C S +EFM++LP++D + AK++ DA+N+GEVLRYVGVVD N+KG VELR+YKK+HPF
Sbjct: 804 KGCTSVEEFMEKLPQYDGDLAKERLDAENSGEVLRYVGVVDAVNQKGTVELRRYKKEHPF 863
Query: 891 AQLSGSDNIIAFTTRRYKNQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 942
AQL+GSDNIIAFTT RYK+ PLIVRGPGAGAQVTAGGIFSDILRLASYLGAP
Sbjct: 864 AQLAGSDNIIAFTTTRYKDHPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 915
>At1g31230 aspartate kinase-homoserine dehydrogenase like protein
Length = 911
Score = 1298 bits (3360), Expect = 0.0
Identities = 654/889 (73%), Positives = 768/889 (85%), Gaps = 31/889 (3%)
Query: 55 RRESPSSGICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDD 114
R E S + S+TD++++ +VE L KGDSW+VHKFGGTC+G+S+RIK+V +V+ DD
Sbjct: 52 RSELQSPRVLGSVTDLALDNSVENGHLPKGDSWAVHKFGGTCVGNSERIKDVAAVVVKDD 111
Query: 115 SERKLVVVSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETLAIFL 174
SERKLVVVSAMSKVTDMMYDLI++A+SRD+SY+S+L VLEKH ATA D+LDG+ L+ FL
Sbjct: 112 SERKLVVVSAMSKVTDMMYDLIHRAESRDDSYLSALSGVLEKHRATAVDLLDGDELSSFL 171
Query: 175 SKLHEDISNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCKWMDTR 234
++L++DI+NLKAMLRAIYIAGH TESF+DFVVGHGELWSAQML+ V+RK+G+DC WMD R
Sbjct: 172 ARLNDDINNLKAMLRAIYIAGHATESFSDFVVGHGELWSAQMLAAVVRKSGLDCTWMDAR 231
Query: 235 EVLIVNPTSSNQVDPDYLESERRLEKWYSLNPCKVIIATGFIASTPENIPTTLKRDGSDF 294
+VL+V PTSSNQVDPD++ESE+RLEKW++ N K+IIATGFIASTP+NIPTTLKRDGSDF
Sbjct: 232 DVLVVIPTSSNQVDPDFVESEKRLEKWFTQNSAKIIIATGFIASTPQNIPTTLKRDGSDF 291
Query: 295 SAAIMGSLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRT 354
SAAIM +LFR+ Q+TIWTDVDGVYSADPRKVSEAV+LKTLSYQEAWEMSYFGANVLHPRT
Sbjct: 292 SAAIMSALFRSHQLTIWTDVDGVYSADPRKVSEAVVLKTLSYQEAWEMSYFGANVLHPRT 351
Query: 355 IIPVMRYGIPILIRNIFNLSAPGTKICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEG 414
IIPVM+Y IPI+IRNIFNLSAPGT IC + D ED L VKGFATIDNLALVNVEG
Sbjct: 352 IIPVMKYDIPIVIRNIFNLSAPGTMICRQI--DDEDGFKLDAPVKGFATIDNLALVNVEG 409
Query: 415 TGMAGVPGTASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQAL 474
TGMAGVPGTASAIF+AVK+VGANVIMISQASSEHSVCFAVPEKEVKAV+EAL SRFRQAL
Sbjct: 410 TGMAGVPGTASAIFSAVKEVGANVIMISQASSEHSVCFAVPEKEVKAVSEALNSRFRQAL 469
Query: 475 DNGRLSQVAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVV 534
GRLSQ+ +IPNCSILAAVGQKMASTPGVSAT FNALAKANIN+RAIAQGCSE+NITVV
Sbjct: 470 AGGRLSQIEIIPNCSILAAVGQKMASTPGVSATFFNALAKANINIRAIAQGCSEFNITVV 529
Query: 535 IKREDSIKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQAKHSHLYISYLLHTR 594
