

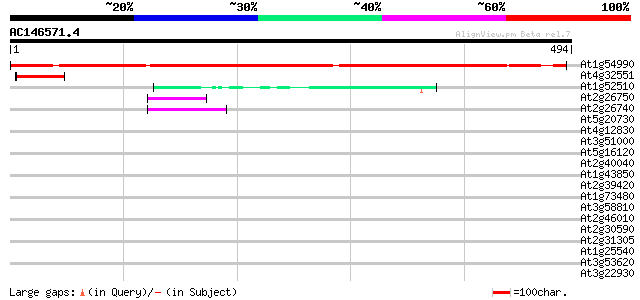
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146571.4 + phase: 0
(494 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g54990 unknown protein 496 e-140
At4g32551 Leunig protein 45 9e-05
At1g52510 unknown protein (At1g52510) 45 9e-05
At2g26750 putative epoxide hydrolase 42 8e-04
At2g26740 epoxide hydrolase (ATsEH) 42 8e-04
At5g20730 auxin response factor 7 (ARF7) 37 0.019
At4g12830 hydrolase like protein 37 0.019
At3g51000 epoxide hydrolase-like protein 37 0.019
At5g16120 lipase-like protein 37 0.033
At2g40040 unknown protein 36 0.056
At1g43850 SEUSS transcriptional co-regulator 36 0.056
At2g39420 phospholipase like protein 35 0.095
At1g73480 lysophospholipase like protein 35 0.095
At3g58810 zinc transporter -like protein 35 0.12
At2g46010 hypothetical protein 35 0.12
At2g30590 unknown protein 35 0.12
At2g31305 unknown protein 34 0.16
At1g25540 hypothetical protein 33 0.28
At3g53620 inorganic pyrophosphatase -like protein 33 0.36
At3g22930 calmodulin, putative 33 0.36
>At1g54990 unknown protein
Length = 473
Score = 496 bits (1277), Expect = e-140
Identities = 257/492 (52%), Positives = 337/492 (68%), Gaps = 25/492 (5%)
Query: 1 MAIITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLSFWFYFTLSIS 60
MAIITEE+E + K K K +++ K KP Q NP FWFYFT+ +S
Sbjct: 1 MAIITEEEEDPKTLNPPKNKPKDSDFTKSESTMKNPKP-----QSQNPFPFWFYFTVVVS 55
Query: 61 LVTLFFLFTSSPLSSSSPQQWFLTLPTTLRQHYSNGRTIKVQIHPNQQPIHLFTFQLDPT 120
L T+ F+ S S + P+ WFL+LP LRQHYSNGRTIKVQ++ N+ PI +F +
Sbjct: 56 LATIIFISLSLFSSQNDPRSWFLSLPPALRQHYSNGRTIKVQVNSNESPIEVFVAESGSI 115
Query: 121 IPNPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKS-VEVSVEGLDG 179
+ETV+I+HG LSS++++ +IQSL ++G+ +AIDLPG+GFSDKS V + + G
Sbjct: 116 ---HTETVVIVHGLGLSSFAFKEMIQSLGSKGIHSVAIDLPGNGFSDKSMVVIGGDREIG 172
Query: 180 IFGRFSYVYSEIKEKGFFWAFDQIVETGQIPYEEVLA-RMSKRKVNKPIDLGPEEIGKVL 238
R VY I+EKG FWAFDQ++ETG +PYEE++ + SKR+ K I+LG EE +VL
Sbjct: 173 FVARVKEVYGLIQEKGVFWAFDQMIETGDLPYEEIIKLQNSKRRSFKAIELGSEETARVL 232
Query: 239 GEVIGTLGLAPVHLVLHDSALGFTANWVSENSDLVSSLTLVDTPVPPSNLGAFPIWVLDV 298
G+VI TLGLAPVHLVLHDSALG +NWVSEN V S+TL+D+ + P A P+WVL+V
Sbjct: 233 GQVIDTLGLAPVHLVLHDSALGLASNWVSENWQSVRSVTLIDSSISP----ALPLWVLNV 288
Query: 299 PLIREVVLGFPYVFAKVVNFYCSKRIGGLDADAHRVLLKSGDGRKAVVAIGKNLNSSFDL 358
P IRE++L F + F K+V+F CSK + D DAHR+LLK +GR+AVVA LN SFD+
Sbjct: 289 PGIREILLAFSFGFEKLVSFRCSKEMTLSDIDAHRILLKGRNGREAVVASLNKLNHSFDI 348
Query: 359 TEWGCSDRLKDMPMQLIWSSDWSEEWSSEGNRVAGALPRAKFVTHSGGRWAQEDVAVEIA 418
+WG SD + +PMQ+IWSS+ S+EWS EG RVA ALP+AKFVTHSG RW QE + E+A
Sbjct: 349 AQWGNSDGINGIPMQVIWSSEASKEWSDEGQRVAKALPKAKFVTHSGSRWPQESKSGELA 408
Query: 419 EKISQFVSSLPKTVRKVEQEPPIPDHVQKMFDEAKSDDGHDHHQGHSHGHDRLGEAHIHE 478
+ IS+FVS LPK++R+V +E PIP+ VQK+ +EAK+ D HDHH GH H H
