

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146571.1 - phase: 0
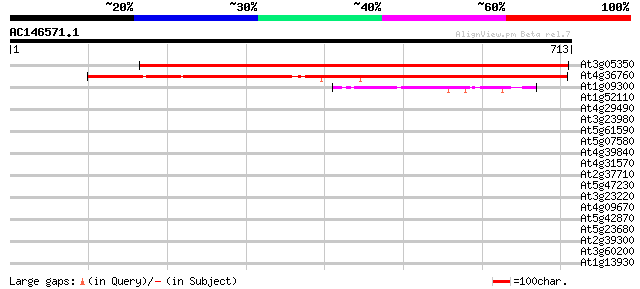
(713 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g05350 aminopeptidase like protein 846 0.0
At4g36760 aminopeptidase-like protein 510 e-144
At1g09300 unknown protein 46 6e-05
At1g52110 37 0.050
At4g29490 putative prolidase, fragment 36 0.066
At3g23980 unknown protein 34 0.33
At5g61590 ethylene responsive element binding factor - like 33 0.42
At5g07580 transcription factor-like protein (emb|CAB87947.1) 33 0.42
At4g39840 unknown protein 33 0.72
At4g31570 putative protein 32 1.2
At2g37710 putative receptor-like protein kinase 32 1.2
At5g47230 ethylene responsive element binding factor 5 (ATERF5) ... 32 1.6
At3g23220 ethylene responsive element binding protein, putative 31 2.1
At4g09670 AX110P like protein 31 2.8
At5g42870 putative protein 30 4.7
At5g23680 unknown protein 30 4.7
At2g39300 hypothetical protein 30 6.1
At3g60200 unknown protein (At3g60200) 29 8.0
At1g13930 hydroxyproline-rich glycoprotein like 29 8.0
>At3g05350 aminopeptidase like protein
Length = 548
Score = 846 bits (2186), Expect = 0.0
Identities = 402/547 (73%), Positives = 476/547 (86%), Gaps = 1/547 (0%)
Query: 165 MRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLV 224
MRAGNPGVPT SEW+ +VLAPG RVGIDPFLF++DAAEELK VI+KKNHELVYLYN NLV
Sbjct: 1 MRAGNPGVPTASEWIADVLAPGGRVGIDPFLFSADAAEELKEVIAKKNHELVYLYNVNLV 60
Query: 225 DEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLN 284
DEIWK++RP+PP++ +R+H LKYAGLDVASKL SLR++++ AG+SAI+++ LDEIAW+LN
Sbjct: 61 DEIWKDSRPKPPSRQIRIHDLKYAGLDVASKLLSLRNQIMDAGTSAIVISMLDEIAWVLN 120
Query: 285 LRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENL 344
LRGSD+PHSPV+YAYLIVE+D A+LF+DNSKVT EV DHLK A IE+RPY+SI+ I++L
Sbjct: 121 LRGSDVPHSPVMYAYLIVEVDQAQLFVDNSKVTVEVKDHLKNAGIELRPYDSILQGIDSL 180
Query: 345 AARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKS 404
AARG+ L +D S++N AI++ YK+AC+RY +N+ES+ K ++K D S + P ++
Sbjct: 181 AARGAQLLMDPSTLNVAIISTYKSACERYSRNFESEAKVKTKFTDSSSGYTANPSGIYMQ 240
Query: 405 SPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRS 464
SP+S AKAIKN+ ELKGM+ HLRDAAALA FW WLE+E+ + LTEV+V+D+LLEFRS
Sbjct: 241 SPISWAKAIKNDAELKGMKNSHLRDAAALAHFWAWLEEEVHKNANLTEVDVADRLLEFRS 300
Query: 465 KQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTV 524
Q GF+DTSFDTISGSG NGAIIHYKPEP SCS VD KLFLLDSGAQYVDGTTDITRTV
Sbjct: 301 MQDGFMDTSFDTISGSGANGAIIHYKPEPESCSRVDPQKLFLLDSGAQYVDGTTDITRTV 360
Query: 525 HFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGV 584
HF +P+ REKECFTRVLQGHIALDQAVFPE TPGFVLD FARS LWK+GLDYRHGTGHGV
Sbjct: 361 HFSEPSAREKECFTRVLQGHIALDQAVFPEGTPGFVLDGFARSSLWKIGLDYRHGTGHGV 420
Query: 585 GAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRF 644
GAALNVHEGPQ IS+RYGN+TPL NGMIVSNEPGYYEDHAFGIRIENLL+VR+ ETPNRF
Sbjct: 421 GAALNVHEGPQSISFRYGNMTPLQNGMIVSNEPGYYEDHAFGIRIENLLHVRDAETPNRF 480
Query: 645 GGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGS-ARQWLWN 703