+KRED I+ALRAVHSRFYLSRTT+A+GIIGPGLIG TLLDQ+RDQA
Sbjct: 530 VKREDCIRALRAVHSRFYLSRTTLAVGIIGPGLIGGTLLDQIRDQA-------------- 575
Query: 595 RLLEICGKLMQQFIQCRLQF*KKNLTLICV*W-A*LAQSQCFSVMSKWKELREERGEVAN 653
L ++F K +L +I + + + S+ +S+W+EL +E GE A+
Sbjct: 576 ------AVLKEEF--------KIDLRVIGITGSSKMLMSESGIDLSRWRELMKEEGEKAD 621
Query: 654 LEKFAQHVHGNNFIPNTALVDCTADSIIAGHYYEWLCKGIHVITPNKKANSGPLEQYLRL 713
+EKF Q+V GN+FIPN+ +VDCTAD+ IA YY+WL +GIHV+TPNKKANSGPL+QYL++
Sbjct: 622 MEKFTQYVKGNHFIPNSVMVDCTADADIASCYYDWLLRGIHVVTPNKKANSGPLDQYLKI 681
Query: 714 RALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRA 773
R LQR+SYTHYFYEATVGAGLPI+STLRGLLETGDKIL+IEGIFSGTLSY+FNNF R+
Sbjct: 682 RDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRIEGIFSGTLSYLFNNFVGTRS 741
Query: 774 FSEVVGEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPEPLRAC 833
FSEVV EAK+AG+TEPDPRDDLSGTDVARKV ILARESGLKL+L +P+++LVP+PL+AC
Sbjct: 742 FSEVVAEAKQAGFTEPDPRDDLSGTDVARKVTILARESGLKLDLEGLPVQNLVPKPLQAC 801
Query: 834 ASAQEFMQQLPKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDHPFAQL 893
ASA+EFM++LP+FD+E +K++E+A+ AGEVLRYVGVVD KKG VEL++YKKDHPFAQL
Sbjct: 802 ASAEEFMEKLPQFDEELSKQREEAEAAGEVLRYVGVVDAVEKKGTVELKRYKKDHPFAQL 861
Query: 894 SGSDNIIAFTTRRYKNQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 942
SG+DNIIAFTT+RYK QPLIVRGPGAGAQVTAGGIFSDILRLA YLGAP
Sbjct: 862 SGADNIIAFTTKRYKEQPLIVRGPGAGAQVTAGGIFSDILRLAFYLGAP 910
>At5g21060 homoserine dehydrogenase-like protein
Length = 376
Score = 190 bits (483), Expect = 3e-48
Identities = 110/267 (41%), Positives = 164/267 (61%), Gaps = 8/267 (2%)
Query: 671 ALVDCTADSIIAGHYYEWLCKGIHVITPNKKANSGPLEQYLRLRALQRQSYTHYFYEATV 730
A+VDC+A + + G ++ NKK + LE Y +L AL + H E+TV
Sbjct: 111 AVVDCSASMETIEILMKAVDLGCCIVLANKKPVTSTLEHYDKL-ALHPRFIRH---ESTV 166
Query: 731 GAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVGEAKEAGYTEPD 790
GAGLP++++L ++ +GD + +I G SGTL Y+ + +DG+ S+VV AK+ GYTEPD
Sbjct: 167 GAGLPVIASLNRIISSGDPVHRIVGSLSGTLGYVMSELEDGKPLSQVVQAAKKLGYTEPD 226
Query: 791 PRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPEPL-RACASAQEFMQQ-LPKFDQ 848
PRDDL G DVARK +ILAR G ++ + +I IESL PE + S +F+ + K DQ
Sbjct: 227 PRDDLGGMDVARKGLILARLLGKRIIMDSIKIESLYPEEMGPGLMSVDDFLHNGIVKLDQ 286
Query: 849 EFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDHPFAQLSGSDNIIAFTTRRYK 908
++ + A + G VLRYV V++ ++ + V +R+ KD P +L GSDNI+ +R YK
Sbjct: 287 NIEERVKKASSKGCVLRYVCVIEGSSVQ--VGIREVSKDSPLGRLRGSDNIVEIYSRCYK 344
