

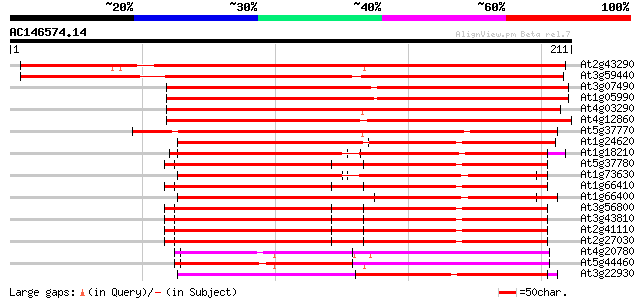
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146574.14 - phase: 0
(211 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g43290 putative calcium binding protein 270 5e-73
At3g59440 calmodulin-like protein 248 1e-66
At3g07490 putative calmodulin 236 5e-63
At1g05990 calcium-binding protein, putative 236 5e-63
At4g03290 putative calmodulin 210 4e-55
At4g12860 putative calmodulin 210 5e-55
At5g37770 CALMODULIN-RELATED PROTEIN 2, TOUCH-INDUCED (TCH2) 115 2e-26
At1g24620 putative calmodulin 114 3e-26
At1g18210 unknown protein 112 1e-25
At5g37780 calmodulin 1 (CAM1) 108 2e-24
At1g73630 calmodulin like protein 108 2e-24
At1g66410 calmodulin-4 108 2e-24
At1g66400 calmodulin-related protein 108 3e-24
At3g56800 calmodulin-3 107 5e-24
At3g43810 calmodulin 7 107 5e-24
At2g41110 calmodulin (cam2) 107 5e-24
At2g27030 calmodulin 107 5e-24
At4g20780 calcium-binding protein - like 100 6e-22
At5g44460 calmodulin-like protein 99 1e-21
At3g22930 calmodulin, putative 99 1e-21
>At2g43290 putative calcium binding protein
Length = 215
Score = 270 bits (689), Expect = 5e-73
Identities = 140/219 (63%), Positives = 171/219 (77%), Gaps = 20/219 (9%)
Query: 5 LLRIFLLYNVVNSFLISLVPKKLITFFPHSWFT---HQT--------LTTPSSTSKRGLV 53
++RIFLLYN++NSFL+SLVPKKL T FP SWF H+ L +PSS+S
Sbjct: 1 MVRIFLLYNILNSFLLSLVPKKLRTLFPLSWFDKTLHKNSPPSPSTMLPSPSSSS----- 55
Query: 54 FTKTITMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRD 113
T +DP+ELKRVFQMFD+N DGRITK+ELNDSLENLGI+IPDK+L+QMI KID N D
Sbjct: 56 -APTKRIDPSELKRVFQMFDKNGDGRITKEELNDSLENLGIYIPDKDLTQMIHKIDANGD 114
Query: 114 GCVDIEEFRELYESIMSER---DEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLK 170
GCVDI+EF LY SI+ E E EEEDM++AFNVFDQ+GDGFI+V+EL+SV+ SLGLK
Sbjct: 115 GCVDIDEFESLYSSIVDEHHNDGETEEEDMKDAFNVFDQDGDGFITVEELKSVMASLGLK 174
Query: 171 QGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMKGGGFTA 209
QG+T++ CKKMI VD DG+G V+YKEF QMMKGGGF++
Sbjct: 175 QGKTLDGCKKMIMQVDADGDGRVNYKEFLQMMKGGGFSS 213
>At3g59440 calmodulin-like protein
Length = 195
Score = 248 bits (634), Expect = 1e-66
Identities = 127/204 (62%), Positives = 155/204 (75%), Gaps = 12/204 (5%)
Query: 5 LLRIFLLYNVVNSFLISLVPKKLITFFPHSWFTHQTLTTPSSTSKRGLVFTKTITMDPNE 64
++R+FLLYN+ NSFL+ LVPKKL FFP SW+ P S+ T P +
Sbjct: 1 MVRVFLLYNLFNSFLLCLVPKKLRVFFPPSWYIDDKNPPPPDESE---------TESPVD 51
Query: 65 LKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFREL 124
LKRVFQMFD+N DGRITK+ELNDSLENLGIF+PDK+L QMI+K+D N DGCVDI EF L
Sbjct: 52 LKRVFQMFDKNGDGRITKEELNDSLENLGIFMPDKDLIQMIQKMDANGDGCVDINEFESL 111
Query: 125 YESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGT 184
Y SI+ +E+EE DMR+AFNVFDQ+GDGFI+V+EL SV+ SLGLKQG+T+E CK+MI
Sbjct: 112 YGSIV---EEKEEGDMRDAFNVFDQDGDGFITVEELNSVMTSLGLKQGKTLECCKEMIMQ 168
Query: 185 VDVDGNGLVDYKEFKQMMKGGGFT 208
VD DG+G V+YKEF QMMK G F+
Sbjct: 169 VDEDGDGRVNYKEFLQMMKSGDFS 192
>At3g07490 putative calmodulin
Length = 153
Score = 236 bits (603), Expect = 5e-63
