

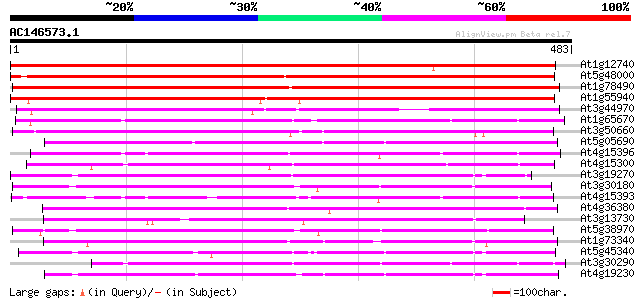
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146573.1 + phase: 0
(483 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g12740 hypothetical protein 731 0.0
At5g48000 cytochrome P450-like protein 387 e-108
At1g78490 unknown protein 380 e-106
At1g55940 cytochrome P450, putative 374 e-104
At3g44970 cytochrome P450 - like protein 305 4e-83
At1g65670 cytochrome P450, putative 298 4e-81
At3g50660 steroid 22-alpha-hydroxylase (DWF4) 295 3e-80
At5g05690 cytochrome P450 90A1 (sp|Q42569) 293 2e-79
At4g15396 unknown protein 289 2e-78
At4g15300 cytochrome P450 like protein 289 3e-78
At3g19270 cytochrome P450, putative 272 3e-73
At3g30180 cytochrome P450 homolog, putative 267 8e-72
At4g15393 putative protein 265 5e-71
At4g36380 cytochrome P450 (ROTUNDIFOLIA3) 259 3e-69
At3g13730 CYP90D 253 1e-67
At5g38970 cytochrome P450 - like protein 253 2e-67
At1g73340 247 9e-66
At5g45340 cytochrome P450 235 4e-62
At3g30290 cytochrome P450, putative 220 1e-57
At4g19230 cytochrome P450 protein like 220 2e-57
>At1g12740 hypothetical protein
Length = 478
Score = 731 bits (1888), Expect = 0.0
Identities = 351/476 (73%), Positives = 402/476 (83%), Gaps = 6/476 (1%)
Query: 1 MWVLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI 60
MW L + + + IT WVY WRNP C GKLPPGSMG PLLGES+QFF PN + DIPPFI
Sbjct: 1 MWALLIWVSLLLISITHWVYSWRNPKCRGKLPPGSMGFPLLGESIQFFKPNKTSDIPPFI 60
Query: 61 RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL 120
++R+K+YGPIFKTNLVGRPV+VSTD DL+YF+F QEG+ FQSWYPDTFT IFG++NVGSL
Sbjct: 61 KERVKKYGPIFKTNLVGRPVIVSTDADLSYFVFNQEGRCFQSWYPDTFTHIFGKKNVGSL 120
Query: 121 HGFMYKYLKNMMLNLFGPESLKKMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTA 180
HGFMYKYLKNM+L LFG + LKKM+ +VE A + L+ S QDSVELK+AT +MIFDLTA
Sbjct: 121 HGFMYKYLKNMVLTLFGHDGLKKMLPQVEMTANKRLELWSNQDSVELKDATASMIFDLTA 180
Query: 181 KKLISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERR 240
KKLIS+DP +SSENLR NFVAFIQGLISFP +IPGTAY+KCLQGR KAMKML+NMLQERR
Sbjct: 181 KKLISHDPDKSSENLRANFVAFIQGLISFPFDIPGTAYHKCLQGRAKAMKMLRNMLQERR 240
Query: 241 EMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNP 300
E PRK DFFDYVIEE++KEGT+LTE IALDLMFVLLFASFETTS ALT AIK LSD+P
Sbjct: 241 ENPRKNPSDFFDYVIEEIQKEGTILTEEIALDLMFVLLFASFETTSLALTLAIKFLSDDP 300
Query: 301 LVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREI 360
V K+L EEHE IL RE+ +SG+TW+EYKSMT+TFQ I ETARLANIVP IFRKALR+I
Sbjct: 301 EVLKRLTEEHETILRNREDADSGLTWEEYKSMTYTFQFINETARLANIVPAIFRKALRDI 360
Query: 361 NFK------GYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFG 414
FK YTIPAGWA+MVCPPAVHLNP Y+DPLVFNPSRWEG + + A+KHF+AFG
Sbjct: 361 KFKEFVNDTDYTIPAGWAVMVCPPAVHLNPEMYKDPLVFNPSRWEGSKVTNASKHFMAFG 420
Query: 415 GGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEK 470
GGMRFCVGT+F K+QMA FLH LVTKYRW IKGGNI RTPGLQFPNG+HV++ +K
Sbjct: 421 GGMRFCVGTDFTKLQMAAFLHSLVTKYRWEEIKGGNITRTPGLQFPNGYHVKLHKK 476
>At5g48000 cytochrome P450-like protein
Length = 477
Score = 387 bits (994), Expect = e-108
Identities = 196/472 (41%), Positives = 298/472 (62%), Gaps = 8/472 (1%)
Query: 1 MWVLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI 60
+WV+ + A+V I++W+YRW NP CNGKLPPGSMGLP++GE+ FF P+ +I PF+
Sbjct: 9 VWVIAVAAVV----ISKWLYRWSNPKCNGKLPPGSMGLPIIGETCDFFEPHGLYEISPFV 64
Query: 61 RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL 120
+KRM +YGP+F+TN+ G VV T+PD+ + +F+QE K F YP+ F + FG++NV
Sbjct: 65 KKRMLKYGPLFRTNIFGSNTVVLTEPDIIFEVFRQENKSFVFSYPEAFVKPFGKENVFLK 124
Query: 121 HGFMYKYLKNMMLNLFGPESL-KKMISEVEQAACRTLQQASCQDSVELKEATETMIFDLT 179
HG ++K++K + L G E+L KKMI E+++ L+ + Q S + KEA E++I
Sbjct: 125 HGNIHKHVKQISLQHLGSEALKKKMIGEIDRVTYEHLRSKANQGSFDAKEAVESVIMAHL 184
Query: 180 AKKLISYDPTESSENLRENFVAFIQGLISFPLNIPG-TAYNKCLQGRKKAMKMLKNMLQE 238
K+IS E+ L +N +A PL + + K R+ A++++K++
Sbjct: 185 TPKIISNLKPETQATLVDNIMALGSEWFQSPLKLTTLISIYKVFIARRYALQVIKDVF-T 243
Query: 239 RREMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSD 298
RR+ R+ DF D ++EE KE + E A++L+F +L + E+TS + AIK L++
Sbjct: 244 RRKASREMCGDFLDTMVEEGEKEDVIFNEESAINLIFAILVVAKESTSSVTSLAIKFLAE 303
Query: 299 NPLVFKQLQEEHEAILERRENPNSGVTWQEYK-SMTFTFQLITETARLANIVPGIFRKAL 357
N +L+ EH AIL+ R +GV+W+EY+ MTFT +I ET R+AN+ P ++RKA+
Sbjct: 304 NHKALAELKREHAAILQNRNGKGAGVSWEEYRHQMTFTNMVINETLRMANMAPIMYRKAV 363
Query: 358 REINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGM 417
++ KGYTIPAGW + V PPAVH N A Y++PL FNP RWEG E +K F+ FGGG+
Sbjct: 364 NDVEIKGYTIPAGWIVAVIPPAVHFNDAIYENPLEFNPWRWEGKELRSGSKTFMVFGGGV 423
Query: 418 RFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITE 469
R CVG EFA++Q+++F+H LVT Y + + +R P FP G ++I++