Query: 909 NQPLIVRGPGAGAQVTAGGIFSDILRL 935
QPL+++G GAG TA G+ +DI+ L
Sbjct: 345 EQPLVIQGAGAGNDTTAAGVLADIIDL 371
>At5g14060 lysine-sensitive aspartate kinase (gb|AAB63104.1)
Length = 544
Score = 182 bits (462), Expect = 8e-46
Identities = 133/479 (27%), Positives = 243/479 (49%), Gaps = 38/479 (7%)
Query: 89 VHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVVVSAMSKVTDMMYDLINKAQSRDESYIS 148
V KFGG+ + S++R+K V +++L+ ER ++V+SAM K T+ + KA + + +
Sbjct: 85 VMKFGGSSVESAERMKEVANLILSFPDERPVIVLSAMGKTTNKLLKAGEKAVTCGVTNVE 144
Query: 149 SLDA---VLEKHSATAHDILDGETLAIFLSKLHEDISNLKAMLRAIYIAGHVTESFTDFV 205
S++ + E H TAH+ L + + + + + L +L+ I + +T D++
Sbjct: 145 SIEELSFIKELHLRTAHE------LGVETTVIEKHLEGLHQLLKGISMMKELTLRTRDYL 198
Query: 206 VGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVNPTSSNQVDPDYLE------SERRLE 259
V GE S ++ S + K G + D E+ + T+ + + D LE S+ +
Sbjct: 199 VSFGECMSTRLFSAYLNKIGHKARQYDAFEIGFI--TTDDFTNADILEATYPAVSKTLVG 256
Query: 260 KWYSLNPCKVIIATGFIASTPENIP-TTLKRDGSDFSAAIMGSLFRARQVTIWTDVDGVY 318
W N V+ TG++ + TTL R GSD +A +G R++ +W DVDGV
Sbjct: 257 DWSKENAVPVV--TGYLGKGWRSCAITTLGRGGSDLTATTIGKALGLREIQVWKDVDGVL 314
Query: 319 SADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGT 378
+ DP A + L++ EA E++YFGA VLHP ++ P IP+ ++N +N +APGT
Sbjct: 315 TCDPNIYPGAQSVPYLTFDEAAELAYFGAQVLHPLSMRPARDGDIPVRVKNSYNPTAPGT 374
Query: 379 KICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGMAGVPGTASAIFAAVKDVGANV 438
V++ D S + + N+ ++++ T M G G + +F +D+G +V
Sbjct: 375 -----VITRSRDMS--KAVLTSIVLKRNVTMLDIASTRMLGQYGFLAKVFTTFEDLGISV 427
Query: 439 IMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDN-----GRLSQVAVIPNCSILAA 493
++ A+SE S+ + K L R + LDN +++ V ++ SI++
Sbjct: 428 DVV--ATSEVSISLTL--DPAKLWGRELIQRVNE-LDNLVEELEKIAVVKLLQRRSIISL 482
Query: 494 VGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKREDSIKALRAVHSRFY 552
+G S+ + +F +NV+ I+QG S+ NI++++ E++ + +RA+HS F+
Sbjct: 483 IGNVQKSSL-ILEKVFQVFRSNGVNVQMISQGASKVNISLIVNDEEAEQCVRALHSAFF 540
>At3g02020 putative aspartate kinase
Length = 559
Score = 181 bits (460), Expect = 1e-45
Identities = 135/494 (27%), Positives = 250/494 (50%), Gaps = 32/494 (6%)
Query: 89 VHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVVVSAMSKVTDMMYDLINKAQS---RDES 145
V KFGG+ + S++R+ V ++L+ E+ +VV+SAM+K T+ + KA +
Sbjct: 86 VMKFGGSSVASAERMIQVAKLILSFPDEKPVVVLSAMAKTTNKLLMAGEKAVCCGVTNVD 145
Query: 146 YISSLDAVLEKHSATAHDILDGETLAIFLSKLHEDISNLKAMLRAIYIAGHVTESFTDFV 205
