

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146574.12 - phase: 0 /pseudo
(951 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
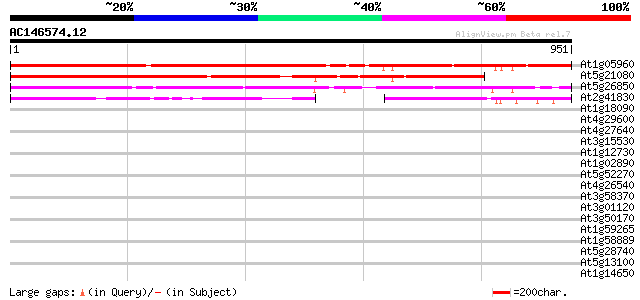
At1g05960 unknown protein 833 0.0
At5g21080 putative protein 599 e-171
At5g26850 unknown protein 438 e-123
At2g41830 hypothetical protein 344 2e-94
At1g18090 unknown protein 37 0.069
At4g29600 cytidine deaminase 7 33 1.00
At4g27640 putative protein 33 1.00
At3g15530 unknown protein 32 2.2
At1g12730 unknown protein 31 3.8
At1g02890 hypothetical protein 31 3.8
At5g52270 putative protein 30 4.9
At4g26540 receptor protein kinase like protein 30 4.9
At3g58370 putative protein 30 4.9
At3g01120 cystathionine gamma-synthase like protein 30 4.9
At3g50170 putative protein 30 6.4
At1g59265 polyprotein, putative 30 6.4
At1g58889 polyprotein, putative 30 6.4
At5g28740 putative protein 30 8.4
At5g13100 unknown protein 30 8.4
At1g14650 splicing factor, putative 30 8.4
>At1g05960 unknown protein
Length = 982
Score = 833 bits (2152), Expect = 0.0
Identities = 485/1007 (48%), Positives = 658/1007 (65%), Gaps = 82/1007 (8%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGVMSRRV+PACGNLC FCPSLRARSR PVKRYKK++AEI PRN+ AE NDRKIGKLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
AS+NPLRIPKITE LEQ+CYK+LRN + GSVKV+LCIY+KLLSSC+EQ+PLF+ SLL I+
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
RTLLEQT+ +EV+ILGCNTLVDFI QT ++MFNLEG IPKLCQLAQE+GDDER+L LR
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGDDERSLQLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQ-SKSNLAKVEKLNSQSQNQLV 239
SAG+Q L+ MV F+GEHS LSMD D IIS ILENY+DL+ + + +V++++
Sbjct: 181 SAGMQALAFMVSFIGEHSQLSMDLDMIISVILENYMDLEKGQEDTKEVDQISD------- 233
Query: 240 QEFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPL 299
+ P VS N T +++E+ +D +K+P+YWS VCL NIAKLAKE TTVRRVLEPL
Sbjct: 234 TKIPNMTKKVSFKPNPVTDYKLEN-MDISKSPSYWSMVCLCNIAKLAKETTTVRRVLEPL 292
Query: 300 FHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQID 359
FD+ ++WS +KGVA VL++LQ L ESG N H+++S L+KHLDHKNV KQ LQI+
Sbjct: 293 LTAFDSGDYWSPQKGVASSVLLFLQSRLEESGENCHVLVSSLIKHLDHKNVIKQQGLQIN 352
Query: 360 IINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIE 419
++N+ T +A + KQQAS A+ I+DLIKHLR+CLQN+AE +D+ D N+ LQ ++E
Sbjct: 353 MVNVATCLALHAKQQASGAMTAVIADLIKHLRKCLQNAAE-SDVSVDKTKQNSDLQHALE 411
Query: 420 MCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKA 479
CI +LSNKVGDAGPI D+ AVVLE +S++ +++RTT SA+ + A +++ VPNV YH K
Sbjct: 412 NCIAELSNKVGDAGPILDMFAVVLETISTNVVLSRTTASAILRAAHIVSVVPNVSYHKKV 471
Query: 480 FPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPWLDQKKISKKVESDGLSIQHE 539
FPDALFHQLLLAM+H D T++ AH+I S+VL+ ++ PW DQ K + + S LS+