Sbjct: 424 RQCVGAEFARLQISIFIHHLVTTYDFSLAQESEFIRAPLPYFPKGLPIKISQ 475
>At1g78490 unknown protein
Length = 479
Score = 380 bits (977), Expect = e-106
Identities = 193/474 (40%), Positives = 298/474 (62%), Gaps = 4/474 (0%)
Query: 3 VLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRK 62
V L + + I+ W+YRW NP C GKLPPGSMG P++GE+L FF P IP F++K
Sbjct: 7 VAMLMVALVVVRISHWLYRWSNPKCPGKLPPGSMGFPIIGETLDFFKPCGVEGIPTFVKK 66
Query: 63 RMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHG 122
RM RYGP+F+TN+ G VVSTDPD+ + IF+QE F+ YPD F ++FG+ N+
Sbjct: 67 RMIRYGPLFRTNIFGSKTVVSTDPDVIHQIFRQENTSFELGYPDIFVKVFGKDNLFLKEV 126
Query: 123 FMYKYLKNMMLNLFGPESLKK-MISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAK 181
F++KYL+ + + + G E LK+ M+ +++A ++ + Q S +++ E ++
Sbjct: 127 FIHKYLQKITMQILGSEGLKQTMLGNMDKATRDHIRSIASQGSFNVRKEVENLVVAYMTP 186
Query: 182 KLISYDPTESSENLRENFVAF-IQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERR 240
KLIS E+ L +N AF + SF A K L+ R++A++++K++L R+
Sbjct: 187 KLISNLKPETQSKLIDNLNAFNLDWFKSFLRLSTWKAVTKALKSREEAIQVMKDVLMMRK 246
Query: 241 EMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNP 300
E R++Q DF + ++EEL K+G+ + A++L+F+L FA E TS A+K +S +P
Sbjct: 247 E-TREKQEDFLNTLLEELEKDGSFFDQGSAINLIFLLAFALREGTSSCTALAVKFISKDP 305
Query: 301 LVFKQLQEEHEAILERRENPNSGVTWQEYK-SMTFTFQLITETARLANIVPGIFRKALRE 359
V +L+ EH+AI++ R++ +GV+W+EY+ +MTFT + E RLAN P +FRKA+++
Sbjct: 306 KVLAELKREHKAIVDNRKDKEAGVSWEEYRHNMTFTNMVSNEVLRLANTTPLLFRKAVQD 365
Query: 360 INFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMRF 419
+ KGYTIPAGW + V P AVH +PA Y++P FNP RWEG E +K F+AFG G+R
Sbjct: 366 VEIKGYTIPAGWIVAVAPSAVHFDPAIYENPFEFNPWRWEGKEMIWGSKTFMAFGYGVRL 425
Query: 420 CVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQK 473
CVG EF+++QMA+FLH LV Y + ++ I+R+P Q+ + I++ K
Sbjct: 426 CVGAEFSRLQMAIFLHHLVAYYDFSMVQDSEIIRSPFHQYTKDLLINISQSPTK 479
>At1g55940 cytochrome P450, putative
Length = 639
Score = 374 bits (960), Expect = e-104
Identities = 196/480 (40%), Positives = 292/480 (60%), Gaps = 12/480 (2%)
Query: 1 MWVLCLGALVTICI--ITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPP 58
MW + L ++ + + I+ W+YRW NP+C+GKLPPGSMG P++GE+++FF P + +I P
Sbjct: 148 MWGVALSFVIALVVVKISLWLYRWANPNCSGKLPPGSMGFPVIGETVEFFKPYSFNEIHP 207
Query: 59 FIRKRM-KRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNV 117
F++KRM K G +F+TN++G +VSTDP++N+ I +QE + F YP+ IFG+ N+
Sbjct: 208 FVKKRMFKHGGSLFRTNILGSKTIVSTDPEVNFEILKQENRCFIMSYPEALVRIFGKDNL 267
Query: 118 GSLHGF-MYKYLKNMMLNLFGPESLK-KMISEVEQAACRTLQQASCQDSVELKEATETMI 175
G ++Y++++ L L GPE LK + I +++ A L+ S Q V++K+ + +I
Sbjct: 268 FFKQGKDFHRYMRHIALQLLGPECLKQRFIQQIDIATSEHLKSVSFQGVVDVKDTSGRLI 327
Query: 176 FDLTAKKLISYDPTESSENLRENFVAFIQGLISFPLNIP--GTAYNKCLQGRKKAMKMLK 233
+ +IS E+ L E+F F L+ P + YN L R MKMLK
Sbjct: 328 LEQMILMIISNIKPETKSKLIESFRDFSFDLVRSPFDPSFWNALYNG-LMARSNVMKMLK 386
Query: 234 NMLQERREMPRKQQM---DFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALT 290
M +ERRE DF + +I E+ KEG + E +++L+ LL AS+ETTS
Sbjct: 387 RMFKERREEATSDDSKYGDFMETMIYEVEKEGDTINEERSVELILSLLIASYETTSTMTA 446
Query: 291 YAIKLLSDNPLVFKQLQEEHEAILERRENPNSGVTWQEYKSM-TFTFQLITETARLANIV 349
+K +++NP V +L+ EHE IL+ R + SGVTW+EY+SM FT +I E+ RL ++
Sbjct: 447 LTVKFIAENPKVLMELKREHETILQNRADKESGVTWKEYRSMMNFTHMVINESLRLGSLS 506
Query: 350 PGIFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKH 409
P +FRKA+ ++ KGYTIPAGW ++V P +H +P Y+ P FNP RWEG E +K
Sbjct: 507 PAMFRKAVNDVEIKGYTIPAGWIVLVVPSLLHYDPQIYEQPCEFNPWRWEGKELLSGSKT 566
Query: 410 FLAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITE 469
F+AFGGG R C G EFA++QMA+FLH LVT Y + I I+R P L+F + I+E
Sbjct: 567 FMAFGGGARLCAGAEFARLQMAIFLHHLVTTYDFSLIDKSYIIRAPLLRFSKPIRITISE 626
>At3g44970 cytochrome P450 - like protein
Length = 455
Score = 305 bits (781), Expect = 4e-83
Identities = 174/475 (36%), Positives = 263/475 (54%), Gaps = 35/475 (7%)
Query: 7 GALVTICIITR---WVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKR 63
G V + ++ R W Y+W NP NGKLPPGSMG P++GE+L FF P +I P+++K+
Sbjct: 8 GFCVIVLVVARVGHWWYQWSNPKSNGKLPPGSMGFPIIGETLDFFKPYGFYEISPYLKKK 67
Query: 64 MKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGF 123
M RYGP+F+TN++G VVSTD D+N I +QE K F YPD + G+ ++ G
Sbjct: 68 MLRYGPLFRTNILGVKTVVSTDKDVNMEILRQENKSFILSYPDGLMKPLGKDSLFLKIGN 127
Query: 124 MYKYLKNMMLNLFGPESLK-KMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAKK 182
++K++K + L+L E LK K++ ++++ L + +++K+A +I K
Sbjct: 128 IHKHIKQITLHLLSSEGLKRKILKDMDRVTREHLSSKAKTGRLDVKDAVSKLIIAHLTPK 187
Query: 183 LISYDPTESSENLRENFVAFIQGLI--SFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERR 240
++S ++ L F AF S+ ++ YN L ++ M+ +K++ R+