Identities = 118/151 (78%), Positives = 134/151 (88%), Gaps = 2/151 (1%)
Query: 60 MDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIE 119
MD EL R+FQMFDRN DG+ITK+ELNDSLENLGI+IPDK+L QMIEKID+N DG VDIE
Sbjct: 1 MDQAELARIFQMFDRNGDGKITKQELNDSLENLGIYIPDKDLVQMIEKIDLNGDGYVDIE 60
Query: 120 EFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCK 179
EF LY++IM ERDEEE DMREAFNVFDQN DGFI+V+ELRSVL SLGLKQGRT+EDCK
Sbjct: 61 EFGGLYQTIMEERDEEE--DMREAFNVFDQNRDGFITVEELRSVLASLGLKQGRTLEDCK 118
Query: 180 KMIGTVDVDGNGLVDYKEFKQMMKGGGFTAL 210
+MI VDVDG+G+V++KEFKQMMKGGGF AL
Sbjct: 119 RMISKVDVDGDGMVNFKEFKQMMKGGGFAAL 149
>At1g05990 calcium-binding protein, putative
Length = 150
Score = 236 bits (603), Expect = 5e-63
Identities = 115/151 (76%), Positives = 136/151 (89%), Gaps = 1/151 (0%)
Query: 60 MDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIE 119
MDP ELKRVFQMFD+N DG IT KEL+++L +LGI+IPDKEL+QMIEKIDVN DGCVDI+
Sbjct: 1 MDPTELKRVFQMFDKNGDGTITGKELSETLRSLGIYIPDKELTQMIEKIDVNGDGCVDID 60
Query: 120 EFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCK 179
EF ELY++IM E DEEEE DM+EAFNVFDQNGDGFI+VDEL++VL SLGLKQG+T++DCK
Sbjct: 61 EFGELYKTIMDEEDEEEE-DMKEAFNVFDQNGDGFITVDELKAVLSSLGLKQGKTLDDCK 119
Query: 180 KMIGTVDVDGNGLVDYKEFKQMMKGGGFTAL 210
KMI VDVDG+G V+YKEF+QMMKGGGF +L
Sbjct: 120 KMIKKVDVDGDGRVNYKEFRQMMKGGGFNSL 150
>At4g03290 putative calmodulin
Length = 154
Score = 210 bits (535), Expect = 4e-55
Identities = 104/149 (69%), Positives = 127/149 (84%), Gaps = 1/149 (0%)
Query: 60 MDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIE 119
MD EL RVFQMFD++ DG+IT KELN+S +NLGI IP+ EL+Q+I+KIDVN DGCVDIE
Sbjct: 1 MDSTELNRVFQMFDKDGDGKITTKELNESFKNLGIIIPEDELTQIIQKIDVNGDGCVDIE 60
Query: 120 EFRELYESIMSE-RDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDC 178
EF ELY++IM E DE EEDM+EAFNVFD+NGDGFI+VDEL++VL SLGLKQG+T+E+C
Sbjct: 61 EFGELYKTIMVEDEDEVGEEDMKEAFNVFDRNGDGFITVDELKAVLSSLGLKQGKTLEEC 120
Query: 179 KKMIGTVDVDGNGLVDYKEFKQMMKGGGF 207
+KMI VDVDG+G V+Y EF+QMMK G F
Sbjct: 121 RKMIMQVDVDGDGRVNYMEFRQMMKKGRF 149
>At4g12860 putative calmodulin
Length = 152
Score = 210 bits (534), Expect = 5e-55
Identities = 100/152 (65%), Positives = 126/152 (82%), Gaps = 2/152 (1%)
Query: 60 MDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIE 119
MD EL RVFQMFD+N DG+I K EL D +++GI +P+ E+++MI K+DVN DG +DI+
Sbjct: 1 MDRGELSRVFQMFDKNGDGKIAKNELKDFFKSVGIMVPENEINEMIAKMDVNGDGAMDID 60
Query: 120 EFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCK 179
EF LY+ ++ E+ EEEEDMREAF VFDQNGDGFI+ +ELRSVL S+GLKQGRT+EDCK
Sbjct: 61 EFGSLYQEMVEEK--EEEEDMREAFRVFDQNGDGFITDEELRSVLASMGLKQGRTLEDCK 118
Query: 180 KMIGTVDVDGNGLVDYKEFKQMMKGGGFTALS 211
KMI VDVDG+G+V++KEFKQMM+GGGF ALS
Sbjct: 119 KMISKVDVDGDGMVNFKEFKQMMRGGGFAALS 150
>At5g37770 CALMODULIN-RELATED PROTEIN 2, TOUCH-INDUCED (TCH2)
Length = 161
Score = 115 bits (288), Expect = 2e-26
Identities = 63/162 (38%), Positives = 105/162 (63%), Gaps = 6/162 (3%)
Query: 47 TSKRGLVFTKTITMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIE 106
+SK G+V + +MD ++K+VFQ FD+N DG+I+ EL + + L +E M++
Sbjct: 2 SSKNGVVRSCLGSMD--DIKKVFQRFDKNGDGKISVDELKEVIRALSPTASPEETVTMMK 59
Query: 107 KIDVNRDGCVDIEEFRELYESIMSE--RDEEEEEDMREAFNVFDQNGDGFISVDELRSVL 164
+ D++ +G +D++EF L++ + + + D++EAF ++D +G+G IS EL SV+
Sbjct: 60 QFDLDGNGFIDLDEFVALFQIGIGGGGNNRNDVSDLKEAFELYDLDGNGRISAKELHSVM 119
Query: 165 VSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMKGGG 206
+LG K +V+DCKKMI VD+DG+G V++ EFK+MM GG
Sbjct: 120 KNLGEKC--SVQDCKKMISKVDIDGDGCVNFDEFKKMMSNGG 159
>At1g24620 putative calmodulin
Length = 186
Score = 114 bits (286), Expect = 3e-26
Identities = 63/142 (44%), Positives = 94/142 (65%), Gaps = 4/142 (2%)
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
EL+ VF+ FD N DG+I+ KEL + +LG +P++EL + I +ID DG ++ EEF E
Sbjct: 37 ELEAVFKKFDVNGDGKISSKELGAIMTSLGHEVPEEELEKAITEIDRKGDGYINFEEFVE 96
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
L M + D E++++AF+V+D +G+G IS +EL VL SLG ++ +C+KMIG
Sbjct: 97 LNTKGMDQND--VLENLKDAFSVYDIDGNGSISAEELHEVLRSLG--DECSIAECRKMIG 152
Query: 184 TVDVDGNGLVDYKEFKQMMKGG 205
VD DG+G +D++EFK MM G
Sbjct: 153 GVDKDGDGTIDFEEFKIMMTMG 174
Score = 44.7 bits (104), Expect = 4e-05
Identities = 24/70 (34%), Positives = 40/70 (56%), Gaps = 2/70 (2%)
Query: 136 EEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDY 195
E ++ F FD NGDG IS EL +++ SLG + E+ +K I +D G+G +++
Sbjct: 34 EIRELEAVFKKFDVNGDGKISSKELGAIMTSLGHEVPE--EELEKAITEIDRKGDGYINF 91
Query: 196 KEFKQMMKGG 205
+EF ++ G
Sbjct: 92 EEFVELNTKG 101
>At1g18210 unknown protein
Length = 170
Score = 112 bits (281), Expect = 1e-25
Identities = 57/142 (40%), Positives = 92/142 (64%), Gaps = 8/142 (5%)
Query: 61 DPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEE 120
+P ELK+VF FD N DG+I+ EL + +G + EL++++E++D +RDG ++++E
Sbjct: 20 NPEELKKVFDQFDSNGDGKISVLELGGVFKAMGTSYTETELNRVLEEVDTDRDGYINLDE 79
Query: 121 FRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKK 180
F L R ++R+AF+++DQ+ +G IS EL VL LG+ +VEDC +
Sbjct: 80 FSTLC------RSSSSAAEIRDAFDLYDQDKNGLISASELHQVLNRLGM--SCSVEDCTR 131
Query: 181 MIGTVDVDGNGLVDYKEFKQMM 202
MIG VD DG+G V+++EF++MM
Sbjct: 132 MIGPVDADGDGNVNFEEFQKMM 153
Score = 45.4 bits (106), Expect = 2e-05
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 2/77 (2%)
Query: 133 DEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGL 192
D E++++ F+ FD NGDG ISV EL V ++G T + +++ VD D +G
Sbjct: 17 DMANPEELKKVFDQFDSNGDGKISVLELGGVFKAMGTSYTET--ELNRVLEEVDTDRDGY 74
Query: 193 VDYKEFKQMMKGGGFTA 209
++ EF + + A
Sbjct: 75 INLDEFSTLCRSSSSAA 91
Score = 44.7 bits (104), Expect = 4e-05
Identities = 20/64 (31%), Positives = 40/64 (62%)
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
E++ F ++D++ +G I+ EL+ L LG+ ++ ++MI +D + DG V+ EEF++
Sbjct: 92 EIRDAFDLYDQDKNGLISASELHQVLNRLGMSCSVEDCTRMIGPVDADGDGNVNFEEFQK 151
Query: 124 LYES 127
+ S
Sbjct: 152 MMTS 155
>At5g37780 calmodulin 1 (CAM1)
Length = 149