GG YLGFEKLT+ PIQ K+VDVSLLS TE+DWLN+YH+ VWEKVSPLL+GS +QWLWN
Sbjct: 481 GGATYLGFEKLTFFPIQTKMVDVSLLSDTEVDWLNSYHAEVWEKVSPLLEGSTTQQWLWN 540
Query: 704 NTQPIIR 710
NT+P+ +
Sbjct: 541 NTRPLAK 547
>At4g36760 aminopeptidase-like protein
Length = 634
Score = 510 bits (1313), Expect = e-144
Identities = 268/636 (42%), Positives = 389/636 (61%), Gaps = 39/636 (6%)
Query: 99 IDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQL 158
+DA ++PS+D HQSE+++ RR+++S F+GS G A++T +A LWTDGRYFLQA +QL
Sbjct: 8 LDALVVPSEDYHQSEYVSARDKRREFVSGFSGSAGLALITKKEARLWTDGRYFLQALQQL 67
Query: 159 NSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYL 218
+ W LMR G P W+++ L A +G+D + + D A +KKN +L+
Sbjct: 68 SDEWTLMRMGED--PLVEVWMSDNLPEEANIGVDSWCVSVDTANRWGKSFAKKNQKLITT 125
Query: 219 YNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIVTALDE 278
++LVDE+WK +RP PV VH L++AG V+ K LR++L Q G+ +++ ALDE
Sbjct: 126 -TTDLVDEVWK-SRPPSEMSPVVVHPLEFAGRSVSHKFEDLRAKLKQEGARGLVIAALDE 183
Query: 279 IAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIV 338
+AWL N+RG+D+ + PVV+A+ I+ D A L++D KV++E + + +E+R Y ++
Sbjct: 184 VAWLYNIRGTDVAYCPVVHAFAILTTDSAFLYVDKKKVSDEANSYFNGLGVEVREYTDVI 243
Query: 339 SEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANS--- 395
S++ LA+ + +V ++AA D +S R S +
Sbjct: 244 SDVALLASDRLISSFASKTVQ------HEAAKD---MEIDSDQPDRLWVDPASCCYALYS 294
Query: 396 --DVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEI--------- 444
D + + SP+SL+KA+KN EL+G++ H+RD AA+ Q+ WL+ ++
Sbjct: 295 KLDAEKVLLQPSPISLSKALKNPVELEGIKNAHVRDGAAVVQYLVWLDNQMQELYGASGY 354
Query: 445 -----------TNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEP 493
+ LTEV VSDKL R+ + F SF TIS G N A+IHY PEP
Sbjct: 355 FLEAEASKKKPSETSKLTEVTVSDKLESLRASKEHFRGLSFPTISSVGSNAAVIHYSPEP 414
Query: 494 GSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFP 553
+C+ +D +K++L DSGAQY+DGTTDITRTVHFGKP+ EKEC+T V +GH+AL A FP
Sbjct: 415 EACAEMDPDKIYLCDSGAQYLDGTTDITRTVHFGKPSAHEKECYTAVFKGHVALGNARFP 474
Query: 554 EDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYR-YGNLTPLVNGMI 612
+ T G+ LD AR+ LWK GLDYRHGTGHGVG+ L VHEGP +S+R PL M
Sbjct: 475 KGTNGYTLDILARAPLWKYGLDYRHGTGHGVGSYLCVHEGPHQVSFRPSARNVPLQATMT 534
Query: 613 VSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLST 672
V++EPGYYED FGIR+EN+L V + ET FG YL FE +T+ P Q+KL+D+ L+
Sbjct: 535 VTDEPGYYEDGNFGIRLENVLVVNDAETEFNFGDKGYLQFEHITWAPYQVKLIDLDELTR 594
Query: 673 TEIDWLNNYHSVVWEKVSPLLDGSARQWLWNNTQPI 708
EIDWLN YHS + ++P ++ + +WL T+P+
Sbjct: 595 EEIDWLNTYHSKCKDILAPFMNQTEMEWLKKATEPV 630
>At1g09300 unknown protein
Length = 493
Score = 46.2 bits (108), Expect = 6e-05
Identities = 66/282 (23%), Positives = 117/282 (41%), Gaps = 52/282 (18%)
Query: 411 KAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFL 470
+ IK+ ELK M R++A++A L K + + + + + +E+ + G
Sbjct: 209 RLIKSPAELKLM-----RESASIA--CQGLLKTMLHSKGFPDEGILSAQVEYECRVRGAQ 261
Query: 471 DTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRT-VHFGKP 529
+F+ + G G N ++IHY + L L+D G + +D+TRT GK