Sbjct: 188 MMSNLKPQTQAKLMGIFKAFTFDWFRTSYLISAGKGLYN-TLWACREGMREIKDIYTMRK 246
Query: 241 EMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNP 300
K DF + IEE K G LL E + L+F L + +TTS+A+ A+K L +NP
Sbjct: 247 TSEEKYD-DFLNTAIEESEKAGELLNENAIITLIFTLSCVTQDTTSKAICLAVKFLLENP 305
Query: 301 LVFKQLQEEHEAILERRENPNSGVTWQEYK-SMTFTFQLITETARLANIVPGIFRKALRE 359
V +L++EHE ILE RE+ GVTW+EY+ MTFT
Sbjct: 306 KVLAELKKEHEVILESREDKEGGVTWEEYRHKMTFT------------------------ 341
Query: 360 INFK-GYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMR 418
N K GYTIPAGW +M+ P VH +P Y++P FNP RWEG E +K F+ FG G+R
Sbjct: 342 -NMKSGYTIPAGWIVMIIPSVVHFDPEIYENPFEFNPWRWEGKELRAGSKTFMVFGTGLR 400
Query: 419 FCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQK 473
C G EFA++Q++VFLH LVT Y + + ++R P PNG + I++ +K
Sbjct: 401 QCAGAEFARLQISVFLHHLVTTYNFSLHQDCEVLRVPAAHLPNGISINISKCPKK 455
>At1g65670 cytochrome P450, putative
Length = 482
Score = 298 bits (763), Expect = 4e-81
Identities = 166/476 (34%), Positives = 266/476 (55%), Gaps = 14/476 (2%)
Query: 6 LGALVTICIIT--RWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKR 63
L +V++ ++ W+Y+ +NP N KLPPGSMG P++GE+ +F P+ + P FI++R
Sbjct: 8 LTVMVSLIVVKLFHWIYQSKNPKPNEKLPPGSMGFPIIGETFEFMKPHDAFQFPTFIKER 67
Query: 64 MKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGF 123
+ RYGPIF+T+L G V++STD +LN I + + ++FG+ N+
Sbjct: 68 IIRYGPIFRTSLFGAKVIISTDIELNMEIAKTN---HAPGLTKSIAQLFGENNLFFQSKE 124
Query: 124 MYKYLKNMMLNLFGPESLKKMISEVEQAACRT-LQQASCQDSVELKEATETMIFDLTAKK 182
+K+++N+ L G + LK + + RT +++ + + +++KE + ++ + AKK
Sbjct: 125 SHKHVRNLTFQLLGSQGLKLSVMQDIDLLTRTHMEEGARRGCLDVKEISSKILIECLAKK 184
Query: 183 LISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREM 242
+ E+++ L + F G FPLN+PGT K ++ RK+ + +LK + ++R
Sbjct: 185 VTGDMEPEAAKELALCWRCFPSGWFRFPLNLPGTGVYKMMKARKRMLHLLKETILKKRAS 244
Query: 243 PRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLV 302
++ +FF + E ++ A++ ++ L + ETT + L IKL+SDNP V
Sbjct: 245 G-EELGEFFKIIFEGAET----MSVDNAIEYIYTLFLLANETTPRILAATIKLISDNPKV 299
Query: 303 FKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINF 362
K+L EHE I+ + + +TW+EYKSMTFT +I E+ R+ + P +FR E
Sbjct: 300 MKELHREHEGIVRGKTEKETSITWEEYKSMTFTQMVINESLRITSTAPTVFRIFDHEFQV 359
Query: 363 KGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSG-ATKHFLAFGGGMRFCV 421
Y IPAGW M P H NP Y DPLVFNP RWEG + ++ ++ FG G R CV
Sbjct: 360 GSYKIPAGWIFMGYPNN-HFNPKTYDDPLVFNPWRWEGKDLGAIVSRTYIPFGAGSRQCV 418
Query: 422 GTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQKKHES 477
G EFAK+QMA+F+H L ++ RW G I+R L FPNG VQ + + + S
Sbjct: 419 GAEFAKLQMAIFIHHL-SRDRWSMKIGTTILRNFVLMFPNGCEVQFLKDTEVDNSS 473
>At3g50660 steroid 22-alpha-hydroxylase (DWF4)
Length = 513
Score = 295 bits (756), Expect = 3e-80
Identities = 172/503 (34%), Positives = 278/503 (55%), Gaps = 41/503 (8%)
Query: 3 VLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRK 62
+L L +L+++ + + R RN LPPG G P LGE++ + P T+ + F+++
Sbjct: 12 LLLLPSLLSLLLFLILLKR-RNRKTRFNLPPGKSGWPFLGETIGYLKPYTATTLGDFMQQ 70
Query: 63 RMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHG 122
+ +YG I+++NL G P +VS D LN FI Q EG++F+ YP + I G+ ++ L G
Sbjct: 71 HVSKYGKIYRSNLFGEPTIVSADAGLNRFILQNEGRLFECSYPRSIGGILGKWSMLVLVG 130
Query: 123 FMYKYLKNMMLNLFGPESLKK-MISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAK 181
M++ ++++ LN L+ ++ +VE+ L ++ + F+L AK
Sbjct: 131 DMHRDMRSISLNFLSHARLRTILLKDVERHTLFVLDSWQQNSIFSAQDEAKKFTFNLMAK 190
Query: 182 KLISYDP-TESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERR 240
++S DP E +E L++ +V F++G++S PLN+PGTAY+K LQ R +K ++ ++ER+
Sbjct: 191 HIMSMDPGEEETEQLKKEYVTFMKGVVSAPLNLPGTAYHKALQSRATILKFIERKMEERK 250
Query: 241 ------------------------EMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFV 276
+ RKQ+ D D ++ + K L TE I LDL+
Sbjct: 251 LDIKEEDQEEEEVKTEDEAEMSKSDHVRKQRTD--DDLLGWVLKHSNLSTEQI-LDLILS 307
Query: 277 LLFASFETTSQALTYAIKLLSDNPLVFKQLQEEH-EAILERRENPNSGVTWQEYKSMTFT 335
LLFA ET+S A+ AI L P ++L+EEH E ++E S + W +YK M FT
Sbjct: 308 LLFAGHETSSVAIALAIFFLQACPKAVEELREEHLEIARAKKELGESELNWDDYKKMDFT 367
Query: 336 FQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNP 395
+I ET RL N+V + RKAL+++ +KGY IP+GW ++ AVHL+ ++Y P +FNP
Sbjct: 368 QCVINETLRLGNVVRFLHRKALKDVRYKGYDIPSGWKVLPVISAVHLDNSRYDQPNLFNP 427
Query: 396 SRWE----GMEPSGA------TKHFLAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRP 445
RW+ G SG+ +++ FGGG R C G+E AK++MAVF+H LV K+ W
Sbjct: 428 WRWQQQNNGASSSGSGSFSTWGNNYMPFGGGPRLCAGSELAKLEMAVFIHHLVLKFNWEL 487
Query: 446 IKGGNIVRTPGLQFPNGFHVQIT 468
+ P + FPNG ++++
Sbjct: 488 AEDDKPFAFPFVDFPNGLPIRVS 510
>At5g05690 cytochrome P450 90A1 (sp|Q42569)