Sbjct: 409 DYISEFVSLLPKSIRRVAEE-PIPEEVQKVLEEAKAGDDHDHHHGHGHAH---------- 457
Query: 479 AGYMDAYGIGHE 490
AGY DAYG+G E
Sbjct: 458 AGYSDAYGLGEE 469
>At4g32551 Leunig protein
Length = 931
Score = 45.1 bits (105), Expect = 9e-05
Identities = 19/42 (45%), Positives = 29/42 (68%)
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNP 48
+Q+ QQQQQQ+Q+Q+Q+QQH N+ S+ Q+ +T Q P
Sbjct: 136 QQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQP 177
Score = 42.0 bits (97), Expect = 8e-04
Identities = 18/43 (41%), Positives = 28/43 (64%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNP 48
++Q QQQQQ+Q+Q+Q+QQ + + + Q PS Q QQ+ P
Sbjct: 129 QQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTP 171
Score = 33.5 bits (75), Expect = 0.28
Identities = 17/43 (39%), Positives = 24/43 (55%), Gaps = 3/43 (6%)
Query: 6 EEQESDQQQQQQKQKQKQ---KQQHSNKTKSKTQKPSTTQPQQ 45
++Q+ QQQQQQ+Q Q Q +QQ T Q+P+ Q Q
Sbjct: 142 QQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTPQQQPQ 184
Score = 32.3 bits (72), Expect = 0.62
Identities = 14/40 (35%), Positives = 22/40 (55%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
++Q+ QQQQ Q Q Q+QQ + + + Q QPQ+
Sbjct: 146 QQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTPQQQPQR 185
>At1g52510 unknown protein (At1g52510)
Length = 380
Score = 45.1 bits (105), Expect = 9e-05
Identities = 61/253 (24%), Positives = 96/253 (37%), Gaps = 53/253 (20%)
Query: 127 TVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVSVEGLDGIFGRFSY 186
T++ +HG S+SYR ++ LS G A D G GFSDK +G F+Y
Sbjct: 127 TIVFVHGAPTQSFSYRTVMSELSDAGFHCFAPDWIGFGFSDKP--------QPGYG-FNY 177
Query: 187 VYSEIKEKGFFWAFDQIVETGQIPYEEVLARMSKRKVNKPIDLGPEEIGKVLGEVIGTLG 246
EK + AFD+++E +V P L V G ++G+ G
Sbjct: 178 T-----EKEYHEAFDKLLEV--------------LEVKSPFFL------VVQGFLVGSYG 212
Query: 247 LAPVHLVLHDSALGFTANWVSENSDLVSSLTLVDTPVPPSNLGAFPIWVLDVPLIREVVL 306
L W +N V L ++++P+ S+ L +PL E
Sbjct: 213 L----------------TWALKNPSKVEKLAILNSPLTVSSPVPGLFKQLRIPLFGEFTC 256
Query: 307 GFPYVFAKVVNFYCSKRIGGLDADAHRV-LLKSGDGRKAVVAIGKNLNSSFDLTEW--GC 363
+ + + + AD +R+ L SG A++ K +N L++ G
Sbjct: 257 QNAILAERFIEGGSPYVLKNEKADVYRLPYLSSGGPGFALLETAKKINFGDTLSQIANGF 316
Query: 364 SDRLKDMPMQLIW 376
S D P L W
Sbjct: 317 SSGSWDKPTLLAW 329
>At2g26750 putative epoxide hydrolase
Length = 320
Score = 42.0 bits (97), Expect = 8e-04
Identities = 22/52 (42%), Positives = 30/52 (57%)
Query: 122 PNPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
P+ VL+LHG YS+R+ I L+ +G R +A DL G G SD E+S
Sbjct: 20 PSDGTIVLLLHGFPELWYSWRHQISGLAARGYRAVAPDLRGYGDSDAPAEIS 71
>At2g26740 epoxide hydrolase (ATsEH)
Length = 321
Score = 42.0 bits (97), Expect = 8e-04
Identities = 26/70 (37%), Positives = 35/70 (49%)
Query: 122 PNPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVSVEGLDGIF 181
