

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.11 + phase: 0
(308 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
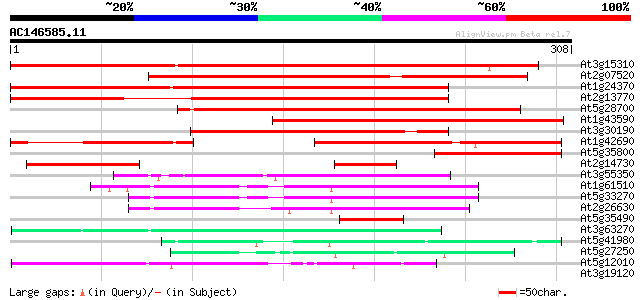
Score E
Sequences producing significant alignments: (bits) Value
At3g15310 unknown protein 364 e-101
At2g07520 pseudogene 350 7e-97
At1g24370 hypothetical protein 313 7e-86
At2g13770 hypothetical protein 241 5e-64
At5g28700 putative protein 202 2e-52
At1g43590 hypothetical protein 176 1e-44
At3g30190 hypothetical protein 158 4e-39
At1g42690 unknown protein 123 1e-28
At5g35800 putative protein 117 1e-26
At2g14730 hypothetical protein 69 4e-12
At3g55350 unknown protein 57 1e-08
At1g61510 hypothetical protein 56 2e-08
At5g33270 putative protein 54 1e-07
At2g26630 En/Spm-like transposon protein 54 1e-07
At5g35490 putative protein 45 4e-05
At3g63270 unknown protein 44 1e-04
At5g41980 unknown protein 43 2e-04
At5g27250 putative protein 42 3e-04
At5g12010 putative protein 42 3e-04
At3g19120 unknown protein 40 0.002
>At3g15310 unknown protein
Length = 415
Score = 364 bits (934), Expect = e-101
Identities = 172/293 (58%), Positives = 219/293 (74%), Gaps = 4/293 (1%)
Query: 1 MLAYGVAADAVDEYIKIGGITALECLRRFCKGIIRLYEQVYLRAPTQDDLQRILHVSEMR 60
+LAYG A+DAVDEY+++G TA+ CL F KGII + YLRAPT +L+R+L++ ++R
Sbjct: 124 LLAYGYASDAVDEYLRMGETTAMSCLENFTKGIISFFGDEYLRAPTATNLRRLLNIGKIR 183
Query: 61 GFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTL 120
GFPGMIGS+DCMHWEWKNCP AW+GQ+TRG G T++LEA+AS DLWIWH FFG PGTL
Sbjct: 184 GFPGMIGSLDCMHWEWKNCPTAWKGQYTRGS-GKPTIVLEAIASQDLWIWHVFFGPPGTL 242
Query: 121 NDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIRLPQSEP 180
NDIN+LDRSP+FDD+ QG+ P V Y VN R Y++ YYL DGIYP + TF++SIRLPQ+
Sbjct: 243 NDINILDRSPIFDDILQGRAPNVKYKVNGREYHLAYYLTDGIYPKWATFIQSIRLPQNRK 302
Query: 181 DKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDER 240
LFA HQE+ RKD+ERAFGVLQARF II+ PA +WD +G IM++CIILHNMIVEDER
Sbjct: 303 ATLFATHQEADRKDVERAFGVLQARFHIIKNPALVWDKEKIGNIMKACIILHNMIVEDER 362
Query: 241 DTYAQRWTDFEQSEGSGSSTQQ---PYSTEVLPTFANHVRARSELRDPNVHHE 290
D Y ++ E + G+ T Q YST + N + R++LRD N+H +
Sbjct: 363 DGYNIQFDVSEFLQVEGNQTPQVDLSYSTGMPLNIENMMGMRNQLRDQNMHQQ 415
>At2g07520 pseudogene
Length = 222
Score = 350 bits (897), Expect = 7e-97
Identities = 168/208 (80%), Positives = 180/208 (85%), Gaps = 6/208 (2%)
Query: 77 KNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVE 136
KNCP AWEGQFTRGDKGT+T+ EAVASHDLWIWH FFGC TLNDIN LDRSPVFDD+E
Sbjct: 16 KNCPIAWEGQFTRGDKGTSTISFEAVASHDLWIWHTFFGCASTLNDINFLDRSPVFDDIE 75
Query: 137 QGKTPRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIRLPQSEPDKLFAKHQESCRKDIE 196
QG TPRVN+FV QR YNM YYLADGIYPSYPTFVKSIRLPQSEPDKLF + QE CRKDIE