Score = 108 bits (270), Expect = 2e-24
Identities = 59/140 (42%), Positives = 88/140 (62%), Gaps = 4/140 (2%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 11 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ ++MI
Sbjct: 71 NLMAKKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVEEMI 126
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 127 READVDGDGQINYEEFVKIM 146
Score = 61.2 bits (147), Expect = 4e-10
Identities = 30/69 (43%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+E+ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 7 DEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 64
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 65 DFPEFLNLM 73
Score = 52.4 bits (124), Expect = 2e-07
Identities = 25/63 (39%), Positives = 40/63 (62%)
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 80 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIREADVDGDGQINY 139
Query: 119 EEF 121
EEF
Sbjct: 140 EEF 142
>At1g73630 calmodulin like protein
Length = 163
Score = 108 bits (270), Expect = 2e-24
Identities = 56/139 (40%), Positives = 92/139 (65%), Gaps = 8/139 (5%)
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
ELK+VF FD N DG+I+ EL + +++G ++EL++++++ID++ DG ++ EEF
Sbjct: 20 ELKKVFDKFDANGDGKISVSELGNVFKSMGTSYTEEELNRVLDEIDIDCDGFINQEEFAT 79
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
+ R ++REAF+++DQN +G IS E+ VL LG+ +VEDC +MIG
Sbjct: 80 IC------RSSSSAVEIREAFDLYDQNKNGLISSSEIHKVLNRLGMT--CSVEDCVRMIG 131
Query: 184 TVDVDGNGLVDYKEFKQMM 202
VD DG+G V+++EF++MM
Sbjct: 132 HVDTDGDGNVNFEEFQKMM 150
Score = 48.5 bits (114), Expect = 3e-06
Identities = 24/73 (32%), Positives = 42/73 (56%), Gaps = 2/73 (2%)
Query: 126 ESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTV 185
ES + ++++ F+ FD NGDG ISV EL +V S+G T E+ +++ +
Sbjct: 7 ESTNKSTTPSTDMELKKVFDKFDANGDGKISVSELGNVFKSMGTSY--TEEELNRVLDEI 64
Query: 186 DVDGNGLVDYKEF 198
D+D +G ++ +EF
Sbjct: 65 DIDCDGFINQEEF 77
Score = 45.8 bits (107), Expect = 2e-05
Identities = 20/64 (31%), Positives = 39/64 (60%)
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
E++ F ++D+N +G I+ E++ L LG+ ++ +MI +D + DG V+ EEF++
Sbjct: 89 EIREAFDLYDQNKNGLISSSEIHKVLNRLGMTCSVEDCVRMIGHVDTDGDGNVNFEEFQK 148
Query: 124 LYES 127
+ S
Sbjct: 149 MMSS 152
>At1g66410 calmodulin-4
Length = 149
Score = 108 bits (270), Expect = 2e-24
Identities = 59/140 (42%), Positives = 88/140 (62%), Gaps = 4/140 (2%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 11 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ ++MI
Sbjct: 71 NLMAKKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVEEMI 126
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 127 READVDGDGQINYEEFVKIM 146
Score = 61.2 bits (147), Expect = 4e-10
Identities = 30/69 (43%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+E+ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 7 DEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 64
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 65 DFPEFLNLM 73