P+ VL+LHG YS+R+ I L+ +G R +A DL G G SD E+S I
Sbjct: 20 PSDGPIVLLLHGFPELWYSWRHQIPGLAARGYRAVAPDLRGYGDSDAPAEISSYTCFNIV 79
Query: 182 GRFSYVYSEI 191
G V S +
Sbjct: 80 GDLIAVISAL 89
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 37.4 bits (85), Expect = 0.019
Identities = 17/39 (43%), Positives = 25/39 (63%)
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
+Q+ QQQ Q+Q+Q Q QQHSN +S++Q+ QQ
Sbjct: 523 QQQQQQQQLLQQQQQLQSQQHSNNNQSQSQQQQQLLQQQ 561
Score = 31.2 bits (69), Expect = 1.4
Identities = 15/45 (33%), Positives = 25/45 (55%)
Query: 5 TEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPL 49
+ +S QQQQQ +Q+Q+QQ + + Q+ + Q +T PL
Sbjct: 544 SNNNQSQSQQQQQLLQQQQQQQLQQQHQQPLQQQTQQQQLRTQPL 588
Score = 30.4 bits (67), Expect = 2.3
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Query: 3 IITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
++ ++Q+ QQQ Q+ Q+Q QQ +T+ Q S QPQQ
Sbjct: 557 LLQQQQQQQLQQQHQQPLQQQTQQQQLRTQ-PLQSHSHPQPQQ 598
Score = 30.0 bits (66), Expect = 3.1
Identities = 17/44 (38%), Positives = 27/44 (60%), Gaps = 2/44 (4%)
Query: 5 TEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPS-TTQPQQTN 47
++ Q+ Q QQQ+Q+Q Q QQH + +TQ+ TQP Q++
Sbjct: 549 SQSQQQQQLLQQQQQQQLQ-QQHQQPLQQQTQQQQLRTQPLQSH 591
>At4g12830 hydrolase like protein
Length = 393
Score = 37.4 bits (85), Expect = 0.019
Identities = 21/44 (47%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Query: 125 SETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDK 168
S V+++HG +YSYR I LS + R IA D G GFSDK
Sbjct: 133 SPPVILIHGFPSQAYSYRKTIPVLS-KNYRAIAFDWLGFGFSDK 175
>At3g51000 epoxide hydrolase-like protein
Length = 323
Score = 37.4 bits (85), Expect = 0.019
Identities = 20/40 (50%), Positives = 26/40 (65%)
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSD 167
VL+LHG + YS+R+ I LS+ G V+A DL G G SD
Sbjct: 30 VLLLHGFPETWYSWRHQIDFLSSHGYHVVAPDLRGYGDSD 69
>At5g16120 lipase-like protein
Length = 351
Score = 36.6 bits (83), Expect = 0.033
Identities = 28/112 (25%), Positives = 43/112 (38%), Gaps = 9/112 (8%)
Query: 83 LTLPTTLRQHYSNGRTIKVQIHPNQQPIHLFTFQLDPTIPNPSETVLILHGQA-LSSYSY 141
L L L + NG K N + + +F+ P P V HG ++ +
Sbjct: 39 LNLDHILFKTPENGIKTKESFEVNSRGVEIFSKSWLPEASKPRALVCFCHGYGDTCTFFF 98
Query: 142 RNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVSVEGLDGIFGRFSYVYSEIKE 193
+ + L+ G V A+D PG G S EGL G F + ++ E
Sbjct: 99 EGIARRLALSGYGVFAMDYPGFGLS--------EGLHGYIPSFDLLVQDVIE 142
>At2g40040 unknown protein
Length = 839
Score = 35.8 bits (81), Expect = 0.056
Identities = 18/45 (40%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query: 7 EQESDQQQQQQKQKQKQKQQHSNK-TKSKTQKPSTTQPQQTNPLS 50
+ +S Q Q Q Q Q Q Q S ++++TQ PS TQ Q +P S
Sbjct: 787 QSQSQSQSQSQSQSQSQSQSQSQSPSQTQTQSPSQTQAQAQSPSS 831
Score = 32.0 bits (71), Expect = 0.80