I L + E H TAH+ L + + + E + L+ +L+ + + +T D++
Sbjct: 146 TIEELSYIKELHIRTAHE------LGVETAVIAEHLEGLEQLLKGVAMMKELTLRSRDYL 199
Query: 206 VGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVNPTSSNQVDPDYLE------SERRLE 259
V GE S ++ + + K G + D E+ I+ T+ + + D LE S++ L
Sbjct: 200 VSFGECMSTRLFAAYLNKIGHKARQYDAFEIGII--TTDDFTNADILEATYPAVSKKLLG 257
Query: 260 KWYSLNPCKVIIATGFIASTPENIP-TTLKRDGSDFSAAIMGSLFRARQVTIWTDVDGVY 318
W N V+ TGF+ + TTL R GSD +A +G R++ +W DVDGV
Sbjct: 258 DWSKENALPVV--TGFLGKGWRSCAVTTLGRGGSDLTATTIGKALGLREIQVWKDVDGVL 315
Query: 319 SADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGT 378
+ DP A + L++ EA E++YFGA VLHP ++ P IP+ ++N +N +APGT
Sbjct: 316 TCDPNIYCGAQPVPHLTFDEAAELAYFGAQVLHPLSMRPAREGNIPVRVKNSYNPTAPGT 375
Query: 379 KICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGMAGVPGTASAIFAAVKDVGANV 438
V++ D S + + N+ ++++ T M G G + +F+ + +G +V
Sbjct: 376 -----VITRSRDMS--KAVLTSIVLKRNVTMLDITSTRMLGQYGFLAKVFSTFEKLGISV 428
Query: 439 IMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDN-GRLSQVAVIPNCSILAAVGQK 497
++ A+SE S+ + + E +Q Q ++ +++ V ++ + SI++ +G
Sbjct: 429 DVV--ATSEVSISLTLDPSKF-CSRELIQHELDQVVEELEKIAVVNLLRHRSIISLIGNV 485
Query: 498 MASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKREDSIKALRAVHSRFYLSRTT 557
S+ + F L INV+ I+QG S+ NI++++ +++ ++A+HS F+ + T
Sbjct: 486 QRSS-FILEKGFRVLRTNGINVQMISQGASKVNISLIVNDDEAEHCVKALHSAFFETDTC 544
Query: 558 IAMGIIGPGLIGST 571
A+ G I ++
Sbjct: 545 EAVSECPTGYIAAS 558
>At5g13280 aspartate kinase
Length = 569
Score = 176 bits (445), Expect = 7e-44
Identities = 135/527 (25%), Positives = 259/527 (48%), Gaps = 35/527 (6%)
Query: 43 FHSLRKGITLPRRRESPSSGICASLTDVSVNVAVEEK------ELSKGDSWSVHKFGGTC 96
F +L+K ++LP S + S + V +EEK E+ + V KFGG+
Sbjct: 38 FPTLKK-LSLPIGDGSSIRKVSGSGSRNIVRAVLEEKKTEAITEVDEKGITCVMKFGGSS 96
Query: 97 MGSSQRIKNVGDIVLNDDSERKLVVVSAMSKVTDMMYDLINKAQS---RDESYISSLDAV 153
+ S++R+K V D++L E ++V+SAM K T+ + KA S + S I L +
Sbjct: 97 VASAERMKEVADLILTFPEESPVIVLSAMGKTTNNLLLAGEKAVSCGVSNASEIEELSII 156
Query: 154 LEKHSATAHDI-LDGETLAIFLSKLHEDISNLKAMLRAIYIAGHVTESFTDFVVGHGELW 212
E H T ++ +D + +L +L + +L+ I + +T D++V GE
Sbjct: 157 KELHIRTVKELNIDPSVILTYLEELEQ-------LLKGIAMMKELTLRTRDYLVSFGECL 209
Query: 213 SAQMLSLVIRKNGIDCKWMDTREVLIVNP---TSSNQVDPDYLESERRLEKWYSLNPCKV 269
S ++ + + G+ + D E+ + T+ + ++ Y +RL + +P V
Sbjct: 210 STRIFAAYLNTIGVKARQYDAFEIGFITTDDFTNGDILEATYPAVAKRLYDDWMHDPA-V 268