Sbjct: 472 FPDALFHQLLLAMSHADCTTRVEAHNIFSVVLLGTLRLPWSDQHKETSEAVSGSLSV--- 528
Query: 540 SLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQLVKCCYKQFLTKYWISEIFRI 599
G + + E++ + SL S + ++ S V Q L+ + + +
Sbjct: 529 --DGICTVRNQEEEKEKVEKSLNSELCKDVN---HISRPSVSGQTSQQLSCQSLDSLKDL 583
Query: 600 *DLLG*AVTK*VSCFLQSGFRQHLLKMALQIMR-------QWHTLIALLYYLLVL----- 647
D + K + S + ++L +L I + +A Y + +L
Sbjct: 584 DDGI-----KSLCSLRLSSHQVNMLLSSLWIQATSTDNTPENFEAMASTYQITLLFSLAK 638
Query: 648 --SYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKV 705
++MALV+CFQLAFSLR++SL+Q+GG+ SRRRS+ T AS+MLIF A+ ++ +L+P +
Sbjct: 639 RSNHMALVQCFQLAFSLRNLSLNQDGGMQHSRRRSIFTFASYMLIFGAKISNILELVPII 698
Query: 706 KASLTEAPVDPFLELVDDNLLRAVCIK-SDKVVFGSVEDEVAAMKSLSAVQLDDRQLKET 764
K SLT VDP+L L D LRAVC + +GS +D+ AA+ S S + DDR+LKE
Sbjct: 699 KESLTAQMVDPYLVLEGDIRLRAVCSGFPQEETYGSDKDDSAALNS-SVIVTDDRRLKEI 757
Query: 765 VISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQIEFP--- 821
VI++F +K L E+E +++ ++ FS DDA+ G LF +TP P SPL Q E P
Sbjct: 758 VITHFTSKLQTLSEEEQLNLRKEIQSDFSLDDAHSLGGQLFTDTPGPSSPLNQTELPAFE 817
Query: 822 -----DVDEMKGVGP--------------------N*VEASQIVEHHFQPTAQM------ 850
D+ +G+ P + + ++++E + Q+
Sbjct: 818 EVELSDIAAFEGISPGASGSQSGHRTSLSTNTNPVDVLSVNELLESVSETARQVASLPVS 877
Query: 851 -----F*VLINSWNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEEVSRQPVKALEYS- 904
+ ++N L TGKQQKM +RSFK Q TKAI S +NE+ + +K E +
Sbjct: 878 SIPVPYDQMMNQCEALVTGKQQKMSVLRSFK-PQATKAIT-SEDNEKDEQYLLKETEEAG 935
Query: 905 KGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
+ D K + Q Q Q+ F SQ+ Q+S RLPPSSPYDKFLKAAG
Sbjct: 936 EDDEKAIIVADVQPQGQLGFFSQEV-PQNSFRLPPSSPYDKFLKAAG 981
>At5g21080 putative protein
Length = 980
Score = 599 bits (1545), Expect = e-171
Identities = 367/829 (44%), Positives = 506/829 (60%), Gaps = 58/829 (6%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGV+SR V P C +LC FCP+LRARSR PVKRYK L+A+I PR++ + NDRKIGKLCEY
Sbjct: 1 MGVVSRTVFPVCESLCCFCPALRARSRHPVKRYKHLLADIFPRSQDEQPNDRKIGKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
A+KNPLRIPKIT +LEQRCYK+LR E F SVK+++ IY+KLL SC EQ+ LFASS LG+I
Sbjct: 61 AAKNPLRIPKITTSLEQRCYKELRMEQFHSVKIVMSIYKKLLVSCNEQMLLFASSYLGLI 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
LL+QTR DE+RILGC L DF+ Q +GTYMFNL+G IPK+C LA E+G+++ L
Sbjct: 121 HILLDQTRYDEMRILGCEALYDFVTSQAEGTYMFNLDGLIPKICPLAHELGEEDSTTNLC 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQ 240
+AGLQ LSS+V FMGE SH+S++FD ++S +LENY S+S+ + V + N +
Sbjct: 181 AAGLQALSSLVWFMGEFSHISVEFDNVVSVVLENYGG-HSQSSTSAVNQDNKVASIDKEL 239
Query: 241 EFPKEEAHVSSMLNVAT--GFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEP 298
+ E ++S + G I + + AKNP +WS+VCL+N+AKLAKEATTVRRVLE
Sbjct: 240 SPAEAETRIASWTRIVDDRGKAIGNNREDAKNPKFWSRVCLHNLAKLAKEATTVRRVLES 299