Sbjct: 76 QGNTPRVNFFVYQRQYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFVQLQEGCRKDIE 135
Query: 197 RAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTYAQRWTDFEQSEGS 256
RAFGVL ARFKII W+IADL IIMRSCIILHNMIVE+ERDTYAQ WTD++QS+ S
Sbjct: 136 RAFGVLHARFKII------WNIADLAIIMRSCIILHNMIVENERDTYAQHWTDYDQSKES 189
Query: 257 GSSTQQPYSTEVLPTFANHVRARSELRD 284
GSST QP+STEVL FANHV ARS+LRD
Sbjct: 190 GSSTPQPFSTEVLHAFANHVHARSKLRD 217
>At1g24370 hypothetical protein
Length = 413
Score = 313 bits (802), Expect = 7e-86
Identities = 143/241 (59%), Positives = 185/241 (76%), Gaps = 1/241 (0%)
Query: 1 MLAYGVAADAVDEYIKIGGITALECLRRFCKGIIRLYEQVYLRAPTQDDLQRILHVSEMR 60
MLAYGV AD+ DEYIKIG TALE L+RFC+ I+ ++ YLR+P +D+ R+LH+ E R
Sbjct: 7 MLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACRYLRSPDANDVARLLHIGESR 66
Query: 61 GFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTL 120
GFP M+GS+DCMHW+WKNCP AW GQ+ G + T+ILEAVA +DLWIWHA+FG PG+
Sbjct: 67 GFPRMLGSLDCMHWKWKNCPTAWGGQYA-GRSRSPTIILEAVADYDLWIWHAYFGLPGSN 125
Query: 121 NDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIRLPQSEP 180
NDINVL+ S +F ++ +G P +Y +N++PYNM+YYLADGIYP + T V++I P+
Sbjct: 126 NDINVLEASHLFANLAEGTAPPASYVINEKPYNMSYYLADGIYPKWSTLVQTIHDPRGPK 185
Query: 181 DKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDER 240
KLFA QE+CRKD+ERAFGVLQ+RF I+ P+RLW+ L IM SCII+HNMI+EDER
Sbjct: 186 KKLFAMKQEACRKDVERAFGVLQSRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMIIEDER 245
Query: 241 D 241
D
Sbjct: 246 D 246
>At2g13770 hypothetical protein
Length = 244
Score = 241 bits (614), Expect = 5e-64
Identities = 121/241 (50%), Positives = 154/241 (63%), Gaps = 36/241 (14%)
Query: 1 MLAYGVAADAVDEYIKIGGITALECLRRFCKGIIRLYEQVYLRAPTQDDLQRILHVSEMR 60
MLAYGV AD+ DEYIKIG TALE L+RFC+ I+ ++ YLR+P +D+ R+LH+ E R
Sbjct: 7 MLAYGVPADSTDEYIKIGESTALESLKRFCRAIVEVFACCYLRSPDANDVARLLHIGESR 66
Query: 61 GFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTL 120
GFP EAVA +DLWIWHA+FG PG+
Sbjct: 67 GFP------------------------------------EAVADYDLWIWHAYFGLPGSN 90
Query: 121 NDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIRLPQSEP 180
N INVL+ S +F ++ +G P +Y +N +PYNM YYLADGIYP + T V++I P+
Sbjct: 91 NGINVLEASHLFANLAEGTAPPASYVINGKPYNMGYYLADGIYPKWSTLVQTIHDPRGPK 150
Query: 181 DKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDER 240
KLFA QE+CRKD+ERAFGVLQ RF I+ P+RLW+ L IM SCII+HNMI+EDER
Sbjct: 151 KKLFAMKQEACRKDVERAFGVLQLRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMIIEDER 210
Query: 241 D 241
D
Sbjct: 211 D 211
>At5g28700 putative protein
Length = 292
Score = 202 bits (513), Expect = 2e-52