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 7/47 (14%)
Query: 7 EQESDQQQQQQKQKQKQKQ-------QHSNKTKSKTQKPSTTQPQQT 46
+ +S Q Q Q Q Q Q Q Q ++T+++ Q PS+ P QT
Sbjct: 791 QSQSQSQSQSQSQSQSQSQSPSQTQTQSPSQTQAQAQSPSSQSPSQT 837
>At1g43850 SEUSS transcriptional co-regulator
Length = 877
Score = 35.8 bits (81), Expect = 0.056
Identities = 16/43 (37%), Positives = 26/43 (60%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNP 48
++Q+ QQQQQQ+Q+Q+Q+QQ ++ + Q Q NP
Sbjct: 593 QQQQQQQQQQQQQQQQQQQQQTVSQNTNSDQSSRQVALMQGNP 635
Score = 34.7 bits (78), Expect = 0.12
Identities = 15/44 (34%), Positives = 27/44 (61%)
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLS 50
+Q QQQQQQ+Q+Q+Q+QQ + + +T +T Q + ++
Sbjct: 586 DQLRQQQQQQQQQQQQQQQQQQQQQQQQTVSQNTNSDQSSRQVA 629
Score = 34.3 bits (77), Expect = 0.16
Identities = 14/36 (38%), Positives = 26/36 (71%)
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQ 42
+Q SDQ +QQQ+Q+Q+Q+QQ + + + Q+ + +Q
Sbjct: 582 QQASDQLRQQQQQQQQQQQQQQQQQQQQQQQQTVSQ 617
Score = 32.3 bits (72), Expect = 0.62
Identities = 13/48 (27%), Positives = 29/48 (60%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLSFWF 53
++Q+ QQQQQQ+Q+Q+Q + + +++ + Q +N +++ F
Sbjct: 596 QQQQQQQQQQQQQQQQQQTVSQNTNSDQSSRQVALMQGNPSNGVNYAF 643
>At2g39420 phospholipase like protein
Length = 317
Score = 35.0 bits (79), Expect = 0.095
Identities = 28/90 (31%), Positives = 38/90 (42%), Gaps = 2/90 (2%)
Query: 106 NQQPIHLFTFQLDPTIPNPSETVLILHGQALS-SYSYRNLIQSLSTQGVRVIAIDLPGSG 164
N + + LFT + P P V I HG A+ S + + + L G V ID G G
Sbjct: 17 NTRGMKLFTCKWVPAKQEPKALVFICHGYAMECSITMNSTARRLVKAGFAVYGIDYEGHG 76
Query: 165 FSDKSVEVSVEGLDGIFGRFSYVYSEIKEK 194
SD + V D + S Y+ I EK
Sbjct: 77 KSD-GLSAYVPNFDHLVDDVSTHYTSICEK 105
>At1g73480 lysophospholipase like protein
Length = 463
Score = 35.0 bits (79), Expect = 0.095
Identities = 23/81 (28%), Positives = 34/81 (41%), Gaps = 8/81 (9%)
Query: 112 LFTFQLDPTIPNPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVE 171
LF+ P PN +++LHG S Y + + L+ G +V ID G G SD
Sbjct: 198 LFSQSWSPLSPNHRGLIVLLHGLNEHSGRYSDFAKQLNANGFKVYGIDWIGHGGSD---- 253
Query: 172 VSVEGLDGIFGRFSYVYSEIK 192
GL Y +++K
Sbjct: 254 ----GLHAYVPSLDYAVTDLK 270
>At3g58810 zinc transporter -like protein
Length = 432
Score = 34.7 bits (78), Expect = 0.12
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 1/25 (4%)
Query: 457 GHDHHQGHSHGHDRLGEAHIHEAGY 481
GHDH GH H HD G H H+ G+
Sbjct: 236 GHDHGHGHGHSHDN-GHGHSHDHGH 259
Score = 28.5 bits (62), Expect = 8.9
Identities = 11/27 (40%), Positives = 15/27 (54%), Gaps = 2/27 (7%)
Query: 455 DDGHDHHQGHSHGHDRLGEAHIHEAGY 481
D+GH H H HGH H H++G+
Sbjct: 248 DNGHGH--SHDHGHGIAATEHHHDSGH 272
>At2g46010 hypothetical protein
Length = 942
Score = 34.7 bits (78), Expect = 0.12