Sbjct: 262 RMAFNPVVGGGSNASVIHYSRND---QRIKDGDLVLMDMGCELHGYVSDLTRTWPPCGKF 318
Query: 530 TTREKECFTRVLQGHIALDQAVFPEDT--------PGFVLDAFARSFLWKVGLDYRH--- 578
++ ++E + +LQ + + P T + D + + K Y
Sbjct: 319 SSVQEELYDLILQTNKECIKQCKPGTTIRQLNTYSTELLCDGLMKMGILKSRRLYHQLNP 378
Query: 579 -GTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAF---------GIR 628
GH +G ++VH+ S G PL G +++ EPG Y +F GIR
Sbjct: 379 TSIGHYLG--MDVHD-----SSAVGYDRPLQPGFVITIEPGVYIPSSFDCPERFQGIGIR 431
Query: 629 IENLLYVRNVETPNRFGGIQYLGFEKLT-YVPIQIKLVDVSL 669
IE+ + + G+E LT +P +IK ++ L
Sbjct: 432 IEDDVLITET------------GYEVLTGSMPKEIKHIETLL 461
>At1g52110
Length = 557
Score = 36.6 bits (83), Expect = 0.050
Identities = 26/82 (31%), Positives = 38/82 (45%), Gaps = 12/82 (14%)
Query: 571 KVGLDYRHGTGHGVGAALNVHEGPQGISY-------RYGNLTPLVNGMIVSNEPGYYEDH 623
KVG + G H ++V GP+GI Y +YG+L +G I + GYY DH
Sbjct: 143 KVGTKWDDGVDHAGFTKIHVRSGPKGIQYIKFLYVDKYGHLK---DGPI--HVEGYYNDH 197
Query: 624 AFGIRIENLLYVRNVETPNRFG 645
I+ L + N++T G
Sbjct: 198 DESGVIQALRFKTNIKTSELMG 219
>At4g29490 putative prolidase, fragment
Length = 326
Score = 36.2 bits (82), Expect = 0.066
Identities = 29/108 (26%), Positives = 47/108 (42%), Gaps = 4/108 (3%)
Query: 450 LTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHY-KPEPGSCSTVDANKLFLLD 508
+ E ++ L G S+ I +G N A++HY + T + L LLD
Sbjct: 215 MKEYQMESMFLHHSYMYGGCRHCSYTCICATGDNSAVLHYGHAAAPNDRTFEDGDLALLD 274
Query: 509 SGAQYVDGTTDITRTVHF---GKPTTREKECFTRVLQGHIALDQAVFP 553
GA+Y +DIT + GK T+ + + VL H ++ A+ P
Sbjct: 275 MGAEYHFYGSDITCSFPVSVNGKFTSDQSLIYNAVLDAHNSVISAMKP 322
>At3g23980 unknown protein
Length = 741
Score = 33.9 bits (76), Expect = 0.33
Identities = 28/111 (25%), Positives = 50/111 (44%), Gaps = 5/111 (4%)
Query: 29 PTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTAL 88
PTL+ + +F S++ ++P FS+ ++S SAK + ++++R S DS A
Sbjct: 215 PTLSSSYLFNSPDTSSRPSEPSDFSVNITSSSPLNSAKSEATVKRSRPSFLDSLNISRAP 274
Query: 89 RRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTN 139
+ P++ D S+ S YIS SNG + +T+
Sbjct: 275 ETQYQHPEIQADLVTSSGSQLSGSDGFGPS-----YISGRRDSNGPSSLTS 320
>At5g61590 ethylene responsive element binding factor - like
Length = 201
Score = 33.5 bits (75), Expect = 0.42
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 2/60 (3%)
Query: 311 IDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDT--SSVNAAIVNAYKA 368
+ N K T+ ++ + RP+ +EI + A +GS +WL T S ++AA Y A
Sbjct: 92 VSNRKKTKRFEETRHYRGVRRRPWGKFAAEIRDPAKKGSRIWLGTFESDIDAARAYDYAA 151
>At5g07580 transcription factor-like protein (emb|CAB87947.1)
Length = 207
Score = 33.5 bits (75), Expect = 0.42
Identities = 20/72 (27%), Positives = 31/72 (42%), Gaps = 13/72 (18%)
Query: 311 IDNSKVTEEVDDHLKKAN-------------IEIRPYNSIVSEIENLAARGSSLWLDTSS 357
+++S T E D +KKA + RP+ +EI + A +GS +WL T
Sbjct: 82 LNSSSSTYETDQSVKKAERFEEEVDARHYRGVRRRPWGKFAAEIRDPAKKGSRIWLGTFE 141
Query: 358 VNAAIVNAYKAA 369
+ AY A
Sbjct: 142 SDVDAARAYDCA 153
>At4g39840 unknown protein
Length = 451
Score = 32.7 bits (73), Expect = 0.72