Identities = 103/190 (54%), Positives = 129/190 (67%), Gaps = 4/190 (2%)
Query: 93 GTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKTPRVNYFVNQRPY 152
G +T+++ VAS DLWIWHA FG PGTLNDINVLDRSPVFDD+ QG+ +V Y VN + Y
Sbjct: 105 GESTLLV--VASQDLWIWHAIFGPPGTLNDINVLDRSPVFDDILQGRASKVKYVVNGKDY 162
Query: 153 NMTYYLADGIYPSYPTFVKSIRLPQSEPDKLFAKHQESCRKDIERAFGVLQARFKIIREP 212
N+ YYL DGIYP + TF++SI PQ + LF Q++CRKD+ERAFGV QARF I++ P
Sbjct: 163 NLAYYLTDGIYPKWATFIQSISNPQGDKASLFVTTQKACRKDVERAFGVFQARFSIVKHP 222
Query: 213 ARLWDIADLGIIMRSCIILHNMIVEDERDTYAQ-RWTDFEQSE-GSGSSTQQPYSTEVLP 270
A D +G IMR+CIILHNMIVEDERD Y Q ++F E S S Y+T +
Sbjct: 223 ALFHDKVKIGNIMRACIILHNMIVEDERDGYTQCDVSEFVHPEFASSSQVDFTYATYMPS 282
Query: 271 TFANHVRARS 280
N + R+
Sbjct: 283 NLGNMMATRA 292
Score = 38.1 bits (87), Expect = 0.006
Identities = 17/23 (73%), Positives = 20/23 (86%)
Query: 1 MLAYGVAADAVDEYIKIGGITAL 23
MLAYG+AADAVDEY++IG T L
Sbjct: 88 MLAYGIAADAVDEYLRIGESTLL 110
>At1g43590 hypothetical protein
Length = 168
Score = 176 bits (447), Expect = 1e-44
Identities = 89/162 (54%), Positives = 110/162 (66%), Gaps = 2/162 (1%)
Query: 145 YFVNQRPYNMTYYLADGIYPSYPTFVKSIRLPQSEPDKLFAKHQESCRKDIERAFGVLQA 204
+ VN YN+ YYL D IYP + TF++SI LPQ E LFA +QE+CRKD+ERAFGVLQA
Sbjct: 3 FVVNGNEYNLAYYLTDEIYPKWATFIQSISLPQDEKASLFATNQEACRKDVERAFGVLQA 62
Query: 205 RFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTYAQ-RWTDFEQSEGSGSS-TQQ 262
RF I++ PA +WD +G IMR+CIILHNMIVEDERD Y Q +DF E + SS
Sbjct: 63 RFAIVKHPALIWDKIKIGNIMRACIILHNMIVEDERDGYTQYDVSDFAHPESASSSQVDF 122
Query: 263 PYSTEVLPTFANHVRARSELRDPNVHHELQADLVKHIWTKFG 304
YST++ N + R+ LRD H L+ADLV+HIW KFG
Sbjct: 123 TYSTDMPSNLGNMMATRTRLRDRTKHEHLKADLVEHIWQKFG 164
>At3g30190 hypothetical protein
Length = 263
Score = 158 bits (399), Expect = 4e-39
Identities = 75/142 (52%), Positives = 99/142 (68%), Gaps = 6/142 (4%)
Query: 100 EAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLA 159
+AVA +D+WIWHA+FG PG+ NDINVL+ +F ++ + P +Y N +PYNM YYLA
Sbjct: 104 QAVADYDMWIWHAYFGLPGSNNDINVLEAYHLFANLAEDTAPPASYVSNGKPYNMGYYLA 163
Query: 160 DGIYPSYPTFVKSIRLPQSEPDKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIA 219
DGIY + T V++I P+ KLFA E+CRKD+E AF VL +RF I+ EP+RLW+
Sbjct: 164 DGIYSKWSTLVQTIHDPRGPKKKLFAMKLETCRKDVEPAFEVLHSRFAIVAEPSRLWN-- 221
Query: 220 DLGIIMRSCIILHNMIVEDERD 241
M SCII+HNMI+EDERD
Sbjct: 222 ----KMTSCIIMHNMIIEDERD 239
>At1g42690 unknown protein
Length = 333
Score = 123 bits (309), Expect = 1e-28
Identities = 68/142 (47%), Positives = 87/142 (60%), Gaps = 10/142 (7%)
Query: 168 TFVKSIRLPQSEPDKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRS 227
TF++SI + Q LFA QE+CRKD+ERAFGVLQARF II+ PA D +G IMR+