Length = 472
Score = 293 bits (749), Expect = 2e-79
Identities = 158/443 (35%), Positives = 252/443 (56%), Gaps = 6/443 (1%)
Query: 31 LPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGRPVVVSTDPDLNY 90
LPPGS+GLPL+GE+ Q + + PFI +R+ RYG +F T+L G P + S DP+ N
Sbjct: 31 LPPGSLGLPLIGETFQLIGAYKTENPEPFIDERVARYGSVFMTHLFGEPTIFSADPETNR 90
Query: 91 FIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGPESLK-KMISEVE 149
F+ Q EGK+F+ YP + + G+ ++ + G ++K + ++ ++ +K ++ +++
Sbjct: 91 FVLQNEGKLFECSYPASICNLLGKHSLLLMKGSLHKRMHSLTMSFANSSIIKDHLMLDID 150
Query: 150 QAACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRENFVAFIQGLISF 209
+ L S V L E + + F+LT K+L+S+DP E SE+LR+ ++ I+G S
Sbjct: 151 RLVRFNLD--SWSSRVLLMEEAKKITFELTVKQLMSFDPGEWSESLRKEYLLVIEGFFSL 208
Query: 210 PLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTEAI 269
PL + T Y K +Q R+K + L ++ +RRE + D + L + E I
Sbjct: 209 PLPLFSTTYRKAIQARRKVAEALTVVVMKRREEEEEGAERKKDMLAALLAADDGFSDEEI 268
Query: 270 ALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERRENPNSGVTWQEY 329
+D + LL A +ETTS +T A+K L++ PL QL+EEHE I + + S + W +Y
Sbjct: 269 -VDFLVALLVAGYETTSTIMTLAVKFLTETPLALAQLKEEHEKIRAMKSDSYS-LEWSDY 326
Query: 330 KSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQD 389
KSM FT ++ ET R+ANI+ G+FR+A+ ++ KGY IP GW + AVHL+P ++D
Sbjct: 327 KSMPFTQCVVNETLRVANIIGGVFRRAMTDVEIKGYKIPKGWKVFSSFRAVHLDPNHFKD 386
Query: 390 PLVFNPSRWEGME-PSGATKHFLAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKG 448
FNP RW+ +G + F FGGG R C G E A+V ++VFLH LVT + W P +
Sbjct: 387 ARTFNPWRWQSNSVTTGPSNVFTPFGGGPRLCPGYELARVALSVFLHRLVTGFSWVPAEQ 446
Query: 449 GNIVRTPGLQFPNGFHVQITEKD 471
+V P + + + + +D
Sbjct: 447 DKLVFFPTTRTQKRYPIFVKRRD 469
>At4g15396 unknown protein
Length = 475
Score = 289 bits (740), Expect = 2e-78
Identities = 159/460 (34%), Positives = 265/460 (57%), Gaps = 11/460 (2%)
Query: 19 VYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGR 78
+Y+WRNP NG+LPPGSMG P++GE+ +F P+ + +P F+++++ R+GP+F+T+L G
Sbjct: 23 IYQWRNPKSNGELPPGSMGYPIIGETFEFMKPHDAIQLPTFVKEKVLRHGPVFRTSLFGG 82
Query: 79 PVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGP 138
V++STD LN I + P + +FG N+ ++ +K+ +++ G
Sbjct: 83 KVIISTDIGLNMEIAKTN---HIPGMPKSLARLFGANNL-FVNKDTHKHARSLTNQFLGS 138
Query: 139 ESLK-KMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRE 197
++LK +M+ +++ L++ + + S+++KE T +I + AKK++ ++++ L
Sbjct: 139 QALKLRMLQDIDFLVRTHLKEGARKGSLDIKETTSKIIIECLAKKVMGEMEPDAAKELTL 198
Query: 198 NFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEE 257
+ F + F NIPGT + ++ R + MK+LK + ++R ++ DFF + +
Sbjct: 199 CWTFFPREWFGFAWNIPGTGVYRMVKARNRMMKVLKETVLKKR-ASGEELGDFFKTIFGD 257
Query: 258 LRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERR 317
+ ++ A + +F L + ETT L IKL+SD+P V ++LQ EHE I+ +
Sbjct: 258 TERGVKTISLESATEYIFTLFLLANETTPAVLAATIKLISDHPKVMQELQREHEGIVRDK 317
Query: 318 --ENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMV 375
+N + +TW++YKSMTFT +I E+ R+ + VP + R E F YTIPAGW I +
Sbjct: 318 IEKNEKADLTWEDYKSMTFTQMVINESLRITSTVPTVLRIIDHEFQFGEYTIPAGW-IFM 376
Query: 376 CPPAVHLNPAKYQDPLVFNPSRWEGMEPSG-ATKHFLAFGGGMRFCVGTEFAKVQMAVFL 434
P VH N KY DPL FNP RW+G + S ++ ++ FG G R CVG EF K++MA+F+
Sbjct: 377 GYPYVHFNAEKYDDPLAFNPWRWKGKDLSAIVSRTYIPFGSGSRLCVGAEFVKLKMAIFI 436
Query: 435 HCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQKK 474
H L ++YRW ++R L P G VQI E + K
Sbjct: 437 HHL-SRYRWSMKTETTLLRRFVLILPRGSDVQILEDTKAK 475
>At4g15300 cytochrome P450 like protein
Length = 487
Score = 289 bits (739), Expect = 3e-78
Identities = 164/479 (34%), Positives = 257/479 (53%), Gaps = 30/479 (6%)
Query: 15 ITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGP----- 69
I WVY+WRNP NGKLPPGSMG P +GE+ +FF P+ + FI+ R+ R+
Sbjct: 9 IFHWVYQWRNPKTNGKLPPGSMGFPFIGETFEFFKPHDALQFSTFIKDRVLRFFADFSSI 68
Query: 70 ---IFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYK 126
F+T+L G ++S D +LN + + + +FG+ N+ +K
Sbjct: 69 HLSFFRTSLFGDKAIISMDMELNLEMAKANSV---PGVTKSVIRLFGENNLFLQSKESHK 125
Query: 127 YLKNMMLNLFGPESLK-KMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAKKLIS 185
+++N+ L GP+ LK +MI +V+ A +++ + +++KE + ++ AKK++
Sbjct: 126 HVRNLTFQLLGPQGLKSRMIEDVDLLARTYMEEGARNGYLDVKETSSKILIGCLAKKVMG 185
Query: 186 YDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCL--------------QGRKKAMKM 231
E+++ L + F G F LN+PGT K + Q RK+ MK+
Sbjct: 186 EMEPEAAKELALCWRYFQSGWFRFFLNLPGTGVYKMMKVLFVQYTEADISWQARKRMMKL 245
Query: 232 LKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTY 291
L+ + +R ++ +FF+ + E+ EG ++ A++ ++ + ETT + L
Sbjct: 246 LRKTVLTKRASG-EELGEFFNIIFGEMEGEGETMSVENAVEYIYTFFLVANETTPRILAA 304
