

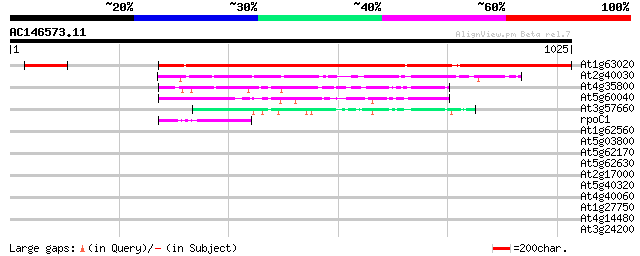
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146573.11 + phase: 2 /pseudo
(1025 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g63020 RNA polymerase IIA largest subunit, putative 811 0.0
At2g40030 putative DNA-directed RNA polymerase II subunit 205 1e-52
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 135 1e-31
At5g60040 DNA-directed RNA polymerase - like protein 110 3e-24
At3g57660 DNA-directed RNA polymerase I 190K chain - like protein 79 1e-14
rpoC1 -chloroplast genome- RNA polymerase beta' subunit-1 47 6e-05
At1g62560 similar to flavin-containing monooxygenase (sp|P36366)... 35 0.28
At5g03800 putative protein 33 0.83
At5g62170 unknown protein 32 1.4
At5g62630 Hedgehog-Interacting-Protein-like (HIPL2) 31 4.1
At2g17000 hypothetical protein 31 4.1
At5g40320 putative protein 30 7.0
At4g40060 homeodomain - like protein 30 7.0
At1g27750 unknown protein 30 7.0
At4g14480 kinase like protein 30 9.2
At3g24200 monooxygenase, putatve 30 9.2
>At1g63020 RNA polymerase IIA largest subunit, putative
Length = 1453
Score = 811 bits (2096), Expect = 0.0
Identities = 426/757 (56%), Positives = 556/757 (73%), Gaps = 13/757 (1%)
Query: 272 LELSEIGIPCQIAEALQVSEYVNRQNRQNLLYCCELRLLEKGQINVRRNGNPVVLYKKED 331
L+L+EIGIP IA+ LQVSE++N+ N++ L+ LL+ +++VRR G+ +V + D
Sbjct: 332 LKLNEIGIPESIAKRLQVSEHLNQCNKERLVTSFVPTLLDNKEMHVRR-GDRLVAIQVND 390
Query: 332 LQIGDIFYRPLVDGDKVLINRPPSIHQHSMIALTVRVLPISSVVSINPICCSPLRGDFDG 391
LQ GD +R L+DGD VL+NRPPSIHQHS+IA+TVR+LP +SVVS+NPICC P RGDFDG
Sbjct: 391 LQTGDKIFRSLMDGDTVLMNRPPSIHQHSLIAMTVRILPTTSVVSLNPICCLPFRGDFDG 450
Query: 392 DCLHGYIPQSVGARVELNELVALDRQLINGQSGGNLLSLSQDSLTAAHLL-LEDGVGFNI 450
DCLHGY+PQS+ A+VEL+ELVALD+QLIN Q+G NLLSL QDSLTAA+L+ +E N
Sbjct: 451 DCLHGYVPQSIQAKVELDELVALDKQLINRQNGRNLLSLGQDSLTAAYLVNVEKNCYLNR 510
Query: 451 YQVQQLEMLCNKELTSPAIFKA-PSSNTPFWSGKQLFSMLLPSKFDYAFPSNDVFVKDGE 509
Q+QQL+M C +L PAI KA PSS P W+G QLF ML P FDY +P N+V V +GE
Sbjct: 511 AQMQQLQMYCPFQLPPPAIIKASPSSTEPQWTGMQLFGMLFPPGFDYTYPLNNVVVSNGE 570
Query: 510 LISCSESSGWLRDSENNVFQSLVEHFQGKILDVLHGAQKALCEWLSMTGFSVSLSDLYLS 569
L+S SE S WLRD E N + L++H +GK+LD+++ AQ+ L +WL M G SVSL+DLYLS
Sbjct: 571 LLSFSEGSAWLRDGEGNFIERLLKHDKGKVLDIIYSAQEMLSQWLLMRGLSVSLADLYLS 630
Query: 570 SDSYARKNMMEEISYGLRVAEQACDFNQLLVDHYSDFLSASPEENEKFVTVDVDRLNHER 629
SD +RKN+ EEISYGLR AEQ C+ QL+V+ + DFL+ + E+ E+ D+ R +ER
Sbjct: 631 SDLQSRKNLTEEISYGLREAEQVCNKQQLMVESWRDFLAVNGEDKEEDSVSDLARFCYER 690
Query: 630 QISAAQSQASVDAFRHVFRNIQSLADKHACKDNSFLAMFKAGSKGNLLKLVQHSMCLGLQ 689
Q SA S+ +V AF+ +R++Q+LA ++ + NSFL M KAGSKGN+ KLVQHSMC+GLQ
Sbjct: 691 QKSATLSELAVSAFKDAYRDVQALAYRYGDQSNSFLIMSKAGSKGNIGKLVQHSMCIGLQ 750
Query: 690 HSLVRLSYRIPRELSCAGWNSQKGLHSMEMSSDTLEPKQSYIAHAVVESSFLTGLNPLEC 749