Identities = 32/117 (27%), Positives = 51/117 (43%), Gaps = 11/117 (9%)
Query: 5 TEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLSFWFYFTLSISLVTL 64
+ S QQQQQQ+Q+Q+Q+QQ +++ + + + S T + + Y + +
Sbjct: 17 SSSSSSVQQQQQQQQQQQQQQQLASRQQQQQHRNSDTNE------NMFAYQPGGVQGMMG 70
Query: 65 FFLFTSSPLSSSSPQQW--FLTLPTTLRQHYSNGRTI---KVQIHPNQQPIHLFTFQ 116
F SSP S PQQ F P +Q G + + +P QQ F Q
Sbjct: 71 GGNFASSPGSMQMPQQSRNFFESPQQQQQQQQQGSSTQEGQQNFNPMQQAYIQFAMQ 127
>At2g30590 unknown protein
Length = 380
Score = 34.7 bits (78), Expect = 0.12
Identities = 15/40 (37%), Positives = 25/40 (62%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
++Q+ QQQQQQ+++Q+Q+QQ + + Q Q QQ
Sbjct: 154 QQQQQQQQQQQQQEQQQQQQQQQQQFHERLQAHHLHQQQQ 193
Score = 32.3 bits (72), Expect = 0.62
Identities = 12/23 (52%), Positives = 20/23 (86%)
Query: 4 ITEEQESDQQQQQQKQKQKQKQQ 26
+ ++Q+ QQQQQQ+Q+Q+Q+QQ
Sbjct: 153 VQQQQQQQQQQQQQEQQQQQQQQ 175
>At2g31305 unknown protein
Length = 107
Score = 34.3 bits (77), Expect = 0.16
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 2/33 (6%)
Query: 444 HVQKMFDE--AKSDDGHDHHQGHSHGHDRLGEA 474
H QK FDE ++ +D ++HH H+H H GEA
Sbjct: 65 HKQKPFDEDDSEEEDDNNHHCDHNHEHSESGEA 97
>At1g25540 hypothetical protein
Length = 809
Score = 33.5 bits (75), Expect = 0.28
Identities = 15/47 (31%), Positives = 25/47 (52%)
Query: 4 ITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLS 50
+ Q+ QQQQQQ+Q+ + Q + + + P QQT+PL+
Sbjct: 710 LQHHQQQQQQQQQQQQQHQLTQLQHHHQQQQQASPLNQMQQQTSPLN 756
Score = 33.1 bits (74), Expect = 0.36
Identities = 15/40 (37%), Positives = 23/40 (57%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
++Q+ QQQQ+Q+Q Q+QQ + + Q P Q QQ
Sbjct: 656 QQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQ 695
Score = 32.3 bits (72), Expect = 0.62
Identities = 18/49 (36%), Positives = 28/49 (56%), Gaps = 4/49 (8%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTK----SKTQKPSTTQPQQTNPLS 50
++ QQQQQQ Q+Q+Q+QQH + + + Q+ Q QQ + LS
Sbjct: 660 QQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLS 708
Score = 32.0 bits (71), Expect = 0.80
Identities = 17/53 (32%), Positives = 27/53 (50%), Gaps = 9/53 (16%)
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQ---------KPSTTQPQQTNPLS 50
+Q+ QQQQQQ+Q Q + QH ++ + + P QQT+PL+
Sbjct: 714 QQQQQQQQQQQQQHQLTQLQHHHQQQQQASPLNQMQQQTSPLNQMQQQTSPLN 766
Score = 31.6 bits (70), Expect = 1.0
Identities = 15/42 (35%), Positives = 23/42 (54%)
Query: 4 ITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
+ ++Q QQQQQ+ +Q+Q+QQH Q+ Q QQ
Sbjct: 682 LQQQQMPQLQQQQQQHQQQQQQQHQLSQLQHHQQQQQQQQQQ 723
Score = 30.8 bits (68), Expect = 1.8
Identities = 20/60 (33%), Positives = 32/60 (53%), Gaps = 10/60 (16%)
Query: 1 MAIITEEQESDQQQQQQK---------QKQKQKQQHSNKTKSKTQ-KPSTTQPQQTNPLS 50
M + ++Q+ QQQQQQ+ Q+Q+Q+QQ + TQ + Q QQ +PL+