Query: 292 AIKLLSDNPLVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPG 351
+K +SD+P V ++LQ EHE I+ + G+TW++YKSM FT +I E+ R+ + P
Sbjct: 305 TVKFISDHPKVKQELQREHEEIVRGKAEKEGGLTWEDYKSMHFTQMVINESLRIISTAPT 364
Query: 352 IFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSG-ATKHF 410
+ R + YTIPAGW M P +H N KY+DP FNP RWEG + +K F
Sbjct: 365 VLRVLEHDFQVGDYTIPAGWTFMGY-PHIHFNSEKYEDPYAFNPWRWEGKDLGAIVSKTF 423
Query: 411 LAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITE 469
+ FG G R CVG EFAK+QMAVF+H L +YRW G I+R+ L FP G VQI+E
Sbjct: 424 IPFGAGRRLCVGAEFAKMQMAVFIHHLF-RYRWSMKSGTTIIRSFMLMFPGGCDVQISE 481
>At3g19270 cytochrome P450, putative
Length = 468
Score = 272 bits (695), Expect = 3e-73
Identities = 145/449 (32%), Positives = 253/449 (56%), Gaps = 9/449 (2%)
Query: 1 MWVLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI 60
+W L + L+ ++ R + + + GKLPPGSMG P LGE+LQ +S N + F
Sbjct: 4 IWFLVVPILILCLLLVRVIVSKKKKNSRGKLPPGSMGWPYLGETLQLYSQNPNV----FF 59
Query: 61 RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL 120
+ KRYG IFKT ++G P V+ P+ F+ +F+ YP + ++ G +
Sbjct: 60 TSKQKRYGEIFKTRILGYPCVMLASPEAARFVLVTHAHMFKPTYPRSKEKLIGPSALFFH 119
Query: 121 HGFMYKYLKNMMLNLFGPESLKKMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTA 180
G + +++ ++ + F PE+++K+I ++E A +LQ + V + + FD+
Sbjct: 120 QGDYHSHIRKLVQSSFYPETIRKLIPDIEHIALSSLQSWANMPIVSTYQEMKKFAFDVGI 179
Query: 181 KKLISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERR 240
+ + + E L+ N+ +G SFP+++PGT+Y+K L RK+ ++ ++ ERR
Sbjct: 180 LAIFGHLESSYKEILKHNYNIVDKGYNSFPMSLPGTSYHKALMARKQLKTIVSEIICERR 239
Query: 241 EMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNP 300
E R Q DF +++ ++G +LT+ D + +LFA+ +TT+ LT+ +K L D+
Sbjct: 240 EK-RALQTDFLGHLLNFKNEKGRVLTQEQIADNIIGVLFAAQDTTASCLTWILKYLHDDQ 298
Query: 301 LVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREI 360
+ + ++ E +AI E +TW++ ++M T ++I E+ R+A+I+ FR+A+ ++
Sbjct: 299 KLLEAVKAEQKAIYEENSREKKPLTWRQTRNMPLTHKVIVESLRMASIISFTFREAVVDV 358
Query: 361 NFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMRFC 420
+KGY IP GW +M +H NP + +P VF+PSR+E + P T F+ FG G+ C
Sbjct: 359 EYKGYLIPKGWKVMPLFRNIHHNPKYFSNPEVFDPSRFE-VNPKPNT--FMPFGSGVHAC 415
Query: 421 VGTEFAKVQMAVFLHCLVTKYRWRPIKGG 449
G E AK+Q+ +FLH LV+ +RW +KGG
Sbjct: 416 PGNELAKLQILIFLHHLVSNFRWE-VKGG 443
>At3g30180 cytochrome P450 homolog, putative
Length = 465
Score = 267 bits (683), Expect = 8e-72
Identities = 155/469 (33%), Positives = 253/469 (53%), Gaps = 17/469 (3%)
Query: 3 VLCLGALVTICIITRWVYRWRNPSCNGK-LPPGSMGLPLLGESLQFFSPNTSCDIPPFIR 61
++ LG LV I + + RW + K LPPG+MG P+ GE+ +F P F++
Sbjct: 5 MMILGLLVIIVCLCTALLRWNQMRYSKKGLPPGTMGWPIFGETTEFLKQG-----PDFMK 59
Query: 62 KRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLH 121
+ RYG FK++++G P +VS D +LN +I E K + YP + +I G N+ ++H
Sbjct: 60 NQRLRYGSFFKSHILGCPTIVSMDAELNRYILMNESKGLVAGYPQSMLDILGTCNIAAVH 119
Query: 122 GFMYKYLKNMMLNLFGPESLKK-MISEVEQAACRTLQQASCQDSVELKEATETMIFDLTA 180
G ++ ++ +L+L P +K ++ +++ L ++V+++E T+ M F +
Sbjct: 120 GPSHRLMRGSLLSLISPTMMKDHLLPKIDDFMRNYLCGWDDLETVDIQEKTKHMAFLSSL 179
Query: 181 KKLISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERR 240
++ E R F + G +S P++IPGT Y +Q R ++L ++QER+
Sbjct: 180 LQIAETLKKPEVEEYRTEFFKLVVGTLSVPIDIPGTNYRSGVQARNNIDRLLTELMQERK 239
Query: 241 EMPRKQQMDFFDYVIEELRKEGT--LLTEAIALDLMFVLLFASFETTSQALTYAIKLLSD 298
E F D + ++KE LLT+ D + +L++ +ET S A+K L D
Sbjct: 240 ESGET----FTDMLGYLMKKEDNRYLLTDKEIRDQVVTILYSGYETVSTTSMMALKYLHD 295
Query: 299 NPLVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALR 358
+P ++L+ EH AI ER+ P+ +T + KSM FT +I ET+RLA IV G+ RK
Sbjct: 296 HPKALEELRREHLAIRERKR-PDEPLTLDDIKSMKFTRAVIFETSRLATIVNGVLRKTTH 354
Query: 359 EINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATK-HFLAFGGGM 417
++ GY IP GW I V ++ + + Y+DP++FNP RW ME S +K +FL FGGG+
Sbjct: 355 DLELNGYLIPKGWRIYVYTREINYDTSLYEDPMIFNPWRW--MEKSLESKSYFLLFGGGV 412
Query: 418 RFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQ 466
R C G E +++ FLH VTKYRW ++ P + P G+H++
Sbjct: 413 RLCPGKELGISEVSSFLHYFVTKYRWEENGEDKLMVFPRVSAPKGYHLK 461
>At4g15393 putative protein
Length = 453
Score = 265 bits (676), Expect = 5e-71
Identities = 158/471 (33%), Positives = 262/471 (55%), Gaps = 29/471 (6%)
Query: 2 WVLCLGALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIR 61
W + + +V + W+Y+W+NP NGKLPPGSMG P++GE+ +F + + +P F++
Sbjct: 8 WAVIVSLIVVK--LCHWIYQWKNPKGNGKLPPGSMGYPIIGETFEFMKLHDAIQLPTFVK 65
Query: 62 KRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLH 121
+++ R T+L G V++STD LN I + P + +FG N+ ++
Sbjct: 66 EKLLR------TSLFGGKVIISTDIGLNMEIAKTN---HIPGMPKSLERLFGATNL-FVN 115