+S V LS+ PREL+CA WN D+ +SY+ + V+E+SFLTGLNPLE
Sbjct: 751 NSAVSLSFGFPRELTCAAWNDPNSPLRGAKGKDS-TTTESYVPYGVIENSFLTGLNPLES 809
Query: 750 FVHSVTNRDSSFSDNADLPGTLTRRLMFFMRDMYDAYDGTVRNLYGNQLIQFSYDAEEDS 809
FVHSVT+RDSSFS NADLPGTL+RRLMFFMRD+Y AYDGTVRN +GNQL+QF+Y+ +
Sbjct: 810 FVHSVTSRDSSFSGNADLPGTLSRRLMFFMRDIYAAYDGTVRNSFGNQLVQFTYETDGPV 869
Query: 810 SCDSYFREDIIGGEPVGALSACAVSEAAYSALSQPISLLEASPLLNLKNVLECGSRKKGG 869
EDI GE +G+LSACA+SEAAYSAL QPISLLE SPLLNLKNVLECGS+K
Sbjct: 870 -------EDIT-GEALGSLSACALSEAAYSALDQPISLLETSPLLNLKNVLECGSKKGQR 921
Query: 870 DQTVSLFLSEKIGKQRNGFEYAALEVKNYLERYMFSNIVSTVMIIFTPQHCSQVKYSPWV 929
+QT+SL+LSE + K+++GFEY +LE+KN+LE+ FS IVST MIIF+P ++V SPWV
Sbjct: 922 EQTMSLYLSEYLSKKKHGFEYGSLEIKNHLEKLSFSEIVSTSMIIFSPSSNTKVPLSPWV 981
Query: 930 CHFHLDKENVTRRKLEVHSIIDSLYQQYDSFRRESKVILPSLKISSNEGGETSVGNEKDG 989
CHFH+ ++ + R++L S++ SL +QY S RE K+ + L I + + KD
Sbjct: 982 CHFHISEKVLKRKQLSAESVVSSLNEQYKSRNRELKLDIVDLDIQNTNHCSSDDQAMKDD 1041
Query: 990 EDCITVAVVEDSKNSV-QLDSVRKSLIPFLLQTAIKG 1025
CITV VVE SK+SV +LD++R LIPFLL + +KG
Sbjct: 1042 NVCITVTVVEASKHSVLELDAIRLVLIPFLLDSPVKG 1078
Score = 107 bits (267), Expect = 3e-23
Identities = 47/79 (59%), Positives = 60/79 (75%)
Query: 27 QEKISALEINAPGQVTCSDLGLPNSSYGCTTCGSKDRKSCEGHFGAIKFPFTILHPYFMA 86
++K+S LE+ AP QVT S LGLPN C TCGSKDRK CEGHFG I F ++I++PYF+
Sbjct: 28 RDKMSVLEVEAPNQVTDSRLGLPNPDSVCRTCGSKDRKVCEGHFGVINFAYSIINPYFLK 87
Query: 87 EIAEILQKICPACKSIRQE 105
E+A +L KICP CK IR++
Sbjct: 88 EVAALLNKICPGCKYIRKK 106
>At2g40030 putative DNA-directed RNA polymerase II subunit
Length = 888
Score = 205 bits (521), Expect = 1e-52
Identities = 194/679 (28%), Positives = 307/679 (44%), Gaps = 94/679 (13%)
Query: 271 YLELSEIGIPCQIAEALQVSEYVNRQNRQNLLYCCELRLL----EKGQINVRRNGNPVVL 326
Y ++E+GIP +IA+ + E V+ NR L + +L + R+G+
Sbjct: 284 YRHVNEVGIPIEIAQRITFEERVSVHNRGYLQKLVDDKLCLSYTQGSTTYSLRDGSK--- 340
Query: 327 YKKEDLQIGDIFYRPLVDGDKVLINRPPSIHQHSMIALTVRVLPISSVVSINPICCSPLR 386
+L+ G + +R ++DGD V INRPP+ H+HS+ AL V V + V INP+ CSPL
Sbjct: 341 -GHTELKPGQVVHRRVMDGDVVFINRPPTTHKHSLQALRVYVHE-DNTVKINPLMCSPLS 398
Query: 387 GDFDGDCLHGYIPQSVGARVELNELVALDRQLINGQSGGNLLSLSQDSLTAAHLLLEDGV 446
DFDGDC+H + PQS+ A+ E+ EL ++++QL++ +G +L + DSL + ++LE V
Sbjct: 399 ADFDGDCVHLFYPQSLSAKAEVMELFSVEKQLLSSHTGQLILQMGSDSLLSLRVMLE-RV 457
Query: 447 GFNIYQVQQLEMLCNKELTSPAIFKAPSSNTPFWSGKQLFSMLLPSKFDYAFPSNDVFVK 506
+ QQL M + L PA+ K+ S + P W+ Q+ + P + D F+
Sbjct: 458 FLDKATAQQLAMYGSLSLPPPALRKS-SKSGPAWTVFQILQLAFPERLSC---KGDRFLV 513
Query: 507 DG-ELISCSESSGWLRDSENNVFQSL-VEHFQGKILDVLHGAQKALCEWLSMTGFSVSLS 564
DG +L+ + N + S+ +E + L Q L E L GFS+SL
Sbjct: 514 DGSDLLKFDFGVDAMGSIINEIVTSIFLEKGPKETLGFFDSLQPLLMESLFAEGFSLSLE 573