Query: 299 LFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQI 358
LF YFD WS+E G+A VL +Q L+ SG+ L HLDHKNV K+P +Q+
Sbjct: 300 LFRYFDFNEVWSTENGLAVYVLQDVQLLIERSGSYC------LRLHLDHKNVLKKPRMQL 353
Query: 359 DIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSI 418
+I+ + T +AQ K SVA+IGA+SD+I+HLR+ + S + +++GN+ N K ++ +
Sbjct: 354 EIVYVATALAQQTKVLPSVAIIGALSDMIRHLRKSIHCSLDDSNLGNEMIQYNLKFEAVV 413
Query: 419 EMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNK 478
E C+LQLS KVGDAGPI D+MAV+LE++S+ T++ART I+A
Sbjct: 414 EQCLLQLSQKVGDAGPILDIMAVMLESMSNITVMARTLIAA------------------- 454
Query: 479 AFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVS---------PWLDQKKISKKV 529
AFPDALFHQLL AM D E+++GAH I S+VL+PS VS P Q+ +S+ V
Sbjct: 455 AFPDALFHQLLQAMVCADHESRMGAHRIFSVVLVPSSVSPSSVLNSRRPADMQRTLSRTV 514
Query: 530 E---SDGLSIQHESLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQLVKCCYKQ 586
S + L ++ ++ E+ S +S + + +F ++ K
Sbjct: 515 SVFSSSAALFRKLKLESDNSVDDTAKMERV---STLSRSTSKFIRGESFDDEEPKNNTSS 571
Query: 587 FLTKYWISEIFRI*DLLG*AVTK*VSCFLQSGFRQHLLKMALQIMRQWHTLIALLYYLLV 646
L++ + + + + V+ SG ++ M Q + IA + L++
Sbjct: 572 VLSR--LKSSYSRSQSVKRNPSSMVADQNSSGSSPEKPSLSPHNMPQNYEAIANTFSLVL 629
Query: 647 L-------SYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFS 699
L S LV FQLAFSLR++SL G L PSRRRSL TLA+ M+IFSA+A +
Sbjct: 630 LFGRTKHSSNEVLVWSFQLAFSLRNLSLG--GPLQPSRRRSLFTLATSMIIFSAKAFNIP 687
Query: 700 DLIPKVKASLTEAPVDPFLELVDDNLLRAVCI-KSDKVV--FGSVEDEVAAMKSLSAV-Q 755
L+ K SL E VDPFL+LV+D L AV ++D+ +GS ED+ A +SL + +
Sbjct: 688 PLVNSAKTSLQEKTVDPFLQLVEDCKLDAVFYGQADQPAKNYGSKEDDDDASRSLVTIEE 747
Query: 756 LDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPL 804
Q +E S M KL + E S+IK QL+ F P D P G L
Sbjct: 748 ASQNQSREHYASMIMKFLGKLSDQESSAIKEQLVSDFIPIDGCPVGTQL 796
>At5g26850 unknown protein
Length = 970
Score = 438 bits (1126), Expect = e-123
Identities = 317/1029 (30%), Positives = 508/1029 (48%), Gaps = 140/1029 (13%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MG +SR V PAC ++CI CP+LR+RSRQPVKRYKKL+ EI P++ N+RKI KLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
A+KNP+RIPKI + LE+RCYKDLR+E + ++ Y K+L C++Q+ FA+SLL ++
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
LL+ ++ D ILGC TL FI Q DGTY ++E F K+C LA+E G++ + LR
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQCLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQ 240
++GLQ LS+MV +MGE SH+ D+I+ + D + ++ E + + + +
Sbjct: 181 ASGLQCLSAMVWYMGEFSHIFATVDEIV----QTNEDREEQNCNWVNEVIRCEGRGTTIC 236
Query: 241 EFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLF 300
P +++ A + + P W+++CL + LAKE+TT+R++L+P+F
Sbjct: 237 NSP---SYMIVRPRTARKDPTLLTKEETEMPKVWAQICLQRMVDLAKESTTLRQILDPMF 293