Sbjct: 687 MPQLQQQQQQHQQQQQQQHQLSQLQHHQQQQQQQQQQQQQHQLTQLQHHHQQQQQASPLN 746
Score = 30.8 bits (68), Expect = 1.8
Identities = 14/40 (35%), Positives = 22/40 (55%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
++Q+ QQQQ Q Q+Q+QQH + + + Q QQ
Sbjct: 676 QQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQHHQQ 715
Score = 29.6 bits (65), Expect = 4.0
Identities = 14/42 (33%), Positives = 24/42 (56%)
Query: 3 IITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQ 44
++ + Q +QQQQQQ+Q +Q+QQ + + Q+ Q Q
Sbjct: 645 VVFKPQIPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQ 686
Score = 29.6 bits (65), Expect = 4.0
Identities = 14/42 (33%), Positives = 23/42 (54%)
Query: 4 ITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
I +Q+ QQQ Q+Q+Q+Q+ Q + + Q+ Q QQ
Sbjct: 651 IPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQ 692
>At3g53620 inorganic pyrophosphatase -like protein
Length = 216
Score = 33.1 bits (74), Expect = 0.36
Identities = 30/95 (31%), Positives = 44/95 (45%), Gaps = 21/95 (22%)
Query: 212 EEVLARMSKRKVNK----PIDLGPE---------EIGK---VLGEVIGTLGLAPVHLVLH 255
E +L+ MS R V +++GPE EIGK V E+ T GL V +L+
Sbjct: 24 ERILSSMSHRSVAAHPWHDLEIGPEAPIIFNCVVEIGKGSKVKYELDKTTGLIKVDRILY 83
Query: 256 DSAL-----GFTANWVSENSDLVSSLTLVDTPVPP 285
S + GF + E+SD + L ++ PV P
Sbjct: 84 SSVVYPHNYGFIPRTLCEDSDPIDVLVIMQEPVIP 118
>At3g22930 calmodulin, putative
Length = 173
Score = 33.1 bits (74), Expect = 0.36
Identities = 14/30 (46%), Positives = 22/30 (72%)
Query: 8 QESDQQQQQQKQKQKQKQQHSNKTKSKTQK 37
+E QQQQQQ+Q+Q+Q+QQ + + TQ+
Sbjct: 2 EEIQQQQQQQQQQQQQQQQQQQQQQELTQE 31
Score = 33.1 bits (74), Expect = 0.36
Identities = 14/26 (53%), Positives = 21/26 (79%)
Query: 1 MAIITEEQESDQQQQQQKQKQKQKQQ 26
M I ++Q+ QQQQQQ+Q+Q+Q+QQ
Sbjct: 1 MEEIQQQQQQQQQQQQQQQQQQQQQQ 26
Score = 32.7 bits (73), Expect = 0.47
Identities = 13/28 (46%), Positives = 21/28 (74%)
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSK 34
+Q+ QQQQQQ+Q+Q+Q+QQ T+ +
Sbjct: 5 QQQQQQQQQQQQQQQQQQQQQQELTQEQ 32
Score = 32.3 bits (72), Expect = 0.62
Identities = 14/31 (45%), Positives = 22/31 (70%)
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQ 36
EE + QQQQQQ+Q+Q+Q+QQ + ++ Q
Sbjct: 2 EEIQQQQQQQQQQQQQQQQQQQQQQELTQEQ 32
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,803,591
Number of Sequences: 26719
Number of extensions: 533211
Number of successful extensions: 4188
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 3258
Number of HSP's gapped (non-prelim): 394
length of query: 494
length of database: 11,318,596
effective HSP length: 103
effective length of query: 391
effective length of database: 8,566,539
effective search space: 3349516749
effective search space used: 3349516749
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146571.4