Query: 270 IIATGFIAST-PENIPTTLKRDGSDFSAAIMGSLFRARQVTIWTDVDGVYSADPRKVSEA 328
I TGF+ TTL R GSD +A +G +++ +W DVDGV + DP A
Sbjct: 269 PIVTGFLGKGWKTGAVTTLGRGGSDLTATTIGKALGLKEIQVWKDVDGVLTCDPTIYKRA 328
Query: 329 VILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGTKICHPVVSDY 388
+ L++ EA E++YFGA VLHP+++ P IP+ ++N +N APGT I
Sbjct: 329 TPVPYLTFDEAAELAYFGAQVLHPQSMRPAREGEIPVRVKNSYNPKAPGTIIT------- 381
Query: 389 EDKSNLQNYVKGFATIDNLALVNVEGTGMAGVPGTASAIFAAVKDVGANVIMISQASSEH 448
+ + ++ + N+ ++++ T M G G + +F+ +++G +V ++ A+SE
Sbjct: 382 KTRDMTKSILTSIVLKRNVTMLDIASTRMLGQVGFLAKVFSIFEELGISVDVV--ATSEV 439
Query: 449 SVCFAVPEKEVKAVAEALQSRFRQALDN-GRLSQVAVIPNCSILAAVGQKMASTPGVSAT 507
S+ + ++ + E +Q ++ +++ V ++ +I++ +G S+ +
Sbjct: 440 SISLTLDPSKLWS-RELIQQELDHVVEELEKIAVVNLLKGRAIISLIGNVQHSSL-ILER 497
Query: 508 LFNALAKANINVRAIAQGCSEYNITVVIKREDSIKALRAVHSRFYLS 554
F+ L +NV+ I+QG S+ NI+ ++ ++ ++A+H F+ S
Sbjct: 498 AFHVLYTKGVNVQMISQGASKVNISFIVNEAEAEGCVQALHKSFFES 544
Score = 35.4 bits (80), Expect = 0.15
Identities = 28/97 (28%), Positives = 47/97 (47%), Gaps = 10/97 (10%)
Query: 390 DKSNLQNYVKGFATIDNLALVNVEGTGMAGVPGTASAIFAAVKDVGANVIMISQASSEHS 449
+K + N +KG A I + NV+ + + F + G NV MISQ +S+ +
Sbjct: 468 EKIAVVNLLKGRAIISLIG--NVQHSSLI-----LERAFHVLYTKGVNVQMISQGASKVN 520
Query: 450 VCFAVPEKEVKAVAEALQSRFRQALDNGRLSQVAVIP 486
+ F V E E + +AL F ++G LS++ + P
Sbjct: 521 ISFIVNEAEAEGCVQALHKSF---FESGDLSELLIQP 554
>At3g18680 uridylate kinase like protein
Length = 339
Score = 41.6 bits (96), Expect = 0.002
Identities = 28/86 (32%), Positives = 44/86 (50%), Gaps = 6/86 (6%)
Query: 292 SDFSAAIMGSLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLH 351
+D +AA+ + A V T+VDGV+ DP++ A +L +L+YQE +V+
Sbjct: 233 TDTAAALRCAEINAEVVLKATNVDGVFDDDPKRNPNARLLDSLTYQEVTSKD---LSVMD 289
Query: 352 PRTIIPVMRYGIPILIRNIFNLSAPG 377
I IP++ +FNLS PG
Sbjct: 290 MTAITLCQENNIPVV---VFNLSEPG 312
>At5g15450 clpB heat shock protein-like
Length = 968
Score = 33.5 bits (75), Expect = 0.58
Identities = 56/234 (23%), Positives = 99/234 (41%), Gaps = 24/234 (10%)
Query: 716 LQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFS 775
L +Q +T +++ V + + ++ET + + +G IF+ K G +
Sbjct: 77 LTQQEFTEMAWQSIVSSPDVAKENKQQIVETEHLMKALLEQKNGLARRIFS--KIGVDNT 134
Query: 776 EVVGEAKEAGYTEPDPRDDLSGTDVARKVIIL---ARESGLKLELSNIPIESLVPEPLRA 832
+V+ ++ +P D +G+ + R + L AR+ L+ S + +E LV A
Sbjct: 135 KVLEATEKFIQRQPKVYGDAAGSMLGRDLEALFQRARQFKKDLKDSYVSVEHLV----LA 190