Query: 565 DLYLSSDSYARKNMMEEISYGLRVAEQACDFNQLLVDHYSDFLSASPEENEKFVTVDVDR 624
DL S +R +M ++ + L + E + ++L R
Sbjct: 574 DL-----SMSRADM--DVIHNLIIREISPMVSRL-------------------------R 601
Query: 625 LNHERQISAAQSQASVDAFRHVFRNIQSLADKHACKDNSFLAMFKAGSKGNLLKLVQHSM 684
L++ ++ S ++ +A K S + S + KLVQ +
Sbjct: 602 LSYRDELQLENS----------IHKVKEVAANFMLKSYSIRNLIDIKSNSAITKLVQQTG 651
Query: 685 CLGLQHSLVRLSYRIPRELSCAGWNSQKGLHSMEMSSDTLEPKQSYIAHAVVESSFLTGL 744
LGLQ S + Y A + +K + S D +V+ F GL
Sbjct: 652 FLGLQLSDKKKFYTKTLVEDMAIFCKRK-YGRISSSGD----------FGIVKGCFFHGL 700
Query: 745 NPLECFVHSVTNRDSSFSDNADL--PGTLTRRLMFFMRDMYDAYDGTVRNLYGNQLIQFS 802
+P E HS+ R+ + L PGTL + LM +RD+ DGTVRN N +IQF
Sbjct: 701 DPYEEMAHSIAAREVIVRSSRGLAEPGTLFKNLMAVLRDIVITNDGTVRNTCSNSVIQFK 760
Query: 803 YDAEEDSSCDSYFREDIIGGEPVGALSACAVSEAAYSALSQPISLLEASPLLN-----LK 857
Y + + F GEPVG L+A A+S AY A +L++SP N +K
Sbjct: 761 YGVDSERGHQGLFE----AGEPVGVLAATAMSNPAYKA------VLDSSPNSNSSWELMK 810
Query: 858 NVLEC--GSRKKGGDQTVSLFLSEKIGKQRNGFEYAALEVKNYLERYMFSNIVSTVMIIF 915
VL C + D+ V L+L+E +R E AA V+N L + + + F
Sbjct: 811 EVLLCKVNFQNTTNDRRVILYLNECHCGKRFCQENAACTVRNKLNKVSLKD----TAVEF 866
Query: 916 TPQHCSQVKYSPWVCHFHL 934
+HC ++ S VC ++
Sbjct: 867 LVEHCCKIGTS--VCRIYI 883
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 135 bits (340), Expect = 1e-31
Identities = 149/571 (26%), Positives = 253/571 (44%), Gaps = 92/571 (16%)
Query: 272 LELSEIGIPCQIAEALQVSEYVNRQNRQNLLYCCELRLLEKG----------QINVRRNG 321
+ + E+G+P IA L E V N + L L++ G + +R +G
Sbjct: 371 INIDELGVPWSIALNLTYPETVTPYNIERLK-----ELVDYGPHPPPGKTGAKYIIRDDG 425
Query: 322 NPVVL-YKKED----LQIGDIFYRPLVDGDKVLINRPPSIHQHSMIALTVRVLPISSVVS 376
+ L Y K+ L++G R L DGD VL NR PS+H+ S++ +R++P S+
Sbjct: 426 QRLDLRYLKKSSDQHLELGYKVERHLQDGDFVLFNRQPSLHKMSIMGHRIRIMPYSTF-R 484
Query: 377 INPICCSPLRGDFDGDCLHGYIPQSVGARVELNELVALDRQLINGQSGGNLLSLSQDSL- 435
+N SP DFDGD ++ ++PQS R E+ EL+ + + +++ Q+ ++ + QD+L
Sbjct: 485 LNLSVTSPYNADFDGDEMNMHVPQSFETRAEVLELMMVPKCIVSPQANRPVMGIVQDTLL 544
Query: 436 -----TAAHLLLEDGVGFNIYQVQQLEMLCNKELTSPAIFKAPSSNTPFWSGKQLFSMLL 490
T +E V N + + ++ +PAI K P W+GKQ+F++++
Sbjct: 545 GCRKITKRDTFIEKDVFMNTLMWWE---DFDGKVPAPAILKP----RPLWTGKQVFNLII 597
Query: 491 PSKFD--------------YAFPSN-DVFVKDGELISCSESSGWLRDSENNVFQSLVEHF 535
P + + + P + V ++ GEL++ + L S ++ + E
Sbjct: 598 PKQINLLRYSAWHADTETGFITPGDTQVRIERGELLAGTLCKKTLGTSNGSLVHVIWEEV 657
Query: 536 QGKILDVLHGAQKALCE-WLSMTGFSVSLSDLYLSSDSYARKNMMEEISYGLRVAEQACD 594
G + L WL GF++ + D+ A + ME+I+ + A+ A
Sbjct: 658 GPDAARKFLGHTQWLVNYWLLQNGFTIGI------GDTIADSSTMEKINETISNAKTA-- 709
Query: 595 FNQLLVDHYSDFLSASPEENEKFVTVDVDRLNHERQISAAQSQASVDAFRHVFRNIQSLA 654
L+ L P R E +++ ++A DA S A
Sbjct: 710 VKDLIRQFQGKELDPEPGRTM--------RDTFENRVNQVLNKARDDA--------GSSA 753