Query: 301 HYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDI 360
YF++ W+ G+A VL +L+ SG + L+LS +V+HLD+K+VA P L+ I
Sbjct: 294 SYFNSRRQWTPPNGLAMIVLSDAVYLMETSG-SQQLVLSTVVRHLDNKHVANDPELKAYI 352
Query: 361 INITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIEM 420
I + +A+ ++ + + I ++DL +HLR+ Q A A IG++ LN +Q+SIE
Sbjct: 353 IQVAGCLAKLIRTSSYLRDISFVNDLCRHLRKSFQ--ATARSIGDEELNLNVMIQNSIED 410
Query: 421 CILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITS-VPNVLYHNKA 479
C+ +++ + + P+FD+MAV +E + SS IV+R + ++ A ++S + + +
Sbjct: 411 CLREIAKGIVNTQPLFDMMAVSVEGLPSSGIVSRAAVGSLLILAHAMSSALSPSMRSQQV 470
Query: 480 FPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPS------------------------- 514
FPD L LL AM HP+ ET++GAH I S++L+ S
Sbjct: 471 FPDTLLDALLKAMLHPNVETRVGAHEIFSVILLQSSGQSQAGLASVRASGYLNESRNWRS 530
Query: 515 -------VVSPWLDQKKISK---KVESDGLSIQHESLSGEDPLNGKPVEEKPIHHSLISM 564
V+ LD+ + K K+E +G + HE L K + P H L S+
Sbjct: 531 DTTSAFTSVTARLDKLRKEKDGVKIEKNGYNNTHEDL--------KNYKSSPKFHKLNSI 582
Query: 565 I-----LVELLCWWAFSNQLVKCCYKQFLTKYWISEIFRI*DLLG*AVTK*VSCFLQSGF 619
I + L + + Q L+ +WI
Sbjct: 583 IDRTAGFINLADMLPSMMKFTEDQIGQLLSAFWIQSAL---------------------- 620
Query: 620 RQHLLKMALQIMRQWHTLIALLYYLLVLSYMALVRCFQLAFSLRSISLD-QEGGLPPSRR 678
+L ++ + +L+ L L +VR FQL FSLR++SLD G LP +
Sbjct: 621 -PDILPSNIEAIAHSFSLVLLSLRLKNPDDGLVVRAFQLLFSLRTLSLDLNNGTLPSVCK 679
Query: 679 RSLLTLASHMLIFSARAADFSDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSDKVVF 738
R +L L++ ML+F+A+ + +KA L VDP+L + D+L V +++ F
Sbjct: 680 RLILALSTSMLMFAAKIYQIPHICEMLKAQL-PGDVDPYL-FIGDDLQLHVRPQANMKDF 737
Query: 739 GSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTK-FSKLPEDELSSIKNQLLQGFSPDDA 797
GS D A L ++ +L T+I+ + K KL + E + +K Q+L+ F+PDDA
Sbjct: 738 GSSSDSQMATSMLFEMR-SKVELSNTIITDIVAKNLPKLSKLEEADVKMQILEQFTPDDA 796
Query: 798 YPSGPPLFMETPRPGSPLAQ------------------------IEFPDVDEMKGVGPN* 833
+ G +E P+P +++ + FP P
Sbjct: 797 FMFGSRPNIE-PQPNQSISKESLSFDEDIPAGSMVEDEVTSELSVRFPPRGSPSPSIPQV 855
Query: 834 VEASQIVEHHFQPTAQM-----------F*VLINSWNRLETGKQQKMLTIRSFKNQQETK 882
+ Q++E + Q+ + + N TG ++K+ + +N+Q
Sbjct: 856 ISIGQLMESALEVAGQVVGSSVSTSPLPYDTMTNRCETFGTGTREKLSRWLATENRQMNG 915
Query: 883 AIVLSSENEEVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSP 942
S E ALE D + +E QD +RLPP+SP
Sbjct: 916 LYGNSLEES-------SALEKVVEDGNIYGRESGMLQD----------SWSMMRLPPASP 958
Query: 943 YDKFLKAAG 951
+D FLKAAG
Sbjct: 959 FDNFLKAAG 967
>At2g41830 hypothetical protein
Length = 961
Score = 344 bits (882), Expect = 2e-94
Identities = 202/523 (38%), Positives = 298/523 (56%), Gaps = 105/523 (20%)
Query: 2 GVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEYA 61
GV+SR+V+P CG+LCI CP+LRARSRQPVKRYKKLIAEI PRN+ +NDRKIGKLCEYA