Query: 122 GFMYKYLKNMMLNLFGPESLK-KMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTA 180
+K+ +++ G ++LK +MI +++ A +++ + + +++KE +
Sbjct: 116 KDTHKHARSLTNQFLGSQALKLRMIQDIDFLARTHMKEGARKGCLDVKET--------AS 167
Query: 181 KKLISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERR 240
KK++ E+++ L + F + F N PGT + ++ R + MK++K + ++R
Sbjct: 168 KKVMGEMEPEAAKELTLCWTFFPRDWFRFAWNFPGTGVYRIVKARNRMMKVIKETVVKKR 227
Query: 241 EMPRKQQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNP 300
+K +FF+ + + E ++ IA + +F L + ETT L IKL+SDNP
Sbjct: 228 ASGKKLG-EFFETIFGD--TESVTMSIEIATEYIFTLFVLANETTPGVLAATIKLISDNP 284
Query: 301 LVFKQLQEEHEAILER--RENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALR 358
V ++L+ EHE I++ +++ + +TW++YKSMTFT +I E+ R+ + VP + R
Sbjct: 285 KVMQELRREHEGIVQDKIKKDETADLTWEDYKSMTFTQMVINESLRITSTVPTVLRIIDH 344
Query: 359 EINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSG-ATKHFLAFGGGM 417
EI F YTIPAGW I + P VH NP KY DPL FNP RW+G + S +K +L FG G
Sbjct: 345 EIQFGDYTIPAGW-IFMGYPYVHFNPEKYDDPLAFNPWRWKGKDLSTIVSKTYLPFGSGT 403
Query: 418 RFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQIT 468
R CVG EF K+QMA+F+H L +YRW ++R L P G VQI+
Sbjct: 404 RLCVGAEFVKLQMAIFIHHLF-RYRWSMKAETTLLRRFILVLPRGSDVQIS 453
>At4g36380 cytochrome P450 (ROTUNDIFOLIA3)
Length = 524
Score = 259 bits (661), Expect = 3e-69
Identities = 146/449 (32%), Positives = 244/449 (53%), Gaps = 8/449 (1%)
Query: 29 GKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGRPVVVSTDPDL 88
G +P GS+G P++GE+L F + S F+ KR YG +FKTN++G P+++STD ++
Sbjct: 67 GMIPNGSLGWPVIGETLNFIACGYSSRPVTFMDKRKSLYGKVFKTNIIGTPIIISTDAEV 126
Query: 89 NYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGPESLKKMIS-E 147
N + Q G F YP + TE+ G+ ++ S++G K L ++ LK I+ +
Sbjct: 127 NKVVLQNHGNTFVPAYPKSITELLGENSILSINGPHQKRLHTLIGAFLRSPHLKDRITRD 186
Query: 148 VEQAACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRENFVAFIQGLI 207
+E + TL + V +++ + M F++ K L+S P E L+ F FI+GLI
Sbjct: 187 IEASVVLTLASWAQLPLVHVQDEIKKMTFEILVKVLMSTSPGEDMNILKLEFEEFIKGLI 246
Query: 208 SFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTE 267
P+ PGT K L+ +++ +KM+K +++ER ++ D V LR G +
Sbjct: 247 CIPIKFPGTRLYKSLKAKERLIKMVKKVVEER-QVAMTTTSPANDVVDVLLRDGGDSEKQ 305
Query: 268 AIALDLM----FVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERRENPNSG 323
+ D + ++ ET A+T A+K LSDNP+ +L EE+ + R+
Sbjct: 306 SQPSDFVSGKIVEMMIPGEETMPTAMTLAVKFLSDNPVALAKLVEENMEMKRRKLELGEE 365
Query: 324 VTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPPAVHLN 383
W +Y S++FT +I ET R+ANI+ G++RKAL+++ KGY IP GW ++ +VH++
Sbjct: 366 YKWTDYMSLSFTQNVINETLRMANIINGVWRKALKDVEIKGYLIPKGWCVLASFISVHMD 425
Query: 384 PAKYQDPLVFNPSRWEGMEPS-GATKHFLAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYR 442
Y +P F+P RW+ + S ++ F FGGG R C G E +K+++++FLH LVT+Y
Sbjct: 426 EDIYDNPYQFDPWRWDRINGSANSSICFTPFGGGQRLCPGLELSKLEISIFLHHLVTRYS 485
Query: 443 WRPIKGGNIVRTPGLQFPNGFHVQITEKD 471
W + IV P ++ +++ D
Sbjct: 486 W-TAEEDEIVSFPTVKMKRRLPIRVATVD 513
>At3g13730 CYP90D
Length = 491
Score = 253 bits (647), Expect = 1e-67
Identities = 142/425 (33%), Positives = 232/425 (54%), Gaps = 20/425 (4%)
Query: 30 KLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGRPVVVSTDPDLN 89
K P GS+G P++GE+++F S S F+ KR YG +FK+++ G +VSTD ++N
Sbjct: 50 KFPHGSLGWPVIGETIEFVSSAYSDRPESFMDKRRLMYGRVFKSHIFGTATIVSTDAEVN 109
Query: 90 YFIFQQEGKIFQSWYPDTFTEIFGQQNV----GSLH----GFMYKYLKNMMLNLFGPESL 141
+ Q + F +YP T E+ G+ ++ GSLH G + +LK+ +L +
Sbjct: 110 RAVLQSDSTAFVPFYPKTVRELMGKSSILLINGSLHRRFHGLVGSFLKSPLLKAQIVRDM 169
Query: 142 KKMISEVEQAACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRENFVA 201
K +SE ++ S V L++ ++T+ F + AK LIS + E E L+ F
Sbjct: 170 HKFLSE-------SMDLWSEDQPVLLQDVSKTVAFKVLAKALISVEKGEDLEELKREFEN 222
Query: 202 FIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQER-REMPRKQQMDFF--DYVIEEL 258
FI GL+S P+N PGT ++ LQ +K +K ++ +++ + R+ K++ D D V L
Sbjct: 223 FISGLMSLPINFPGTQLHRSLQAKKNMVKQVERIIEGKIRKTKNKEEDDVIAKDVVDVLL 282
Query: 259 RKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERRE 318
+ LT + + M ++ ++ +T A+K LSD+P L EE+ + +E
Sbjct: 283 KDSSEHLTHNLIANNMIDMMIPGHDSVPVLITLAVKFLSDSPAALNLLTEENMKLKSLKE 342
Query: 319 NPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPP 378
+ W +Y S+ FT ++ITET R+ N++ G+ RKA++++ KGY IP GW +
Sbjct: 343 LTGEPLYWNDYLSLPFTQKVITETLRMGNVIIGVMRKAMKDVEIKGYVIPKGWCFLAYLR 402
Query: 379 AVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMRFCVGTEFAKVQMAVFLHCLV 438
+VHL+ Y+ P FNP RW+ E T F FGGG R C G + A+++ +VFLH LV
Sbjct: 403 SVHLDKLYYESPYKFNPWRWQ--ERDMNTSSFSPFGGGQRLCPGLDLARLETSVFLHHLV 460
Query: 439 TKYRW 443
T++RW
Sbjct: 461 TRFRW 465