Query: 655 DKHACKDNSFLAMFKAGSKGNLLKLVQHSMCLGLQHSLVRLSYRIPRELSCAGWNSQKGL 714
K + N+ AM AGSKG+ + + Q + C+G Q+ + RIP G++ +
Sbjct: 754 QKSLAETNNLKAMVTAGSKGSFINISQMTACVGQQNVEGK---RIP-----FGFDGRTLP 805
Query: 715 HSMEMSSDTLEPKQSYIAHAVVESSFLTGLNPLECFVHSVTNRDSSFSD--NADLPGTLT 772
H + D P+ + VE+S+L GL P E F H++ R+ G +
Sbjct: 806 H---FTKDDYGPE----SRGFVENSYLRGLTPQEFFFHAMGGREGLIDTAVKTSETGYIQ 858
Query: 773 RRLMFFMRDMYDAYDGTVRNLYGNQLIQFSY 803
RRL+ M D+ YDGTVRN G+ +IQF Y
Sbjct: 859 RRLVKAMEDIMVKYDGTVRNSLGD-VIQFLY 888
Score = 35.0 bits (79), Expect = 0.22
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 1/64 (1%)
Query: 39 GQVTCSDLGLPNSSYGCTTCGSKDRKSCEGHFGAIKFPFTILHPYFMAEIAEILQKICPA 98
G ++ + LG + C TC + + C GHFG ++ + H FM + I++ +C
Sbjct: 50 GGLSDTRLGTIDRKVKCETCMA-NMAECPGHFGYLELAKPMYHVGFMKTVLSIMRCVCFN 108
Query: 99 CKSI 102
C I
Sbjct: 109 CSKI 112
Score = 33.9 bits (76), Expect = 0.49
Identities = 52/253 (20%), Positives = 103/253 (40%), Gaps = 62/253 (24%)
Query: 812 DSYFREDIIG-GEPVGALSACAVSEAA---------YSALSQPISLLEASPLLNLKNVLE 861
+S F + ++ GE +G ++A ++ E A Y+ +S L L + NV
Sbjct: 1071 ESRFLQSLVAPGEMIGCVAAQSIGEPATQMTLNTFHYAGVSAKNVTLGVPRLREIINVA- 1129
Query: 862 CGSRKKGGDQTVSLFLSEKIGKQRNGFEYAALEVKNYLERYMFSNIVSTVMIIFTPQHCS 921
K+ ++S++L+ + K + G A V+ LE ++ + + P S
Sbjct: 1130 ----KRIKTPSLSVYLTPEASKSKEG----AKTVQCALEYTTLRSVTQATEVWYDPDPMS 1181
Query: 922 QV----------------------KYSPWVCHFHLDKENVTRRKLEVHSIIDSLYQQYDS 959
+ K SPW+ L++E + +KL + I + + ++D
Sbjct: 1182 TIIEEDFEFVRSYYEMPDEDVSPDKISPWLLRIELNREMMVDKKLSMADIAEKINLEFDD 1241
Query: 960 -----FRRES--KVILPSLKISSNEGGETSVGNEKDGEDCITVAVVEDSK---------- 1002
F ++ K+IL ++I ++EG + + +E ED + + +E +
Sbjct: 1242 DLTCIFNDDNAQKLIL-RIRIMNDEGPKGELQDE-SAEDDVFLKKIESNMLTEMALRGIP 1299
Query: 1003 --NSVQLDSVRKS 1013
N V + VRKS
Sbjct: 1300 DINKVFIKQVRKS 1312
>At5g60040 DNA-directed RNA polymerase - like protein
Length = 1328
Score = 110 bits (276), Expect = 3e-24
Identities = 146/581 (25%), Positives = 238/581 (40%), Gaps = 126/581 (21%)
Query: 272 LELSEIGIPCQIAEALQVSEYVNRQNRQNLLYCCELRLLE-KGQINVRR-NGNPVVL--- 326
L+++E+GIP +A+ L E V+R N + L C + G NVR +G+ L
Sbjct: 376 LKITEVGIPILMAQILTFPECVSRHNIEKLRQCVRNGPNKYPGARNVRYPDGSSRTLVGD 435
Query: 327 YKK---EDLQIGDIFYRPLVDGDKVLINRPPSIHQHSMIALTVRVLPISSVVSINPICCS 383
Y+K ++L IG I R L +GD VL NR PS+H+ S++ R++P + + N C+
Sbjct: 436 YRKRIADELAIGCIVDRHLQEGDVVLFNRQPSLHRMSIMCHRARIMPWRT-LRFNESVCN 494
Query: 384 PLRGDFDGDCLHGYIPQSVGARVELNELVALDRQLINGQSGGNLLSLSQDSLTAAHLLLE 443
P DFDGD ++ ++PQ+ AR E L+ G + SL ++ +
Sbjct: 495 PYNADFDGDEMNMHVPQTEEARTEAITLM-----------GDTFYDRAAFSLICSY--MG 541
Query: 444 DGVGFNIYQVQQLEMLCNKELTSPAIFKAPSSNTPFWSGKQLFSMLLPSK---------- 493
DG+ + +L +P I K W+GKQ+FS+LL
Sbjct: 542 DGMD-------------SIDLPTPTILKP----IELWTGKQIFSVLLRPNASIRVYVTLN 584