Sbjct: 6 GVISRQVLPVCGSLCILCPALRARSRQPVKRYKKLIAEIFPRNQEEGINDRKIGKLCEYA 65
Query: 62 SKNPLRIPK---ITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLG 118
+KN +R+PK I+++LE RCYK+LRNE+F S K+ +CIYR+LL +C+EQIPLF+S L
Sbjct: 66 AKNAVRMPKFFQISDSLEHRCYKELRNENFHSAKIAMCIYRRLLVTCKEQIPLFSSGFLR 125
Query: 119 IIRTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALL 178
++ LL+QTR DE++I+GC +L +F+I Q LCQL E GDD+R+
Sbjct: 126 TVQALLDQTRQDEMQIVGCQSLFEFVINQ---------------LCQLGLEGGDDDRSRS 170
Query: 179 LRSAGLQTLSSMVKFMGEHSHLSMDFDKIISAI-LENYVDLQSKSNLA-KVEKLNSQSQN 236
LR+AGLQ LS+M+ MGE+SH+ +FD ++ + Y DL S + V N QS+
Sbjct: 171 LRAAGLQALSAMIWLMGEYSHIPSEFDNVLLGFRVPIYSDLLSFIEINWFVVSNNRQSEK 230
Query: 237 QLVQEFPKEEAHVSSML-NVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRV 295
V + VS++L N + + D+ + W L N +A E + +
Sbjct: 231 HSV-----GDKVVSAVLENYGHPKILTNANDSGRK---WVDEVLKNEGHVAYEDSLI--- 279
Query: 296 LEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPI 355
W + ++ + G +H +LS+L+KHLDHK+V K P
Sbjct: 280 ---------NVPSWRT--------------VVNDKGQRTHFLLSMLIKHLDHKSVLKHPS 316
Query: 356 LQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQ 415
+Q++I+ +T+ +++ K + S ++ AISD+++HLR+C+ +S + ++G DA +
Sbjct: 317 MQLNILEVTSSLSETAKVEHSATIVSAISDIMRHLRKCMHSSLDEANLGTDAANCIRMVS 376
Query: 416 SSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLY 475
+++ C++QL+ K
Sbjct: 377 VAVDKCLVQLTKK----------------------------------------------- 389
Query: 476 HNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSP 518
AFP+ALFHQLL AM HPD +T+IGAH I S+VL+P+ V P
Sbjct: 390 ---AFPEALFHQLLQAMVHPDHKTRIGAHRIFSVVLVPTSVCP 429
Score = 131 bits (330), Expect = 2e-30
Identities = 107/364 (29%), Positives = 172/364 (46%), Gaps = 51/364 (14%)
Query: 635 HTLIALLYYLLVLSYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSAR 694
++L+ L + S+ AL+R FQ+A SLR ISL + G LPPSRRRSL TLA+ M++FS++
Sbjct: 601 YSLVLLFSRVKNSSHDALIRSFQMALSLRDISLMEGGPLPPSRRRSLFTLAASMVLFSSK 660
Query: 695 AADFSDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAV 754
A + L K +L +DPFL LVDD+ L+AV KV +G +D+ +A+ +LS +
Sbjct: 661 AFNLFSLADFTKVTLQGPRLDPFLNLVDDHKLKAVNSDQLKVAYGCEKDDASALDTLSNI 720
Query: 755 QLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSP 814
L + T++ + + E+ ++ QLL F PDDA P G +T +
Sbjct: 721 ALSTEHSRGTLVYEIVKSLEDMCNSEMDKMREQLLTEFMPDDACPLGTRFLEDTHK---- 776
Query: 815 LAQIEFPDV--------DEMKGVG---------------PN*VEASQIVEHHFQPTAQMF 851
QI+ DV D+ G G P+ + +QI+E + T Q+
Sbjct: 777 TYQIDSGDVKPRKEDAEDQEFGDGTETVTKNNHVTFSEIPDLLTVNQILESVVETTRQVG 836
Query: 852 *VLINS------------WNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEE----VSR 895
+ ++ L GKQQK+ ++ + + + E+ +++E S
Sbjct: 837 RISFHTAADASYKEMTLHCENLLMGKQQKISSLLNSQLRHESSVNCSPRQHDEEIKIASF 896