Score = 52.4 bits (124), Expect = 2e-07
Identities = 25/63 (39%), Positives = 40/63 (62%)
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 80 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVEEMIREADVDGDGQINY 139
Query: 119 EEF 121
EEF
Sbjct: 140 EEF 142
>At1g66400 calmodulin-related protein
Length = 157
Score = 108 bits (269), Expect = 3e-24
Identities = 54/143 (37%), Positives = 94/143 (64%), Gaps = 2/143 (1%)
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
++K+VFQ FD+N+DG+I+ EL D + L +E M+++ D++ +G +D++EF
Sbjct: 15 DIKKVFQRFDKNNDGKISIDELKDVIGALSPNASQEETKAMMKEFDLDGNGFIDLDEFVA 74
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
L++ + D++EAF+++D + +G IS +EL SV+ +LG K +++DC++MI
Sbjct: 75 LFQISDQSSNNSAIRDLKEAFDLYDLDRNGRISANELHSVMKNLGEK--CSIQDCQRMIN 132
Query: 184 TVDVDGNGLVDYKEFKQMMKGGG 206
VD DG+G VD++EFK+MM G
Sbjct: 133 KVDSDGDGCVDFEEFKKMMMING 155
Score = 53.5 bits (127), Expect = 8e-08
Identities = 26/61 (42%), Positives = 39/61 (63%), Gaps = 2/61 (3%)
Query: 138 EDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKE 197
ED+++ F FD+N DG IS+DEL+ V+ + L + E+ K M+ D+DGNG +D E
Sbjct: 14 EDIKKVFQRFDKNNDGKISIDELKDVIGA--LSPNASQEETKAMMKEFDLDGNGFIDLDE 71
Query: 198 F 198
F
Sbjct: 72 F 72
>At3g56800 calmodulin-3
Length = 149
Score = 107 bits (267), Expect = 5e-24
Identities = 59/140 (42%), Positives = 87/140 (62%), Gaps = 4/140 (2%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 11 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 71 NLMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMI 126
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 127 KEADVDGDGQINYEEFVKVM 146
Score = 60.1 bits (144), Expect = 8e-10
Identities = 29/69 (42%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 7 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 64
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 65 DFPEFLNLM 73
Score = 52.8 bits (125), Expect = 1e-07
Identities = 25/63 (39%), Positives = 41/63 (64%)
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI++ DV+ DG ++
Sbjct: 80 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINY 139
Query: 119 EEF 121
EEF
Sbjct: 140 EEF 142
>At3g43810 calmodulin 7
Length = 149
Score = 107 bits (267), Expect = 5e-24
Identities = 59/140 (42%), Positives = 87/140 (62%), Gaps = 4/140 (2%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 11 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 71 NLMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMI 126
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 127 READVDGDGQINYEEFVKVM 146
Score = 60.1 bits (144), Expect = 8e-10
Identities = 29/69 (42%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 7 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 64
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 65 DFPEFLNLM 73
Score = 52.4 bits (124), Expect = 2e-07
Identities = 25/63 (39%), Positives = 40/63 (62%)