Query: 494 ------------FDYAFPSND--VFVKDGELISCSESSGWL--------RDSENNVFQSL 531
FD ND V+ ++ ELIS L +++ ++ L
Sbjct: 585 VKEKNFKKGEHGFDETMCINDGWVYFRNSELISGQLGKATLALDIFPLGNGNKDGLYSIL 644
Query: 532 VEHFQGKILDV-LHGAQKALCEWLSMTGFSVSLSDLYLSSDSYARKNMMEEISYGLRVAE 590
+ + V ++ K W+ + GFS+ + D+ + K + I +G
Sbjct: 645 LRDYNSHAAAVCMNRLAKLSARWIGIHGFSIGIDDVQPGEE--LSKERKDSIQFG----- 697
Query: 591 QACDFNQLLVDHYSDFLSASPEENEKFVTVDVDRLNHERQISAAQS-QASVDAFRHVFRN 649
++Q + K + L + + A+S +A + + R
Sbjct: 698 ----YDQC---------------HRKIEEFNRGNLQLKAGLDGAKSLEAEITGILNTIRE 738
Query: 650 IQSLADKHACKD-----NSFLAMFKAGSKGNLLKLVQHSMCLGLQHSLVRLSYRIPRELS 704
A AC NS L M + GSKG+ + + Q C+G Q +R P
Sbjct: 739 ----ATGKACMSGLHWRNSPLIMSQCGSKGSPINISQMVACVGQQ---TVNGHRAP---- 787
Query: 705 CAGWNSQKGLHSMEMSSDTLEPKQSYIAHAVVESSFLTGLNPLECFVHSVTNRDSSFSDN 764
G+ + H MS +S A V +SF +GL E F H++ R+
Sbjct: 788 -DGFIDRSLPHFPRMS-------KSPAAKGFVANSFYSGLTATEFFFHTMGGREGLVDTA 839
Query: 765 ADLPGT--LTRRLMFFMRDMYDAYDGTVRNLYGNQLIQFSY 803
T ++RRLM + D+ YD TVRN G ++QF+Y
Sbjct: 840 VKTASTGYMSRRLMKALEDLLVHYDNTVRNASG-CILQFTY 879
Score = 38.9 bits (89), Expect = 0.015
Identities = 18/57 (31%), Positives = 31/57 (53%), Gaps = 1/57 (1%)
Query: 46 LGLPNSSYGCTTCGSKDRKSCEGHFGAIKFPFTILHPYFMAEIAEILQKICPACKSI 102
+G PN CTTC + ++C GH+G +K + + + I +IL+ IC C ++
Sbjct: 66 MGPPNKKSICTTCEG-NFQNCPGHYGYLKLDLPVYNVGYFNFILDILKCICKRCSNM 121
>At3g57660 DNA-directed RNA polymerase I 190K chain - like protein
Length = 1670
Score = 79.0 bits (193), Expect = 1e-14
Identities = 147/605 (24%), Positives = 234/605 (38%), Gaps = 130/605 (21%)
Query: 335 GDIFYRPLVDGDKVLINRPPSIHQHSMIALTVRVLPISSVVSINPICCSPLRGDFDGDCL 394
G +R + DGD VL+NR P++H+ S++A VRVL + ++ CS DFDGD +
Sbjct: 549 GKTVHRHMRDGDIVLVNRQPTLHKPSLMAHKVRVLKGEKTLRLHYANCSTYNADFDGDEM 608
Query: 395 HGYIPQSVGARVELNELVALDRQLINGQSGGNLLSLSQDSLTAAHLLLE-----DGVGFN 449
+ + PQ +R E +V + Q +G L +L QD + ++ LL + D FN
Sbjct: 609 NVHFPQDEISRAEAYNIVNANNQYARPSNGEPLRALIQDHIVSSVLLTKRDTFLDKDHFN 668
Query: 450 --IYQVQQLEML----------------CNKEL--TSPAIFKAPSSNTPFWSGKQLFSML 489
++ +M+ + EL +PAI K P W+GKQ+ + +
Sbjct: 669 QLLFSSGVTDMVLSTFSGRSGKKVMVSASDAELLTVTPAILKP----VPLWTGKQVITAV 724
Query: 490 ------------------LPSKFDYAFPSNDVFVKDGELISCSE-SSGWLRDSENNVFQS 530
LP F + S +V G+L E W ++ +
Sbjct: 725 LNQITKGHPPFTVEKATKLPVDF-FKCRSREVKPNSGDLTKKKEIDESWKQNLNEDKLHI 783
Query: 531 LVEHFQGKILD--------------VLHGAQKA----------LCEWLSMTGFSVSLSDL 566
F ++D L+G+ A +L GF+ + DL
Sbjct: 784 RKNEFVCGVIDKAQFADYGLVHTVHELYGSNAAGNLLSVFSRLFTVFLQTHGFTCGVDDL 843
Query: 567 YL--SSDSYARKNMMEEISYGLRVAEQACDFNQLLVDHYSDFLSASPEENEKFVTVDVDR 624
+ D K + E + G RV + + VD + P++ + ++R
Sbjct: 844 IILKDMDEERTKQLQECENVGERVLRKTFGID---VD-----VQIDPQD----MRSRIER 891