Query: 896 QPVKALEYSKGDLKLVTQEQFQAQD--------QIRFRSQDTRKQHSLRLPPSSPYDKFL 947
P+ + G + ++F + Q ++ + RLP SSPYD FL
Sbjct: 897 HPMINSAFHTGVEVPLLSKEFDMKSPRTPVGTIQSPCYAELQNNPQAFRLPASSPYDNFL 956
Query: 948 KAAG 951
KAAG
Sbjct: 957 KAAG 960
>At1g18090 unknown protein
Length = 577
Score = 36.6 bits (83), Expect = 0.069
Identities = 72/302 (23%), Positives = 123/302 (39%), Gaps = 41/302 (13%)
Query: 657 QLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFS-DLIPKVKASLTE---A 712
QLA+ L S+ L+Q GG+ +++T S +L + +A F D K + + +
Sbjct: 157 QLAY-LSSLELEQ-GGIA-----AVITEDSDLLAYGCKAVIFKMDRYGKGEELVLDNVFQ 209
Query: 713 PVD--PFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFM 770
VD P + D L A+C+ + SV S + + Q E V+S+
Sbjct: 210 AVDQKPSFQNFDQELFTAMCVLAGCDFLPSVP---GVGISRAHAFISKYQSVELVLSFLK 266
Query: 771 TKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGS------PLAQIEFPDVD 824
TK KL D+ SS + + F Y +P S P+ Q+EF D
Sbjct: 267 TKKGKLVPDDYSSSFTEAVSVFQHARVYDFDAKKLKHL-KPLSHNLLNLPVEQLEFLGPD 325
Query: 825 EMKGVGPN*VEASQIVEHHFQPTAQMF*VLINSWNRLETGKQQKMLTIRSFKNQQETKAI 884
V A I E + P + + ++N ++ +RSFK Q++ +
Sbjct: 326 LSPSV------AVAIAEGNVDP------ITMKAFNHFSVSRKPLKTPVRSFKEQEKGSSF 373
Query: 885 VLS--SENEEVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFR--SQDTRKQHSLRLPPS 940
++ S++EE+ A E +V ++ + QD ++ +Q R Q R PS
Sbjct: 374 LVCSLSKSEEIIELKRTADEAMIDPEAIVKKKMYSKQDSDLYKLLAQPNRDQVVTR--PS 431
Query: 941 SP 942
+P
Sbjct: 432 NP 433
>At4g29600 cytidine deaminase 7
Length = 307
Score = 32.7 bits (73), Expect = 1.00
Identities = 24/80 (30%), Positives = 34/80 (42%), Gaps = 9/80 (11%)
Query: 728 AVCIKSDKVVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTKFSKLPEDELSSIKNQ 787
AV I SD + FG+ ++ +LD + + K E SS+K
Sbjct: 97 AVAISSDCIEFGAPCGNCRQFLMETSNELDIK---------ILLKSKHEAEGSFSSLKLL 147
Query: 788 LLQGFSPDDAYPSGPPLFME 807
L F+PDD P G PL +E
Sbjct: 148 LPYRFTPDDVLPKGSPLLLE 167
>At4g27640 putative protein
Length = 651
Score = 32.7 bits (73), Expect = 1.00
Identities = 32/156 (20%), Positives = 68/156 (43%), Gaps = 9/156 (5%)
Query: 92 KVILCIYRKLLSSCREQIPLFASSLLGIIRTLLEQTRADEVRILGCNTLVDFIIFQTDGT 151
+V L ++ L + + + L ++ ++ + VR+ + F+ F DG
Sbjct: 136 EVALILFSSLTETIGNTFRPYFADLQALLLKCMQDESSSRVRVAALKAVGSFLEFTNDGD 195
Query: 152 YMFNLEGFIPKLCQLAQEV---GDDERALLLRSAGLQTLSSMVKFMGEHSHLSMDFDKII 208
+ FIP + ++++ G+++ A+L + + S +G+ + F +
Sbjct: 196 EVVKFRDFIPSILDVSRKCIASGEEDVAILAFEIFDELIESPAPLLGDSVKAIVQFSLEV 255
Query: 209 SA--ILEN---YVDLQSKSNLAKVEKLNSQSQNQLV 239
S LE+ + +Q S LAK K NS +++LV
Sbjct: 256 SCNQNLESSTRHQAIQIVSWLAKY-KYNSLKKHKLV 290
>At3g15530 unknown protein
Length = 288
Score = 31.6 bits (70), Expect = 2.2