Sbjct: 191 TFIQSISILQGNKASLFATTQEACRKDVERAFGVLQARFAIIKHPALFHDKVKIGNIMRA 250
Query: 228 CIILHNMIVEDERDTYAQRWTDFEQSE------GSGSSTQQPYSTEVLPTFANHVRARSE 281
CIILHNMIVED+RD Y T F+ SE S S Y+T + N + +
Sbjct: 251 CIILHNMIVEDKRDGY----TQFDVSEFVHPESASSSQVDFTYATVMPSNLGNMMATGAR 306
Query: 282 LRDPNVHHELQADLVKHIWTKF 303
+RD H EL+ADLV+H+W +
Sbjct: 307 VRDRIKHEELKADLVEHVWQHY 328
Score = 108 bits (270), Expect = 4e-24
Identities = 54/101 (53%), Positives = 62/101 (60%), Gaps = 31/101 (30%)
Query: 1 MLAYGVAADAVDEYIKIGGITALECLRRFCKGIIRLYEQVYLRAPTQDDLQRILHVSEMR 60
MLAYG+AADA YLR PT+DDL R+LH+ E R
Sbjct: 120 MLAYGIAADA------------------------------YLRRPTRDDLIRLLHIGEQR 149
Query: 61 GFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEA 101
GFPGMIGSIDCMHWEWKNCP AW+GQ+TRG G T++LEA
Sbjct: 150 GFPGMIGSIDCMHWEWKNCPTAWKGQYTRG-SGKPTIVLEA 189
>At5g35800 putative protein
Length = 73
Score = 117 bits (292), Expect = 1e-26
Identities = 54/70 (77%), Positives = 61/70 (87%)
Query: 234 MIVEDERDTYAQRWTDFEQSEGSGSSTQQPYSTEVLPTFANHVRARSELRDPNVHHELQA 293
MIV DER+TYAQ W+D+ QSE SGSST QP+S EVLP FA+HVRARSELRD N+HHELQA
Sbjct: 1 MIVNDERETYAQHWSDYNQSEASGSSTPQPFSIEVLPEFASHVRARSELRDSNLHHELQA 60
Query: 294 DLVKHIWTKF 303
LVK+IWTKF
Sbjct: 61 GLVKYIWTKF 70
>At2g14730 hypothetical protein
Length = 117
Score = 68.6 bits (166), Expect = 4e-12
Identities = 31/62 (50%), Positives = 44/62 (70%)
Query: 10 AVDEYIKIGGITALECLRRFCKGIIRLYEQVYLRAPTQDDLQRILHVSEMRGFPGMIGSI 69
AVD+Y++IG T + C+ + II L+ + YLR PT+ DL+R+L + E+RGF GM GSI
Sbjct: 19 AVDKYLRIGENTLMSCMIHSVEAIIYLFGKEYLRRPTRQDLKRLLRIGELRGFLGMTGSI 78
Query: 70 DC 71
DC
Sbjct: 79 DC 80
Score = 52.0 bits (123), Expect = 4e-07
Identities = 23/34 (67%), Positives = 27/34 (78%)
Query: 179 EPDKLFAKHQESCRKDIERAFGVLQARFKIIREP 212
+ D LFA +QE CRKD+ERAFGVLQARF I+ P
Sbjct: 81 DKDSLFATNQEVCRKDVERAFGVLQARFAIVTNP 114
>At3g55350 unknown protein
Length = 406
Score = 57.4 bits (137), Expect = 1e-08
Identities = 57/195 (29%), Positives = 86/195 (43%), Gaps = 18/195 (9%)
Query: 58 EMRGFPGMIGSIDCMHWEWKNCP------KAWEGQFTRGDKGTTTVILEAVASHDLWIWH 111
++ G P G+ID H N P K W G+K + + L+AV D+
Sbjct: 176 KISGLPNCCGAIDITHIVM-NLPAVEPSNKVW----LDGEKNFS-MTLQAVVDPDMRFLD 229
Query: 112 AFFGCPGTLNDINVLDRSPVFDDVEQGKTPRVN---YFVNQRPYNMTYYLADGIYPSYPT 168
G PG+LND VL S + VE+GK R+N +++R Y + D +P P
Sbjct: 230 VIAGWPGSLNDDVVLKNSGFYKLVEKGK--RLNGEKLPLSERTELREYIVGDSGFPLLPW 287
Query: 169 FVKSIR-LPQSEPDKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRS 227
+ + P S P F K K + A L+ R++II + D L I+
Sbjct: 288 LLTPYQGKPTSLPQTEFNKRHSEATKAAQMALSKLKDRWRIINGVMWMPDRNRLPRIIFV 347
Query: 228 CIILHNMIVEDERDT 242
C +LHN+I++ E T
Sbjct: 348 CCLLHNIIIDMEDQT 362
>At1g61510 hypothetical protein