Query: 625 LNHERQISAAQSQASVDAFRHVFRNIQSLADKHACKD------------NSFLAMFKAGS 672
+ +E SA AS+D R + + + K D N M +G+
Sbjct: 892 ILYEDGESAL---ASLD--RSIVNYLNQCSSKGVMNDLLSDGLLKTPGRNCISLMTISGA 946
Query: 673 KGNLLKLVQHSMCLGLQHSLVRLSYRIPRELSCAGWNSQKGLHSMEMSSDTLEPKQSYIA 732
KG+ + Q S LG Q + R+PR +S + H + S P+ A
Sbjct: 947 KGSKVNFQQISSHLGQQDLEGK---RVPRMVS---GKTLPCFHPWDWS-----PR----A 991
Query: 733 HAVVESSFLTGLNPLECFVHSVTNRDS--SFSDNADLPGTLTRRLMFFMRDMYDAYDGTV 790
+ FL+GL P E + H + R+ + G L R LM + + YD TV
Sbjct: 992 GGFISDRFLSGLRPQEYYFHCMAGREGLVDTAVKTSRSGYLQRCLMKNLESLKVNYDCTV 1051
Query: 791 RNLYGNQLIQFSYDAE----EDSSCDSYFREDIIGGEPVGALSACAVSEAAYSALSQPIS 846
R+ G+ +IQF Y + SS F+E I + V L C SE S S IS
Sbjct: 1052 RDADGS-IIQFQYGEDGVDVHRSSFIEKFKELTINQDMV--LQKC--SEDMLSGASSYIS 1106
Query: 847 LLEAS 851
L S
Sbjct: 1107 DLPIS 1111
Score = 38.5 bits (88), Expect = 0.020
Identities = 20/62 (32%), Positives = 28/62 (44%), Gaps = 1/62 (1%)
Query: 38 PGQVTCSDLGLPNSSYGCTTCGSKDRKSCEGHFGAIKFPFTILHPYFMAEIAEILQKICP 97
PG + LG + C +CG + +C GH G I+ F I HP + LQ+ C
Sbjct: 62 PGGLYDLKLGPKDDKQACNSCGQL-KLACPGHCGHIELVFPIYHPLLFNLLFNFLQRACF 120
Query: 98 AC 99
C
Sbjct: 121 FC 122
>rpoC1 -chloroplast genome- RNA polymerase beta' subunit-1
Length = 680
Score = 47.0 bits (110), Expect = 6e-05
Identities = 45/170 (26%), Positives = 77/170 (44%), Gaps = 17/170 (10%)
Query: 272 LELSEIGIPCQIAEALQVSEYVNRQNRQNLLYCCELRLLEKGQINVRRNGNPVVLYKKED 331
L L G+P +IA L + + RQ+L + K QI R P+V E
Sbjct: 390 LSLHRCGLPREIAIELFQTFVIRGLIRQHLASNIGVA---KSQI---REKKPIVW---EI 440
Query: 332 LQIGDIFYRPLVDGDKVLINRPPSIHQHSMIALTVRVLPISSVVSINPICCSPLRGDFDG 391
LQ ++ G VL+NR P++H+ + + +L + ++P+ C DFDG
Sbjct: 441 LQ-------EVMQGHPVLLNRAPTLHRLGIQSFQP-ILVEGRTICLHPLVCKGFNADFDG 492
Query: 392 DCLHGYIPQSVGARVELNELVALDRQLINGQSGGNLLSLSQDSLTAAHLL 441
D + ++P S+ A+ E L+ L++ G + +QD L ++L
Sbjct: 493 DQMAVHVPLSLEAQAEARLLMFSHMNLLSPAIGDPISVPTQDMLIGLYVL 542
>At1g62560 similar to flavin-containing monooxygenase (sp|P36366);
similar to ESTs gb|R30018, gb|H36886, gb|N37822, and
gb|T88100
Length = 462
Score = 34.7 bits (78), Expect = 0.28
Identities = 49/214 (22%), Positives = 88/214 (40%), Gaps = 34/214 (15%)
Query: 317 VRRNGNPVVLYKKEDLQIGDIF-YRPLVDGDKVLINRPPSIHQHSMIALTVRVLPISSVV 375
+RR G+ VV++++E Q+G ++ Y P D D + ++ P HS I ++R +
Sbjct: 29 LRREGHSVVVFEREK-QVGGLWVYTPKSDSDPLSLD-PTRSKVHSSIYESLRTNVPRESM 86
Query: 376 SINPICCSPLRGDFDGDC--------LHGYIPQSVGARVELNELVALDRQLINGQ--SGG 425
+ P D D + YI Q ++ E++ + +++ + G
Sbjct: 87 GVRDFPFLPRFDDESRDARRYPNHREVLAYI-QDFAREFKIEEMIRFETEVVRVEPVDNG 145
Query: 426 NLLSLSQDSLTAAHLLLEDGVGFNIYQVQQLEMLCNKELTSPAIFKAPSSNTPFWSGKQL 485
N S++S GF ++ ++CN T P I P + W GKQ+
Sbjct: 146 NWRVQSKNS-----------GGFLEDEIYDAVVVCNGHYTEPNIAHIPGIKS--WPGKQI 192