Identities = 22/78 (28%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Query: 627 ALQIMRQWHTLIALL---YYLLVLSYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLT 683
A+ + QW TL+ LL ++ +LS + L+ Q+ F L S L L ++
Sbjct: 15 AIYGIEQWQTLVFLLFHAFFFSLLSLLFLIYFDQICFFLDSFFLSGAARLAAGFTGAVTA 74
Query: 684 LASHMLIFSARAADFSDL 701
L++ L+F+A +SD+
Sbjct: 75 LSAVCLLFAAANFVYSDV 92
>At1g12730 unknown protein
Length = 474
Score = 30.8 bits (68), Expect = 3.8
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 8/61 (13%)
Query: 620 RQHLLKMALQIMRQWHTLIALLYYLLVLSYMALVRCFQLAFSLRSISLDQEGGLPPSRRR 679
+Q L Q+ W T++ ++++LV S LV C S+SL Q GGL +R
Sbjct: 252 KQEKLTPTTQLPFSWRTVLHFVFWVLVWSSYVLVLC--------SLSLKQYGGLEEMFKR 303
Query: 680 S 680
+
Sbjct: 304 T 304
>At1g02890 hypothetical protein
Length = 1240
Score = 30.8 bits (68), Expect = 3.8
Identities = 20/54 (37%), Positives = 25/54 (46%), Gaps = 3/54 (5%)
Query: 893 VSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKF 946
+S Q V+ E S K T F+A D++RF T SLR PP P F
Sbjct: 575 LSSQAVRRQEVSTATSKSYT---FKAGDRVRFLGPSTSSLASLRAPPRGPATGF 625
>At5g52270 putative protein
Length = 214
Score = 30.4 bits (67), Expect = 4.9
Identities = 22/80 (27%), Positives = 38/80 (47%), Gaps = 10/80 (12%)
Query: 162 KLCQLAQEVGDDERALLLRSAGLQTLSSMVKFMGEHSHLS--------MDFDKIISAILE 213
K+C +A R LL LQ L+ + + E + + + F KII I +
Sbjct: 71 KICYIALSDSSYPRKLLFNY--LQNLNKELDKLDEKALIQKISKPYSFIRFGKIIGRIRK 128
Query: 214 NYVDLQSKSNLAKVEKLNSQ 233
Y+D ++++NL+K+ L Q
Sbjct: 129 QYIDTRTQANLSKLNALRKQ 148
>At4g26540 receptor protein kinase like protein
Length = 926
Score = 30.4 bits (67), Expect = 4.9
Identities = 26/96 (27%), Positives = 43/96 (44%), Gaps = 19/96 (19%)
Query: 667 LDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKVKASLTEAPVDPFLELVDDNLL 726
+D +G LP + RSL +L S + + + + +IPK TE LEL+D
Sbjct: 78 MDLQGSLPVTSLRSLKSLTS----LTLSSLNLTGVIPKEIGDFTE------LELLD---- 123
Query: 727 RAVCIKSDKVVFGSVEDEVAAMKSLSAVQLDDRQLK 762
SD + G + E+ +K L + L+ L+
Sbjct: 124 -----LSDNSLSGDIPVEIFRLKKLKTLSLNTNNLE 154
>At3g58370 putative protein
Length = 219
Score = 30.4 bits (67), Expect = 4.9
Identities = 37/116 (31%), Positives = 55/116 (46%), Gaps = 18/116 (15%)
Query: 151 TYMFNLEGFIPKLCQLAQEVGDDERALLLRSAGLQTLSSMVKFMGEHSHLSMDF-DKIIS 209
TYM L G I LCQL +E+ DD+ L S V ++ E+ L +D+ +K ++
Sbjct: 113 TYMTVLLGLIKTLCQLPEELTDDD---------LDEASVAVSYV-ENGGLRLDWLEKKLA 162
Query: 210 AILENYVDLQS-KSNLAKVE----KLNSQSQNQLVQEFPKEEAHVSSMLNVATGFE 260
+ +++ K+ L + E KLN Q +L KE A VS NV FE
Sbjct: 163 EVKAKKKKVETGKARLQRAEEELQKLN-QKCLELKAFLEKENADVSE-ANVPLSFE 216
>At3g01120 cystathionine gamma-synthase like protein
Length = 563
Score = 30.4 bits (67), Expect = 4.9
Identities = 14/34 (41%), Positives = 19/34 (55%)
Query: 704 KVKASLTEAPVDPFLELVDDNLLRAVCIKSDKVV 737