>At5g38970 cytochrome P450 - like protein
Length = 465
Score = 253 bits (645), Expect = 2e-67
Identities = 148/471 (31%), Positives = 251/471 (52%), Gaps = 17/471 (3%)
Query: 3 VLCLGALVTICIITRWVYRWRNP--SCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFI 60
++ +G L+ I + + RW + NG LPPG+MG P+ GE+ +F P F+
Sbjct: 5 MVMMGLLLIIVSLCSALLRWNQMRYTKNG-LPPGTMGWPIFGETTEFLKQG-----PNFM 58
Query: 61 RKRMKRYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSL 120
R + RYG FK++L+G P ++S D ++N +I + E K YP + +I G N+ ++
Sbjct: 59 RNQRLRYGSFFKSHLLGCPTLISMDSEVNRYILKNESKGLVPGYPQSMLDILGTCNMAAV 118
Query: 121 HGFMYKYLKNMMLNLFGPESLKK-MISEVEQAACRTLQQASCQDSVELKEATETMIFDLT 179
HG ++ ++ +L+L ++ ++ +V+ L Q + + +++++ T+ M F +
Sbjct: 119 HGSSHRLMRGSLLSLISSTMMRDHILPKVDHFMRSYLDQWNELEVIDIQDKTKHMAFLSS 178
Query: 180 AKKLISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQER 239
++ E + F + G +S P+++PGT Y +Q R ++L+ ++QER
Sbjct: 179 LTQIAGNLRKPFVEEFKTAFFKLVVGTLSVPIDLPGTNYRCGIQARNNIDRLLRELMQER 238
Query: 240 REMPRKQQMDFFDYVIEELRKEGTL--LTEAIALDLMFVLLFASFETTSQALTYAIKLLS 297
R+ F D + ++KEG LT+ D + +L++ +ET S A+K L
Sbjct: 239 RDSGET----FTDMLGYLMKKEGNRYPLTDEEIRDQVVTILYSGYETVSTTSMMALKYLH 294
Query: 298 DNPLVFKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKAL 357
D+P ++L+ EH A ER+ + + ++ KSM FT +I ET+RLA IV G+ RK
Sbjct: 295 DHPKALQELRAEHLAFRERKRQ-DEPLGLEDVKSMKFTRAVIYETSRLATIVNGVLRKTT 353
Query: 358 REINFKGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGM 417
R++ GY IP GW I V ++ + Y+DPL+FNP RW F+ FGGG
Sbjct: 354 RDLEINGYLIPKGWRIYVYTREINYDANLYEDPLIFNPWRWMKKSLESQNSCFV-FGGGT 412
Query: 418 RFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQIT 468
R C G E V+++ FLH VT+YRW I G ++ P + P GFH++I+
Sbjct: 413 RLCPGKELGIVEISSFLHYFVTRYRWEEIGGDELMVFPRVFAPKGFHLRIS 463
>At1g73340
Length = 512
Score = 247 bits (631), Expect = 9e-66
Identities = 150/476 (31%), Positives = 244/476 (50%), Gaps = 45/476 (9%)
Query: 30 KLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMK------------------------ 65
+LPPGS G PL+G++ + + F+ K++K
Sbjct: 42 RLPPGSRGWPLIGDTFAWLNAVAGSHPSSFVEKQIKKFVSLLCSVLLLILKRPDNSGFNE 101
Query: 66 -RYGPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFM 124
RYG IF +L G+ VVS DPD N FI Q EGK+FQS YP +F ++ G+ V ++HG
Sbjct: 102 IRYGRIFSCSLFGKWAVVSADPDFNRFIMQNEGKLFQSSYPKSFRDLVGKDGVITVHGDQ 161
Query: 125 YKYLKNMMLNLFGPESLKKMISEV-EQAACRTLQQASCQDSVELKEATETMIFDLTAKKL 183
+ L ++ ++ + LK EV +TL + V L++ + L +L
Sbjct: 162 QRRLHSIASSMMRHDQLKTHFLEVIPVVMLQTLSNFKDGEVVLLQDICRKVAIHLMVNQL 221
Query: 184 ISYDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMP 243
+ + + + F F+ G +S P+++PG YNK ++ RK+ ++ + +++R +
Sbjct: 222 LGVSSESEVDEMSQLFSDFVDGCLSVPIDLPGFTYNKAMKARKEIIRKINKTIEKR--LQ 279
Query: 244 RKQQMDFF-DYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLV 302
K D + V+ L +E +L E++A D + LLFA ETTS+ + +A+ L+ P
Sbjct: 280 NKAASDTAGNGVLGRLLEEESLPNESMA-DFIINLLFAGNETTSKTMLFAVYFLTHCPKA 338
Query: 303 FKQLQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINF 362
QL EEH+ + +TWQ+YK+M FT +I ET RL I + R+A ++++
Sbjct: 339 MTQLLEEHDRL------AGGMLTWQDYKTMDFTQCVIDETLRLGGIAIWLMREAKEDVSY 392
Query: 363 KGYTIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKH-------FLAFGG 415
+ Y IP G ++ AVHL+ + Y++ L FNP RW ++P K + FGG
Sbjct: 393 QDYVIPKGCFVVPFLSAVHLDESYYKESLSFNPWRW--LDPETQQKRNWRTSPFYCPFGG 450
Query: 416 GMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKD 471
G RFC G E A++Q+A+FLH +T Y+W +K I P + NGF +Q+ +D
Sbjct: 451 GTRFCPGAELARLQIALFLHYFITTYKWTQLKEDRISFFPSARLVNGFKIQLNRRD 506
>At5g45340 cytochrome P450
Length = 463
Score = 235 bits (600), Expect = 4e-62
Identities = 140/466 (30%), Positives = 251/466 (53%), Gaps = 19/466 (4%)
Query: 8 ALVTICIITRWVYRWRNPSCNGKLPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRY 67
A + +C++ R+ S LPPG+MG P +GE+ Q +S + + F + +RY
Sbjct: 13 AALFLCLLRFIAGVRRSSSTKLPLPPGTMGYPYVGETFQLYSQDPNV----FFAAKQRRY 68
Query: 68 GPIFKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKY 127
G +FKT+++G P V+ + P+ F+ + +F+ +P + + G+Q + G +
Sbjct: 69 GSVFKTHVLGCPCVMISSPEAAKFVLVTKSHLFKPTFPASKERMLGKQAIFFHQGDYHSK 128
Query: 128 LKNMMLNLFGPESLKKMISEVEQAACRTLQQASCQDSVELKEATE--TMIFDLTAKKLIS 185
L+ ++L F P++++ M+ +E A +L D +L E T F++ ++
Sbjct: 129 LRKLVLRAFMPDAIRNMVPHIESIAQESLNS---WDGTQLNTYQEMKTYTFNVALISILG 185
Query: 186 YDPTESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRK 245
D E+L+ + +G S P+N+PGT ++K ++ RK+ ++L N+L +RR+ P
Sbjct: 186 KDEVYYREDLKRCYYILEKGYNSMPINLPGTLFHKAMKARKELAQILANILSKRRQNP-S 244
Query: 246 QQMDFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQ 305
D +E+ K G LT+ D + ++FA+ +TT+ LT+ +K L+DNP V +