Query: 486 FS--MLLPSKFDYAFPSNDVFVKDGELISCSESS 517
S +P F+ N+V V G S ++ S
Sbjct: 193 HSHNYRVPDPFE-----NEVVVVIGNFASGADIS 221
>At5g03800 putative protein
Length = 1388
Score = 33.1 bits (74), Expect = 0.83
Identities = 63/260 (24%), Positives = 101/260 (38%), Gaps = 35/260 (13%)
Query: 451 YQVQQLEMLCNKELTSPAIFKAPSSNTPFWSGKQLFSMLLPSKFDYAFPSNDVFVKDGEL 510
+++++ ++ E T AI A + F G Q+ +++ S F N VFV + +
Sbjct: 169 FRMRKAGLVQPNEYTFVAILTACVRVSRFSLGIQIHGLIVKSGF-----LNSVFVSNSLM 223
Query: 511 ISCSESSGWLRD---------------SENNVFQSLVEHFQGKILDVLHGAQKALCEWLS 555
+ SG D S N V SLV+ +GK H A E
Sbjct: 224 SLYDKDSGSSCDDVLKLFDEIPQRDVASWNTVVSSLVK--EGKS----HKAFDLFYEMNR 277
Query: 556 MTGFSV---SLSDLYLS-SDSYARKNMMEEISYGLRVA-EQACDFNQLLVDHYSDFLSAS 610
+ GF V +LS L S +DS E +R+ Q N L+ YS F
Sbjct: 278 VEGFGVDSFTLSTLLSSCTDSSVLLRGRELHGRAIRIGLMQELSVNNALIGFYSKFWDMK 337
Query: 611 PEENEKFVTVDVDRLNHERQISAAQSQASVDAFRHVFRNIQSLADKHACKDNSFLAMFKA 670
E+ + + D + I+A S VD+ +F N+ +K+ N+ +A F
Sbjct: 338 KVESLYEMMMAQDAVTFTEMITAYMSFGMVDSAVEIFANV---TEKNTITYNALMAGFCR 394
Query: 671 GSKG-NLLKLVQHSMCLGLQ 689
G LKL + G++
Sbjct: 395 NGHGLKALKLFTDMLQRGVE 414
>At5g62170 unknown protein
Length = 703
Score = 32.3 bits (72), Expect = 1.4
Identities = 21/95 (22%), Positives = 44/95 (46%), Gaps = 8/95 (8%)
Query: 901 RYMFSNIVSTVMIIFTPQHCSQVKYSPWVCHFHLDKENVTRRKLEVHSIIDSLYQQYDSF 960
R + ++V ++ H + PWVCH+ + ++R L+ +ID L ++ + F
Sbjct: 550 RKLLFHLVDEILADILKPH---INLKPWVCHYPIR----SQRNLKGSELIDELSRRIERF 602
Query: 961 RRESKVILPSL-KISSNEGGETSVGNEKDGEDCIT 994
++L + + + + E E+DGE +T
Sbjct: 603 PLAKCLVLEDIDALVAGDFPEIESAFEEDGEGIVT 637
>At5g62630 Hedgehog-Interacting-Protein-like (HIPL2)
Length = 696
Score = 30.8 bits (68), Expect = 4.1
Identities = 23/77 (29%), Positives = 36/77 (45%), Gaps = 7/77 (9%)
Query: 420 NGQSGGNLLSLSQDSLTAAHLLLEDGVGFNIYQVQQLEMLCNKELTSPAIFK------AP 473
+G + G + S QD+ HLL GV + I + + + C+KE T+ + K AP
Sbjct: 608 SGPALGYIYSFGQDNNKDIHLLTSSGV-YRIVRPSRCNLACSKENTTASAGKQNPAGSAP 666
Query: 474 SSNTPFWSGKQLFSMLL 490
P + K FS+ L
Sbjct: 667 PQPLPSSARKLCFSVFL 683
>At2g17000 hypothetical protein
Length = 849
Score = 30.8 bits (68), Expect = 4.1
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Query: 483 KQLFSMLLPSKFDYAFPSNDVFVKDGELISCSESSGWLRDSENNVFQSLVEHFQGKIL 540
KQ S + SK +Y +P DV VKD E ++ + WL N+ Q++ E F + L
Sbjct: 740 KQRISSYIDSKPEYWYPKADVIVKDVEDLNIVRIAIWLCHKINH--QNMGERFTRRAL 795
>At5g40320 putative protein
Length = 594
Score = 30.0 bits (66), Expect = 7.0
Identities = 25/115 (21%), Positives = 44/115 (37%), Gaps = 11/115 (9%)
Query: 53 YGCTTCGSKDRKSCEGHFGAIKFPFTILHPYFMAEIAEILQKICPACKSIRQEIRVKVCL 112
Y C TCG K C I P HP L + ++ + ++C
Sbjct: 80 YNCNTCGLTVHKECAESSPEINHPSHQRHP---------LMLLTHGLPQEAEDDKCRLCG 130