Query: 833 CASAQEFMQQLPKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVE-LRKYKKDHPFA 891
A + F +QL K +F + +A E +R V + +G E L KY KD
Sbjct: 191 FADDKRFGKQLFK---DFQISERSLKSAIESIRGKQSVIDQDPEGKYEALEKYGKDLTAM 247
Query: 892 QLSGS--------DNI---IAFTTRRYKNQPLIVRGPGAGAQVTAGGIFSDILR 934
G D I I +RR KN P+++ PG G + G+ I++
Sbjct: 248 AREGKLDPVIGRDDEIRRCIQILSRRTKNNPVLIGEPGVGKTAISEGLAQRIVQ 301
>At1g35770 hypothetical protein
Length = 1010
Score = 33.5 bits (75), Expect = 0.58
Identities = 38/150 (25%), Positives = 67/150 (44%), Gaps = 21/150 (14%)
Query: 170 LAIFLSKLHEDISNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCK 229
+ F++ H IS L +L+ SF F + HG LW S+ +R+
Sbjct: 735 MRFFINVSHFRISQLHPLLKKRCPPSRHLTSFFFFTLAHGALW-----SVPVRQENEIVS 789
Query: 230 WMDTREVLIVNPTSSNQVDPD--YLESERRLEKWYSLNPCKVIIATGFIASTPENIPTTL 287
W + + VN VDP+ + S ++L+ S++ C G IA + ++I +
Sbjct: 790 WHPSPVINQVN------VDPEERFQNSMKKLKAISSISIC------GGIALSNKDILDLI 837
Query: 288 KRDGSDFSAAIMGSLFR-ARQVTIWTDVDG 316
R + ++ +M +L R +R + DVDG
Sbjct: 838 DRK-KNLTSKVMDALIRFSRHLLRTDDVDG 866
>At4g03260 protein phosphatase regulatory subunit like
Length = 677
Score = 32.3 bits (72), Expect = 1.3
Identities = 34/126 (26%), Positives = 55/126 (42%), Gaps = 12/126 (9%)
Query: 689 LCKGIHVITPNKKANS--GPLEQYLRLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLET 746
L +G+H + +K + S L + RLR L SY +G GL S+L+ L
Sbjct: 439 LPRGLHALNLSKNSISVIEGLRELTRLRVLDL-SYNRIL---RLGHGLASCSSLKELYLA 494
Query: 747 GDKILQIEGIFS----GTLSYIFNNFKDGRAFSEVVG--EAKEAGYTEPDPRDDLSGTDV 800
G+KI +IEG+ L FN F + ++ + +A E +P G +
Sbjct: 495 GNKISEIEGLHRLLKLTVLDLRFNKFSTTKCLGQLAANYSSLQAISLEGNPAQKNVGDEQ 554
Query: 801 ARKVII 806
RK ++
Sbjct: 555 LRKYLL 560
>At5g01660 putative protein
Length = 621
Score = 30.8 bits (68), Expect = 3.7
Identities = 22/80 (27%), Positives = 33/80 (40%), Gaps = 5/80 (6%)
Query: 175 SKLHEDISNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCKWMDTR 234
S+ H D+ N K + + HV E D V H E + V + N CK ++ R
Sbjct: 180 SENHPDVQNPKQIDK-----DHVLEKLKDLVFSHDEHGDNSLTETVEQANIPTCKNLEDR 234
Query: 235 EVLIVNPTSSNQVDPDYLES 254
+ L S ++D L S
Sbjct: 235 DTLEEETCSEGKIDGSCLVS 254
>At5g23240 unknown protein
Length = 465
Score = 30.4 bits (67), Expect = 4.9
Identities = 23/69 (33%), Positives = 37/69 (53%), Gaps = 5/69 (7%)
Query: 118 KLVVVSAMSKVTDM-MYDLINKAQSRDESYISSLDAVLEK--HSATAHDILDGETLAIFL 174
K S+ S +TD +YDL+ +S D+S I S L+K H A D G +AI L