Length = 608
Score = 56.2 bits (134), Expect = 2e-08
Identities = 59/231 (25%), Positives = 98/231 (41%), Gaps = 32/231 (13%)
Query: 45 PTQDDLQRI--LHVSEMRGFP---GMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVIL 99
PT+DD+ I V++ R P IG++D H + P + + + RG K T+ +
Sbjct: 356 PTRDDVTGISPFLVNDKRYMPYFIDCIGALDGTHVSVR--PPSGDVERYRGRKSEATMNI 413
Query: 100 EAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLA 159
AV + + A+ G PG +D VL + + P P YYL
Sbjct: 414 LAVCNFSMKFNIAYVGVPGRAHDTKVL----TYCATHEASFPH--------PPAGKYYLV 461
Query: 160 DGIYPSYPTFVKSIRL------------PQSEPDKLFAKHQESCRKDIERAFGVLQARFK 207
D YP+ ++ R P + +LF + S R IER FGV +A+++
Sbjct: 462 DSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWR 521
Query: 208 IIREPARLWDIADLGIIMRSCIILHNMIVEDER-DTYAQRWTDFEQSEGSG 257
I+ +++ I+ S + LHN I + ++ D+ + W E E G
Sbjct: 522 ILDRKHPKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWEIVESYEQHG 572
>At5g33270 putative protein
Length = 343
Score = 53.9 bits (128), Expect = 1e-07
Identities = 50/205 (24%), Positives = 87/205 (42%), Gaps = 27/205 (13%)
Query: 66 IGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINV 125
IG++D H + P + + + RG K T+ + A+ + + +A+ G PG +D V
Sbjct: 117 IGALDGTHVSVR--PPSGDVERYRGRKSEATMNILALCNFSMKFTYAYVGVPGRAHDTKV 174
Query: 126 LDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIRL---------- 175
L + + P P YYL D YP+ ++ R
Sbjct: 175 L----TYCATHEASFPH--------PPAGKYYLVDSGYPTRSGYLGPHRRTRYHLELFNR 222
Query: 176 --PQSEPDKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHN 233
P + +LF + S R IER FGV +A+++I+ +++ I+ S + LHN
Sbjct: 223 GGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWRILDRKHLKYEVKKWIKIVTSTMALHN 282
Query: 234 MIVEDER-DTYAQRWTDFEQSEGSG 257
I + ++ D+ + W E E G
Sbjct: 283 YIRDSQQEDSDFRHWEIVESYEQHG 307
>At2g26630 En/Spm-like transposon protein
Length = 292
Score = 53.5 bits (127), Expect = 1e-07
Identities = 49/205 (23%), Positives = 84/205 (40%), Gaps = 37/205 (18%)
Query: 66 IGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINV 125
+G++D H + P + + +G K T+ + A+ + D+ +A+ G PG +D V
Sbjct: 45 VGALDGTHVPVR--PPSQTAKKYKGRKLEPTMNVLAICNFDMKFIYAYVGVPGRAHDTKV 102
Query: 126 LDRSPVFDDVEQGKTPRVNYFVNQRPY-----NMTYYLADGIYPSYPTFVKSIRL----- 175
L NY PY N YYL D YP+ ++ R
Sbjct: 103 L-----------------NYCATNEPYFSHPPNGKYYLVDSGYPTRTGYLGPHRRMRYHL 145
Query: 176 -------PQSEPDKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSC 228
P +LF + R IER FGV +A+++I+ + +A I+ +
Sbjct: 146 GQFGRGGPPVTARELFNRKHSGLRSVIERTFGVWKAKWRIVDRKHPKYGLAKWIKIVTAT 205
Query: 229 IILHNMIVEDERDTY-AQRWTDFEQ 252
+ LHN I + R+ + +W E+
Sbjct: 206 MALHNFIRDSHREDHDFLQWQSIEE 230
>At5g35490 putative protein