KV TE+P +PFL VD L+ +C K +V
Sbjct: 318 KVSLFFTESPTNPFLRCVDIELVSKICHKRGTLV 351
>At3g50170 putative protein
Length = 541
Score = 30.0 bits (66), Expect = 6.5
Identities = 15/66 (22%), Positives = 34/66 (50%), Gaps = 3/66 (4%)
Query: 223 NLAKVEKLNSQSQNQLVQEFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNI 282
N+ ++ +S+S ++V+E P+E S ++++ E + D + W K+C+Y +
Sbjct: 62 NVQNHKQSHSESGKEVVEERPEETTGDSWVISIRDKLE---QADRDDDTTIWGKLCIYRV 118
Query: 283 AKLAKE 288
+E
Sbjct: 119 PHYLQE 124
>At1g59265 polyprotein, putative
Length = 1466
Score = 30.0 bits (66), Expect = 6.5
Identities = 13/28 (46%), Positives = 16/28 (56%)
Query: 784 IKNQLLQGFSPDDAYPSGPPLFMETPRP 811
+ N LQG DD Y S PP F++ RP
Sbjct: 1066 VNNAFLQGTLTDDVYMSQPPGFIDKDRP 1093
>At1g58889 polyprotein, putative
Length = 1466
Score = 30.0 bits (66), Expect = 6.5
Identities = 13/28 (46%), Positives = 16/28 (56%)
Query: 784 IKNQLLQGFSPDDAYPSGPPLFMETPRP 811
+ N LQG DD Y S PP F++ RP
Sbjct: 1066 VNNAFLQGTLTDDVYMSQPPGFIDKDRP 1093
>At5g28740 putative protein
Length = 917
Score = 29.6 bits (65), Expect = 8.4
Identities = 22/80 (27%), Positives = 36/80 (44%), Gaps = 13/80 (16%)
Query: 772 KFSKLPEDELSSIKNQLLQGFSPDD---------AYPSGPPLFMETPRPGSPLA----QI 818
K + LPEDE+++++ QLL +P + + S + G P+ I
Sbjct: 794 KRAGLPEDEMAALERQLLSTTTPTEPAKDGGRRVGFVSAGVISQSGENEGKPVTGNGEDI 853
Query: 819 EFPDVDEMKGVGPN*VEASQ 838
E PD + + G + VE SQ
Sbjct: 854 ELPDESDDESDGDDHVEISQ 873
>At5g13100 unknown protein
Length = 354
Score = 29.6 bits (65), Expect = 8.4
Identities = 14/50 (28%), Positives = 25/50 (50%)
Query: 746 AAMKSLSAVQLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPD 795
A+ K++ A +D R L+ + F+ F E+E I + + +SPD
Sbjct: 117 ASQKAMEARGIDARDLRRRTVRRFLEDFRSASEEEKEVITDAIRHMYSPD 166
>At1g14650 splicing factor, putative
Length = 785
Score = 29.6 bits (65), Expect = 8.4
Identities = 20/84 (23%), Positives = 35/84 (40%), Gaps = 5/84 (5%)
Query: 737 VFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQG---FS 793
+FG+ E+EV+ + D Q K+ + + LS N QG +
Sbjct: 475 IFGTTEEEVSNAVKAEIEKKKDEQPKQVIWDGHTGSIGRTANQALSQNANGEEQGDGVYG 534
Query: 794 PDDAYPSGPPLFMETPRPGSPLAQ 817
+++P P + PRPG P+ +
Sbjct: 535 DPNSFPG--PAALPPPRPGVPIVR 556
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,081,740
Number of Sequences: 26719
Number of extensions: 765516
Number of successful extensions: 2940
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 2899
Number of HSP's gapped (non-prelim): 34
length of query: 951
length of database: 11,318,596
effective HSP length: 109
effective length of query: 842
effective length of database: 8,406,225
effective search space: 7078041450
effective search space used: 7078041450
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146574.12