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 80 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY 139
Query: 119 EEF 121
EEF
Sbjct: 140 EEF 142
>At2g41110 calmodulin (cam2)
Length = 149
Score = 107 bits (267), Expect = 5e-24
Identities = 59/140 (42%), Positives = 87/140 (62%), Gaps = 4/140 (2%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 11 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 71 NLMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMI 126
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 127 KEADVDGDGQINYEEFVKVM 146
Score = 60.1 bits (144), Expect = 8e-10
Identities = 29/69 (42%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 7 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 64
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 65 DFPEFLNLM 73
Score = 52.8 bits (125), Expect = 1e-07
Identities = 25/63 (39%), Positives = 41/63 (64%)
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI++ DV+ DG ++
Sbjct: 80 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINY 139
Query: 119 EEF 121
EEF
Sbjct: 140 EEF 142
>At2g27030 calmodulin
Length = 149
Score = 107 bits (267), Expect = 5e-24
Identities = 59/140 (42%), Positives = 87/140 (62%), Gaps = 4/140 (2%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 11 SEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 70
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 71 NLMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMI 126
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 127 KEADVDGDGQINYEEFVKVM 146
Score = 60.1 bits (144), Expect = 8e-10
Identities = 29/69 (42%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 7 DDQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 64
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 65 DFPEFLNLM 73
Score = 52.8 bits (125), Expect = 1e-07
Identities = 25/63 (39%), Positives = 41/63 (64%)
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI++ DV+ DG ++
Sbjct: 80 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIKEADVDGDGQINY 139
Query: 119 EEF 121
EEF
Sbjct: 140 EEF 142
>At4g20780 calcium-binding protein - like
Length = 191
Score = 100 bits (249), Expect = 6e-22
Identities = 59/155 (38%), Positives = 91/155 (58%), Gaps = 16/155 (10%)
Query: 65 LKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFREL 124
L+R+F +FD+N DG IT +EL+ +L LG+ +L +E + ++ ++F L
Sbjct: 30 LQRIFDLFDKNGDGFITVEELSQALTRLGLNADLSDLKSTVESYIQPGNTGLNFDDFSSL 89
Query: 125 YESI------------MSERDE----EEEEDMREAFNVFDQNGDGFISVDELRSVLVSLG 168
++++ +E D E E D+ EAF VFD+NGDGFIS EL++VL LG
Sbjct: 90 HKTLDDSFFGGACGGGENEDDPSSAAENESDLAEAFKVFDENGDGFISARELQTVLKKLG 149
Query: 169 LKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMK 203
L +G +E +KMI +VD + +G VD+ EFK MM+
Sbjct: 150 LPEGGEMERVEKMIVSVDRNQDGRVDFFEFKNMMR 184
Score = 46.2 bits (108), Expect = 1e-05
Identities = 24/71 (33%), Positives = 43/71 (59%), Gaps = 6/71 (8%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPD----KELSQMIEKIDVNRDGCVDI 118