Query: 113 LPIVKVLFHAYSVLNHSLDTIISPKVANIAVGSLSCHE-IQSFIQ*FLQENCNHC 166
+ K+++H S+ + SLD + ++ V HE + + ++ CN C
Sbjct: 131 EKVGKLVYHC-SICDFSLDLFCARNPLSLVVYFRKGHEHTLTLMPRLIRFTCNAC 184
>At4g40060 homeodomain - like protein
Length = 294
Score = 30.0 bits (66), Expect = 7.0
Identities = 23/69 (33%), Positives = 34/69 (48%), Gaps = 7/69 (10%)
Query: 951 DSLYQQYDSFRRESKVILPSL-KISSNEGGETSVGNEKDGEDCITVAVVEDSKNSVQLDS 1009
DSL +DS RR++ +L + KI + GE N K IT V E+ + + DS
Sbjct: 128 DSLRHNFDSLRRDNDSLLQEISKIKAKVNGEEDNNNNK----AITEGVKEEEVH--KTDS 181
Query: 1010 VRKSLIPFL 1018
+ S + FL
Sbjct: 182 IPSSPLQFL 190
>At1g27750 unknown protein
Length = 1975
Score = 30.0 bits (66), Expect = 7.0
Identities = 30/114 (26%), Positives = 46/114 (40%), Gaps = 3/114 (2%)
Query: 353 PPSIHQHSMIALTVRVLPISSVVSINPICCSPLRGDFDGDCLHGYIPQSVGARVELNELV 412
P Q + + P S+ P+ S D + S G+RV + +
Sbjct: 1117 PKGHQQQKFVPKPMNPTPTSNSTPF-PVSLSSSLRQSDSSGASSRVSASGGSRVRIGDQG 1175
Query: 413 ALDRQLINGQSGGNLLS-LSQDSLTAAHLLLEDGVGFNIYQVQQLEMLCNKELT 465
L Q GG+ ++ L QD AA L +DG G + + Q + L N+ELT
Sbjct: 1176 QLVSSKSPAQGGGSFVNYLPQDEAVAAGLGPDDG-GLDPVESQGVVDLLNRELT 1228
>At4g14480 kinase like protein
Length = 487
Score = 29.6 bits (65), Expect = 9.2
Identities = 26/118 (22%), Positives = 52/118 (44%), Gaps = 12/118 (10%)
Query: 368 VLPISSVVSINPICCSPLRGDFDGDCLHGYIPQSVGARVELNELVALDRQLINGQSGGNL 427
V+P S S++ I S +C+ ++ +++ A L++ L R + GN+
Sbjct: 90 VMPFMSCGSLHSIVSSSFPSGLPENCISVFLKETLNAISYLHDQGHLHRDI----KAGNI 145
Query: 428 LSLSQDSLTAAHLLLEDGVGFNIYQVQQLEMLCNKELTSPAIFKAPSSNTPFWSGKQL 485
L S S+ L + GV +IY+ + + TS ++ + TP+W ++
Sbjct: 146 LVDSDGSVK----LADFGVSASIYE----PVTSSSGTTSSSLRLTDIAGTPYWMAPEV 195
>At3g24200 monooxygenase, putatve
Length = 509
Score = 29.6 bits (65), Expect = 9.2
Identities = 25/100 (25%), Positives = 50/100 (50%), Gaps = 8/100 (8%)
Query: 819 IIGGEPVG-ALSACAVSEAAYSALSQPISLLEASPLLNLKNVLECGSRKKGGDQTVSLFL 877
I+GG VG AL+A S+ L+ +++++ +PLL +N++E G + D VS
Sbjct: 58 IVGGGMVGIALAASLASKPLTKHLN--VAIIDNNPLLGRRNIIEKGHQP---DPRVSTVT 112
Query: 878 SEKIG--KQRNGFEYAALEVKNYLERYMFSNIVSTVMIIF 915
I K ++Y + Y ++ +I T+++++
Sbjct: 113 PATISFLKDIGAWKYIEQQRHAYFDKMQALDIQDTMLMMY 152
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,192,953
Number of Sequences: 26719
Number of extensions: 949195
Number of successful extensions: 3016
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 2970
Number of HSP's gapped (non-prelim): 33
length of query: 1025
length of database: 11,318,596
effective HSP length: 109
effective length of query: 916
effective length of database: 8,406,225
effective search space: 7700102100
effective search space used: 7700102100
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146573.11