Length = 106
Score = 45.4 bits (106), Expect = 4e-05
Identities = 19/35 (54%), Positives = 25/35 (71%)
Query: 182 KLFAKHQESCRKDIERAFGVLQARFKIIREPARLW 216
K+FA QE+CRKD+ER F VLQ++F II + W
Sbjct: 70 KMFAAKQEACRKDVERVFRVLQSKFAIIARSSNCW 104
>At3g63270 unknown protein
Length = 396
Score = 43.5 bits (101), Expect = 1e-04
Identities = 55/239 (23%), Positives = 92/239 (38%), Gaps = 5/239 (2%)
Query: 2 LAYGVAADAVDEYIKIGGITALECLRRFCKGIIRLYEQVYLRAPTQDDLQRILH-VSEMR 60
LA G + +V +G T + RF + + + +LR P D ++ I EM
Sbjct: 116 LASGDSQVSVGAAFGVGQSTVSQVTWRFIEALEERAKH-HLRWPDSDRIEEIKSKFEEMY 174
Query: 61 GFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTL 120
G P G+ID H P + ++ L+ V H++ + G PG +
Sbjct: 175 GLPNCCGAIDTTHIIM-TLPAVQASDDWCDQEKNYSMFLQGVFDHEMRFLNMVTGWPGGM 233
Query: 121 NDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLADGI-YPSYPTFVKSIRLPQ-S 178
+L S F E + N + + Y+ GI YP P + S
Sbjct: 234 TVSKLLKFSGFFKLCENAQILDGNPKTLSQGAQIREYVVGGISYPLLPWLITPHDSDHPS 293
Query: 179 EPDKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVE 237
+ F + E R AF L+ ++I+ + D L I+ C +LHN+I++
Sbjct: 294 DSMVAFNERHEKVRSVAATAFQQLKGSWRILSKVMWRPDRRKLPSIILVCCLLHNIIID 352
>At5g41980 unknown protein
Length = 374
Score = 43.1 bits (100), Expect = 2e-04
Identities = 48/227 (21%), Positives = 89/227 (39%), Gaps = 27/227 (11%)
Query: 84 EGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDD---VEQGKT 140
+G F G+ G T + A +S DL + G G+ +D VL+ + + V QGK
Sbjct: 161 QGPFRNGN-GLLTQNVLAASSFDLRFNYVLAGWEGSASDQQVLNAALTRRNKLQVPQGK- 218
Query: 141 PRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIR----LPQSEPDKLFAKHQESCRKDIE 196
YY+ D YP+ P F+ + E ++F + + + I
Sbjct: 219 ---------------YYIVDNKYPNLPGFIAPYHGVSTNSREEAKEMFNERHKLLHRAIH 263
Query: 197 RAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTYAQRWTDFEQSEGS 256
R FG L+ RF I+ + +++ +C LHN + ++ D R + E +
Sbjct: 264 RTFGALKERFPILLSAPPYPLQTQVKLVIAAC-ALHNYVRLEKPDDLVFRMFEEETLAEA 322
Query: 257 GSSTQQPYSTEVLPTFANHVRARSELRDPNVHHELQADLVKHIWTKF 303
G + E + R E + ++ L+ ++ +W +
Sbjct: 323 GEDREVALEEEQVEIVGQEHGFRPEEVEDSL--RLRDEIASELWNHY 367
>At5g27250 putative protein
Length = 348
Score = 42.4 bits (98), Expect = 3e-04
Identities = 50/206 (24%), Positives = 79/206 (38%), Gaps = 30/206 (14%)
Query: 89 RGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVEQGKTPRVNYFVN 148
R KG + + A + DL + G G+ +D VL + T R N
Sbjct: 135 RNRKGVISQNVLAACNFDLEFIYVLSGWEGSAHDSKVL---------QDALTRRTNRL-- 183
Query: 149 QRPYNMTYYLADGIYPSYPTFVKSIRLPQ-------------SEPDKLFAKHQESCRKDI 195
Q P YYLAD +P+ F+ +R + + ++LF S R I
Sbjct: 184 QVPEGK-YYLADCGFPNRRNFLAPLRSTRYHLQDFRGEGRDPTNQNELFNLRHASLRNVI 242