++L F++FD N DG I+ +EL L+ LG +P+ + + +MI +D N+DG VD
Sbjct: 119 SDLAEAFKVFDENGDGFISARELQTVLKKLG--LPEGGEMERVEKMIVSVDRNQDGRVDF 176
Query: 119 EEFRELYESIM 129
EF+ + +++
Sbjct: 177 FEFKNMMRTVV 187
>At5g44460 calmodulin-like protein
Length = 181
Score = 99.4 bits (246), Expect = 1e-21
Identities = 56/147 (38%), Positives = 85/147 (57%), Gaps = 8/147 (5%)
Query: 65 LKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFREL 124
L RVF +FD+N+DG IT +EL+ +L LG+ +L ++ + ++F L
Sbjct: 29 LHRVFDLFDKNNDGFITVEELSQALSRLGLDADFSDLKSTVDSFIKPDKTGLRFDDFAAL 88
Query: 125 YESIMSER--------DEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVE 176
++++ D E D+ EAFNVFD++GDGFIS EL+ VL LGL + +E
Sbjct: 89 HKTLDESFFGGEGSCCDGSPESDLEEAFNVFDEDGDGFISAVELQKVLKKLGLPEAGEIE 148
Query: 177 DCKKMIGTVDVDGNGLVDYKEFKQMMK 203
+KMI +VD + +G VD+ EFK MM+
Sbjct: 149 QVEKMIVSVDSNHDGRVDFFEFKNMMQ 175
Score = 44.7 bits (104), Expect = 4e-05
Identities = 23/71 (32%), Positives = 43/71 (60%), Gaps = 6/71 (8%)
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPD----KELSQMIEKIDVNRDGCVDI 118
++L+ F +FD + DG I+ EL L+ LG+ P+ +++ +MI +D N DG VD
Sbjct: 110 SDLEEAFNVFDEDGDGFISAVELQKVLKKLGL--PEAGEIEQVEKMIVSVDSNHDGRVDF 167
Query: 119 EEFRELYESIM 129
EF+ + ++++
Sbjct: 168 FEFKNMMQTVV 178
>At3g22930 calmodulin, putative
Length = 173
Score = 99.4 bits (246), Expect = 1e-21
Identities = 57/143 (39%), Positives = 87/143 (59%), Gaps = 4/143 (2%)
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
E K F +FD++ DG IT EL + +L ++EL MI +ID + +G ++ EF
Sbjct: 35 EFKEAFCLFDKDGDGCITADELATVIRSLDQNPTEQELQDMITEIDSDGNGTIEFSEFLN 94
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
L + + E D +EE ++EAF VFD++ +G+IS ELR V+++LG K T E+ +MI
Sbjct: 95 LMANQLQETDADEE--LKEAFKVFDKDQNGYISASELRHVMINLGEK--LTDEEVDQMIK 150
Query: 184 TVDVDGNGLVDYKEFKQMMKGGG 206
D+DG+G V+Y EF +MM G
Sbjct: 151 EADLDGDGQVNYDEFVRMMMING 173
Score = 58.2 bits (139), Expect = 3e-09
Identities = 29/72 (40%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Query: 131 ERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGN 190
E +E+ + +EAF +FD++GDG I+ DEL +V+ S L Q T ++ + MI +D DGN
Sbjct: 27 ELTQEQIMEFKEAFCLFDKDGDGCITADELATVIRS--LDQNPTEQELQDMITEIDSDGN 84
Query: 191 GLVDYKEFKQMM 202
G +++ EF +M
Sbjct: 85 GTIEFSEFLNLM 96
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,896,054
Number of Sequences: 26719
Number of extensions: 222893
Number of successful extensions: 2190
Number of sequences better than 10.0: 210
Number of HSP's better than 10.0 without gapping: 146
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 1405
Number of HSP's gapped (non-prelim): 424
length of query: 211
length of database: 11,318,596
effective HSP length: 95
effective length of query: 116
effective length of database: 8,780,291
effective search space: 1018513756
effective search space used: 1018513756
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146574.14