Identities = 30/87 (34%), Positives = 39/87 (44%), Gaps = 11/87 (12%)
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSIS 64
S SSSSS + TKL I SS T N + H N T S+S +
Sbjct: 82 SSSSSSSGTKKNQTKLLKPISSSSSTKNQTKLAKTTTMGTSHK-------LNSTKSSSNT 134
Query: 65 AKPSSQLRK----NRSSNSDSDPKLTA 87
K SS+L+K +S+NS S K +A
Sbjct: 135 TKTSSELKKLNSGTKSTNSTSSIKKSA 161
>At4g31570 putative protein
Length = 2712
Score = 32.0 bits (71), Expect = 1.2
Identities = 45/186 (24%), Positives = 78/186 (41%), Gaps = 15/186 (8%)
Query: 310 FIDNSKVT--EEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYK 367
F++N++ T EEV ++ + + ++ ++NL + D SSV+ +
Sbjct: 819 FLENTQYTNLEEVREYTSEFSALMKNLEKGEKMVQNLEEAIKQILTD-SSVSKSSDKGAT 877
Query: 368 AACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHL 427
A + Q +ESK K + + D+ A VS+ I+N L+G+ + L
Sbjct: 878 PAVSKLIQAFESKRKPEEPESENAQLTDDLSEA---DQFVSVNVQIRN---LRGLLDQLL 931
Query: 428 RDAA-ALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAI 486
+A A QF ++ +DR T + + +EF S Q DTI A+
Sbjct: 932 LNARKAGIQF-----NQLNDDRTSTNQRLEELNVEFASHQDHINVLEADTIESKVSFEAL 986
Query: 487 IHYKPE 492
HY E
Sbjct: 987 KHYSYE 992
>At2g37710 putative receptor-like protein kinase
Length = 675
Score = 32.0 bits (71), Expect = 1.2
Identities = 22/105 (20%), Positives = 46/105 (42%), Gaps = 6/105 (5%)
Query: 543 GHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYG 602
GH + + +D+P + +F+ SF++ + +GHG+ + S YG
Sbjct: 64 GHAFYTKPIRFKDSPNGTVSSFSTSFVFAIHSQIAILSGHGIAFVV-----APNASLPYG 118
Query: 603 NLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGI 647
N + + ++N G +H F + ++ +L +T + GI
Sbjct: 119 NPSQYIGLFNLANN-GNETNHVFAVELDTILSTEFNDTNDNHVGI 162
>At5g47230 ethylene responsive element binding factor 5 (ATERF5)
(sp|O80341)
Length = 300
Score = 31.6 bits (70), Expect = 1.6
Identities = 19/60 (31%), Positives = 27/60 (44%), Gaps = 2/60 (3%)
Query: 315 KVTEEVDDHLKK--ANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDR 372
+VT+ V + KK + RP+ +EI + RGS +WL T AY A R
Sbjct: 143 EVTKPVSEEEKKHYRGVRQRPWGKFAAEIRDPNKRGSRVWLGTFDTAIEAARAYDEAAFR 202
>At3g23220 ethylene responsive element binding protein, putative
Length = 128
Score = 31.2 bits (69), Expect = 2.1
Identities = 19/77 (24%), Positives = 33/77 (42%), Gaps = 5/77 (6%)
Query: 324 LKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAAC-----DRYYQNYE 378
+K + RP+ +EI + A G+ +WL T + AY A R N+
Sbjct: 1 MKYRGVRKRPWGKYAAEIRDSARHGARVWLGTFNTAEDAARAYDRAAFGMRGQRAILNFP 60
Query: 379 SKHKTRSKGFDGSIANS 395
+++ G +GS N+
Sbjct: 61 HEYQMMKDGPNGSHENA 77
>At4g09670 AX110P like protein
Length = 362
Score = 30.8 bits (68), Expect = 2.8
Identities = 14/45 (31%), Positives = 23/45 (51%)
Query: 331 IRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQ 375
+R + +V EI+N A+ W S +V+A K + D+ YQ
Sbjct: 311 VREFARLVGEIKNNGAKPDGYWPSISRKTQLVVDAVKESVDKNYQ 355
>At5g42870 putative protein
Length = 930
Score = 30.0 bits (66), Expect = 4.7
Identities = 23/97 (23%), Positives = 45/97 (45%), Gaps = 5/97 (5%)