Query: 196 ERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVE----DERDTYAQRWTDFE 251
ER FG+ ++RF I + A + I+ SC LHN + + DE + + TD +
Sbjct: 243 ERIFGIFKSRFLIFKS-APPFSFKTQAEIVLSCAALHNFLRQKCRSDEFSSDEEDETDVD 301
Query: 252 QSEGSGSSTQQPYSTEVLPTFANHVR 277
+ + + E H R
Sbjct: 302 NANQNSEENGGEENVETQEQEREHAR 327
>At5g12010 putative protein
Length = 502
Score = 42.4 bits (98), Expect = 3e-04
Identities = 51/245 (20%), Positives = 102/245 (40%), Gaps = 29/245 (11%)
Query: 2 LAYGVAADAVDEYIKIGGITALECLRRFCKGIIRLYEQVYLRAPTQDDLQRILHVSE-MR 60
LA G V + +G T + + CK I + YL+ P + L+ I E +
Sbjct: 221 LATGEPLRLVSKKFGLGISTCHKLVLEVCKAIKDVLMPKYLQWPDDESLRNIRERFESVS 280
Query: 61 GFPGMIGSIDCMHWEWKNCPKAWEGQF------TRGDKGTTTVILEAVASHDLWIWHAFF 114
G P ++GS+ H PK + R K + ++ ++AV +
Sbjct: 281 GIPNVVGSMYTTHIP-IIAPKISVASYFNKRHTERNQKTSYSITIQAVVNPKGVFTDLCI 339
Query: 115 GCPGTLNDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIR 174
G PG++ D VL++S ++ G + ++A G P +P + +
Sbjct: 340 GWPGSMPDDKVLEKSLLYQRANNGGLLK------------GMWVAGG--PGHP-LLDWVL 384
Query: 175 LPQSEPDKLFAKH-----QESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCI 229
+P ++ + + +H + + AFG L+ R+ +++ + + DL ++ +C
Sbjct: 385 VPYTQQNLTWTQHAFNEKMSEVQGVAKEAFGRLKGRWACLQKRTEV-KLQDLPTVLGACC 443
Query: 230 ILHNM 234
+LHN+
Sbjct: 444 VLHNI 448
>At3g19120 unknown protein
Length = 446
Score = 39.7 bits (91), Expect = 0.002
Identities = 44/199 (22%), Positives = 80/199 (40%), Gaps = 7/199 (3%)
Query: 58 EMRGFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCP 117
E+ P + G+ID + + K G V+L+ VA H W P
Sbjct: 217 ELTSLPNICGAIDSTPVKLRRRTKLNPRNIYGCKYGYDAVLLQVVADHKKIFWDVCVKAP 276
Query: 118 GTLNDINVLDRSPVFDDVEQGKTPRVNYFVNQRPYNMTYYLADGIYPSYPTFVKSIRLPQ 177
G +D + S ++ + G + + Y + D YP +F+ + P
Sbjct: 277 GGEDDSSHFRDSLLYKRLTSGDIVWEKVINIRGHHVRPYIVGDWCYPLL-SFLMTPFSPN 335
Query: 178 SE---PDKLFAKHQESCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNM 234
P+ LF R + A G+L+AR+KI++ + A I+ +C +LHN
Sbjct: 336 GSGTPPENLFDGMLMKGRSVVVEAIGLLKARWKILQSLNVGVNHAPQTIV--ACCVLHN- 392
Query: 235 IVEDERDTYAQRWTDFEQS 253
+ + R+ + W D +++
Sbjct: 393 LCQIAREPEPEIWKDPDEA 411
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,345,496
Number of Sequences: 26719
Number of extensions: 313101
Number of successful extensions: 767
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 732
Number of HSP's gapped (non-prelim): 33
length of query: 308
length of database: 11,318,596
effective HSP length: 99
effective length of query: 209
effective length of database: 8,673,415
effective search space: 1812743735
effective search space used: 1812743735
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146585.11