Sbjct: 36 KCRATSSSSSITDFDLYDLLGIDRSSDKSQIKSAYRALQKRCHPDIAGD--PGHDMAIIL 93
Query: 175 SKLHEDISN 183
++ ++ +S+
Sbjct: 94 NEAYQLLSD 102
>At5g07820 putative protein
Length = 561
Score = 30.4 bits (67), Expect = 4.9
Identities = 35/137 (25%), Positives = 57/137 (41%), Gaps = 6/137 (4%)
Query: 65 ASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVVVSA 124
A+ ++SV E +GD +H T S +K VL+ + + V
Sbjct: 18 ANPMEISVETIPVVDEEVQGDDDVLHPAAETNHAS---LKRYSTRVLDKKTGKAQVQTRY 74
Query: 125 MSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETLAIFLSKLHE-DISN 183
T +DL + R+E + V +K + + D+ GETL L + + D S+
Sbjct: 75 RGNQTSSTHDLCKHGKRREEDLVIKPWKVAKKKNVDSGDLGKGETLRKSLGNVSKPDKSS 134
Query: 184 LKAMLRAIYIAGHVTES 200
L+A R AG V +S
Sbjct: 135 LQAAKRE--AAGEVVKS 149
>At4g21100 UV-damaged DNA-binding protein- like
Length = 1102
Score = 30.4 bits (67), Expect = 4.9
Identities = 30/124 (24%), Positives = 53/124 (42%), Gaps = 10/124 (8%)
Query: 4 SLSSSLSHFSRISVTSLQHDYNNKIPADS-QCRHFLLSRRF-------HSLRKGITLPRR 55
S S SHF R+ + + ++ + P D+ +C +LS F + + LP
Sbjct: 751 SAEESESHFVRL-LDAQSFEFLSSYPLDAFECGCSILSCSFTDDKNVYYCVGTAYVLPEE 809
Query: 56 RESPSSGICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDS 115
E I + + + EKE +KG +S++ F G + S + + +L DD
Sbjct: 810 NEPTKGRILVFIVEEGRLQLITEKE-TKGAVYSLNAFNGKLLASINQKIQLYKWMLRDDG 868
Query: 116 ERKL 119
R+L
Sbjct: 869 TREL 872
>At1g63740 disease resistance like protein
Length = 992
Score = 29.6 bits (65), Expect = 8.3
Identities = 29/115 (25%), Positives = 50/115 (43%), Gaps = 17/115 (14%)
Query: 70 VSVNVAVEEKELSKGDSWSVHKFGGTCMGS--------SQRIKNVGDIV----LNDDSER 117
++V V+ ++ K + F TC G SQ + +VG+I LN D+E
Sbjct: 60 MTVFYGVDPSDVRKQTGDILKVFKKTCSGKTEEKRRRWSQALNDVGNIAGEHFLNWDNES 119
Query: 118 KLVVVSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETLAI 172
K+ M K+ + + +N SRD + ++ LEK + H D E + +
Sbjct: 120 KM-----MEKIARDISNKVNTTISRDFEDMVGVETHLEKIQSLLHLDNDDEAMIV 169
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,789,218
Number of Sequences: 26719
Number of extensions: 828139
Number of successful extensions: 2037
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 2011
Number of HSP's gapped (non-prelim): 19
length of query: 942
length of database: 11,318,596
effective HSP length: 109
effective length of query: 833
effective length of database: 8,406,225
effective search space: 7002385425
effective search space used: 7002385425
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146585.12