Sbjct: 245 SHTDLLGSFMED--KAG--LTDEQIADNIIGVIFAARDTTASVLTWILKYLADNPTVLEA 300
Query: 306 LQEEHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGY 365
+ EE AI + ++ S +TW++ K M T+++I ET R A I+ FR+A+ ++ ++GY
Sbjct: 301 VTEEQMAIRKDKKEGES-LTWEDTKKMPLTYRVIQETLRAATILSFTFREAVEDVEYEGY 359
Query: 366 TIPAGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSGATKHFLAFGGGMRFCVGTEF 425
IP GW ++ +H N + DP F+PSR+E + P T F+ FG G+ C G E
Sbjct: 360 LIPKGWKVLPLFRNIHHNADIFSDPGKFDPSRFE-VAPKPNT--FMPFGSGIHSCPGNEL 416
Query: 426 AKVQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFP-NGFHVQITEK 470
AK++++V +H L TKYRW + + ++ P NG + + K
Sbjct: 417 AKLEISVLIHHLTTKYRWSIVGPSDGIQYGPFALPQNGLPIALERK 462
>At3g30290 cytochrome P450, putative
Length = 408
Score = 220 bits (561), Expect = 1e-57
Identities = 134/411 (32%), Positives = 220/411 (52%), Gaps = 11/411 (2%)
Query: 71 FKTNLVGRPVVVSTDPDLNYFIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGF-MYKYLK 129
F+T+L G V++S D +LN + + + +FG+ N L +K+++
Sbjct: 5 FRTSLFGGKVIISMDNELNMEMAKTNRT---PGITKSIARLFGEDNNLFLQSTESHKHVR 61
Query: 130 NMMLNLFGPESLKKMISEVEQAACRTLQQASCQD-SVELKEATETMIFDLTAKKLISYDP 188
N+ + + G +SLK I E RT + +D S+++KE T ++ + AKK++
Sbjct: 62 NLTVQMLGSQSLKLRIMENIDLLTRTHMEEGARDGSLDVKETTSKILIECLAKKVMGEME 121
Query: 189 TESSENLRENFVAFIQGLISFPLNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRKQQM 248
E+++ L + F G P N+PG ++ RK+ +LK + ++RE ++
Sbjct: 122 PEAAKKLALCWRYFPSGWFRLPFNLPGIGVYNMMKARKRMKTLLKEEVLKKREAG-EEFG 180
Query: 249 DFFDYVIEELRKEGTLLTEAIALDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQE 308
+F + E E ++ ++ ++ + ETT + L +K +S+NP V ++LQ
Sbjct: 181 EFSKIIFGEKEGEKETMSMKNVIEYIYTFFVIANETTPRILAATVKFISENPKVMQELQR 240
Query: 309 EHEAILERRENPNSGVTWQEYKSMTFTFQLITETARLANIVPGIFRKALREINFKGYTIP 368
EH I E + +G+TW++YKSMTFT +I E+ R++ VP I RK + YTIP
Sbjct: 241 EHAMIFENKSE-EAGLTWEDYKSMTFTNMVINESLRISTTVPVILRKPDHDTKVGDYTIP 299
Query: 369 AGWAIMVCPPAVHLNPAKYQDPLVFNPSRWEGMEPSG-ATKHFLAFGGGMRFCVGTEFAK 427
AGW M P A H +P KY+DPL FNP RW+G + + +++ FG G R CVG FAK
Sbjct: 300 AGWNFMGYPSA-HFDPTKYEDPLEFNPWRWKGNDLDAIVSTNYIPFGAGPRLCVGAYFAK 358
Query: 428 VQMAVFLHCLVTKYRWRPIKGGNIVRTPGLQFPNGFHVQITEKDQKKHESE 478
+ MA+F+H L +YRW + R+ L FP G VQI+ KD ++H+S+
Sbjct: 359 LLMAIFIHHL-CRYRWSMKAEVTVTRSYMLMFPRGCDVQIS-KDTQEHDSD 407
>At4g19230 cytochrome P450 protein like
Length = 467
Score = 220 bits (560), Expect = 2e-57
Identities = 129/443 (29%), Positives = 239/443 (53%), Gaps = 15/443 (3%)
Query: 31 LPPGSMGLPLLGESLQFFSPNTSCDIPPFIRKRMKRYGPIFKTNLVGRPVVVSTDPDLNY 90
LPPG+MG P +GE+ Q +S + + F + + KRYG +FKT+++G P V+ + P+
Sbjct: 36 LPPGTMGWPYVGETFQLYSQDPNV----FFQSKQKRYGSVFKTHVLGCPCVMISSPEAAK 91
Query: 91 FIFQQEGKIFQSWYPDTFTEIFGQQNVGSLHGFMYKYLKNMMLNLFGPESLKKMISEVEQ 150
F+ + +F+ +P + + G+Q + G + L+ ++L F PES++ M+ ++E
Sbjct: 92 FVLVTKSHLFKPTFPASKERMLGKQAIFFHQGDYHAKLRKLVLRAFMPESIRNMVPDIES 151
Query: 151 AACRTLQQASCQDSVELKEATETMIFDLTAKKLISYDPTESSENLRENFVAFIQGLISFP 210
A +L+ + + +T F++ + D E+L+ + +G S P
Sbjct: 152 IAQDSLRSWE-GTMINTYQEMKTYTFNVALLSIFGKDEVLYREDLKRCYYILEKGYNSMP 210
Query: 211 LNIPGTAYNKCLQGRKKAMKMLKNMLQERREMPRKQQMDFFDYVIEELRKEGTLLTEAIA 270
+N+PGT ++K ++ RK+ ++L +L ERR+ D + + + L E IA
Sbjct: 211 VNLPGTLFHKSMKARKELSQILARILSERRQNGSSHN----DLLGSFMGDKEELTDEQIA 266
Query: 271 LDLMFVLLFASFETTSQALTYAIKLLSDNPLVFKQLQEEHEAILERRENPNSGVTWQEYK 330
D + ++FA+ +TT+ +++ +K L++NP V + + EE AI + +E S +TW + K
Sbjct: 267 -DNIIGVIFAARDTTASVMSWILKYLAENPNVLEAVTEEQMAIRKDKEEGES-LTWGDTK 324
Query: 331 SMTFTFQLITETARLANIVPGIFRKALREINFKGYTIPAGWAIMVCPPAVHLNPAKYQDP 390
M T ++I ET R+A+I+ FR+A+ ++ ++GY IP GW ++ +H + + +P
Sbjct: 325 KMPLTSRVIQETLRVASILSFTFREAVEDVEYEGYLIPKGWKVLPLFRNIHHSADIFSNP 384
Query: 391 LVFNPSRWEGMEPSGATKHFLAFGGGMRFCVGTEFAKVQMAVFLHCLVTKYRWRPIKGGN 450
F+PSR+E + P T F+ FG G C G E AK++M++ +H L TKY W + +
Sbjct: 385 GKFDPSRFE-VAPKPNT--FMPFGNGTHSCPGNELAKLEMSIMIHHLTTKYSWSIVGASD 441
Query: 451 IVRTPGLQFP-NGFHVQITEKDQ 472
++ P NG + + K +
Sbjct: 442 GIQYGPFALPQNGLPIVLARKPE 464
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,171,651
Number of Sequences: 26719
Number of extensions: 479675
Number of successful extensions: 1814
Number of sequences better than 10.0: 249
Number of HSP's better than 10.0 without gapping: 186
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 1139
Number of HSP's gapped (non-prelim): 317
length of query: 483
length of database: 11,318,596
effective HSP length: 103
effective length of query: 380
effective length of database: 8,566,539
effective search space: 3255284820
effective search space used: 3255284820
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146573.1