Query: 58 TTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKPDV---SIDAYIIPSQDAHQSEF 114
T+ + SS + S ++S PKL A + + + P+V S+ +++ + +E
Sbjct: 500 TSKLDLREDESSSGGLDAESVAESSPKLKAFKHVIANPEVVELSLCKHLL--SEGMGAEA 557
Query: 115 IAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYF 151
+Q++ K S G +++ NDK + G YF
Sbjct: 558 ASQAFNSEKLDMEKFASLGPSILENDKLVVKIGGCYF 594
>At5g23680 unknown protein
Length = 295
Score = 30.0 bits (66), Expect = 4.7
Identities = 22/79 (27%), Positives = 41/79 (51%), Gaps = 3/79 (3%)
Query: 2 NMRSLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSN 61
N +S ++SSS T+T L+ + + TL+ +P+ ++ K K TT+N
Sbjct: 93 NNQSGKTTSSSRTRTMTNLSSGGYENTGTLDEDPVSIGSWRVKKWVKSSGGETAATTTTN 152
Query: 62 SISAKPSSQLRKNRSSNSD 80
+ SAK ++R N ++ +D
Sbjct: 153 TASAK---RVRSNWATRND 168
>At2g39300 hypothetical protein
Length = 768
Score = 29.6 bits (65), Expect = 6.1
Identities = 32/153 (20%), Positives = 63/153 (40%), Gaps = 34/153 (22%)
Query: 313 NSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGS------SLWLDTSSVNAAIVNAY 366
N ++ ++ DDH ++ + N SE+ A R S + +D +S N + +
Sbjct: 18 NKQLHKQKDDHFQRYLNSPKGLNKSQSEVSGAALRRSRSLSSAAFVIDGTSSNQHRLRNH 77
Query: 367 KAAC---DRYYQNYES--------------------KHKTRSKGFDGSIANSDVP----- 398
+ C +R ++ Y S +H RSK GS+ +S +
Sbjct: 78 SSRCLTPERQFKEYGSMSTCSSNVSSQVLDRYIDGEEHLERSKQKSGSLHSSSLSGSRRR 137
Query: 399 IAVHKSSPVSLAKAIKNETELKGMQECHLRDAA 431
+ SP L+++ K++ + KG+++ R A
Sbjct: 138 LPPRAQSPSPLSESGKDKRKSKGLRDASARSLA 170
>At3g60200 unknown protein (At3g60200)
Length = 305
Score = 29.3 bits (64), Expect = 8.0
Identities = 19/87 (21%), Positives = 35/87 (39%), Gaps = 7/87 (8%)
Query: 42 KSNKHNKPPFFSIRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKPDVSIDA 101
KSN +N PP S P RK+ + DP +++ RR + P + +
Sbjct: 45 KSNNNNHPPLLIFPR-------SVSPYVTRRKSDAGAGGGDPLVSSNRRFITTPQIDLVG 97
Query: 102 YIIPSQDAHQSEFIAQSYARRKYISAF 128
Y ++++S Q ++ + F
Sbjct: 98 YSCKDFESNRSNKSKQGKKVSRFSNLF 124
>At1g13930 hydroxyproline-rich glycoprotein like
Length = 155
Score = 29.3 bits (64), Expect = 8.0
Identities = 25/95 (26%), Positives = 37/95 (38%), Gaps = 19/95 (20%)
Query: 312 DNSKVTEEVDDHLKKA-NIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAA- 369
D++K E + + A N E+ +V+E AAR S LD V A + AA
Sbjct: 20 DHNKPVEGTETATRPATNAELMASAKVVAEAAQAAARNESDKLDKGKVAGASADILDAAE 79
Query: 370 -----------------CDRYYQNYESKHKTRSKG 387
++Y +YES H T + G
Sbjct: 80 KYGKFDEKSSTGQYLDKAEKYLNDYESSHSTGAGG 114
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,538,561
Number of Sequences: 26719
Number of extensions: 724950
Number of successful extensions: 2049
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 2033
Number of HSP's gapped (non-prelim): 21
length of query: 713
length of database: 11,318,596
effective HSP length: 106
effective length of query: 607
effective length of database: 8,486,382
effective search space: 5151233874
effective search space used: 5151233874
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146571.1


